Abstract
Idiosyncratic adverse drug reactions are unpredictable, dose-independent and potentially life threatening; this makes them a major factor contributing to the cost and uncertainty of drug development. Clinical data suggest that many such reactions involve immune mechanisms, and genetic association studies have identified strong linkages between drug hypersensitivity reactions to several drugs and specific HLA alleles. One of the strongest such genetic associations found has been for the antiviral drug abacavir, which causes severe adverse reactions exclusively in patients expressing the HLA molecular variant B*57:01. Abacavir adverse reactions were recently shown to be driven by drug-specific activation of cytokine-producing, cytotoxic CD8+ T cells that required HLA-B*57:01 molecules for their function; however, the mechanism by which abacavir induces this pathologic T-cell response remains unclear. Here we show that abacavir can bind within the F pocket of the peptide-binding groove of HLA-B*57:01, thereby altering its specificity. This provides an explanation for HLA-linked idiosyncratic adverse drug reactions, namely that drugs can alter the repertoire of self-peptides presented to T cells, thus causing the equivalent of an alloreactive T-cell response. Indeed, we identified specific self-peptides that are presented only in the presence of abacavir and that were recognized by T cells of hypersensitive patients. The assays that we have established can be applied to test additional compounds with suspected HLA-linked hypersensitivities in vitro. Where successful, these assays could speed up the discovery and mechanistic understanding of HLA-linked hypersensitivities, and guide the development of safer drugs.
Keywords: 3D structure, small molecule, binding site
Abacavir is a nucleoside analog that suppresses HIV replication. In approximately 8% of recipients, abacavir is associated with significant immune-mediated drug hypersensitivity, which is strongly associated with the presence of the HLA-B*57:01 allele (1, 2). Three complementary models for the mechanism of immune-mediated severe adverse drug reactions have traditionally been discussed (3, 4). The hapten (or prohapten) model states that drugs and their metabolites are too small to be immunogenic on their own, but rather act like haptens and modify certain self-proteins in the host that lead to immune recognition of the resulting hapten–self-peptide complexes as de novo antigens (5–7). The pharmacologic interaction with immune receptors (p-i) model states that drugs can induce the formation of HLA–drug complexes that can activate T-cell immune responses directly without requiring a specific peptide ligand (8). The danger model, which is in principle compatible with other models, states that danger signals other than the drug itself (e.g., chemical, physical, or viral stress) are required to overcome immune tolerance barriers that otherwise suppress drug hypersensitivity reactions (7).
None of these existing models provides a convincing mechanism explaining how abacavir induces adverse reactions through the activation of CD8+ cells in a strictly HLA-B*57:01–restricted manner, as was described in a groundbreaking paper by the McCluskey group (2). For the hapten hypothesis to apply, abacavir would need to modify one or more self-ligands that are presented solely by HLA-B*57:01 (Fig. 1), an unlikely proposition given that HLA molecules are known to fall in groups of overlapping binding specificity and that HLA-B*57:01 has a similar binding motif to the abacavir-insensitive HLA-B*58:01 (9). In addition, no natural HLA-specific drug haptenated peptide has been identified to date, although this has been attempted, at least for carbamazepine (10, 11). For the p-i model to apply, abacavir would need to bind to a unique surface patch of HLA-B*57:01 that is capable of inducing TCR recognition. However, the two residues that distinguish abacavir-sensitive HLA-B*57:01 from abacavir-insensitive HLA-B*57:03 are located at the bottom of the HLA-binding groove and are unlikely to contact the T-cell receptor. Finally, the danger model might well be relevant for adverse reactions to abacavir, but does not explain its HLA restriction. We find an alternative hypothesis more attractive, namely that the binding groove of HLA-B*57:01 can accommodate abacavir (12), thereby altering the repertoire of self-peptide ligands that are bound and presented (2, 10). This could lead to a primary and polyclonal immune response, which is in line with observations that abacavir can induce a relatively diverse response in T cells from abacavir-naïve individuals after in vitro stimulation for 11 d (2), with broad use of V beta receptors by the responding T cells (2). However, to date there has been no experimental evidence supporting this “altered self-repertoire” hypothesis as a mechanism for drug hypersensitivity.
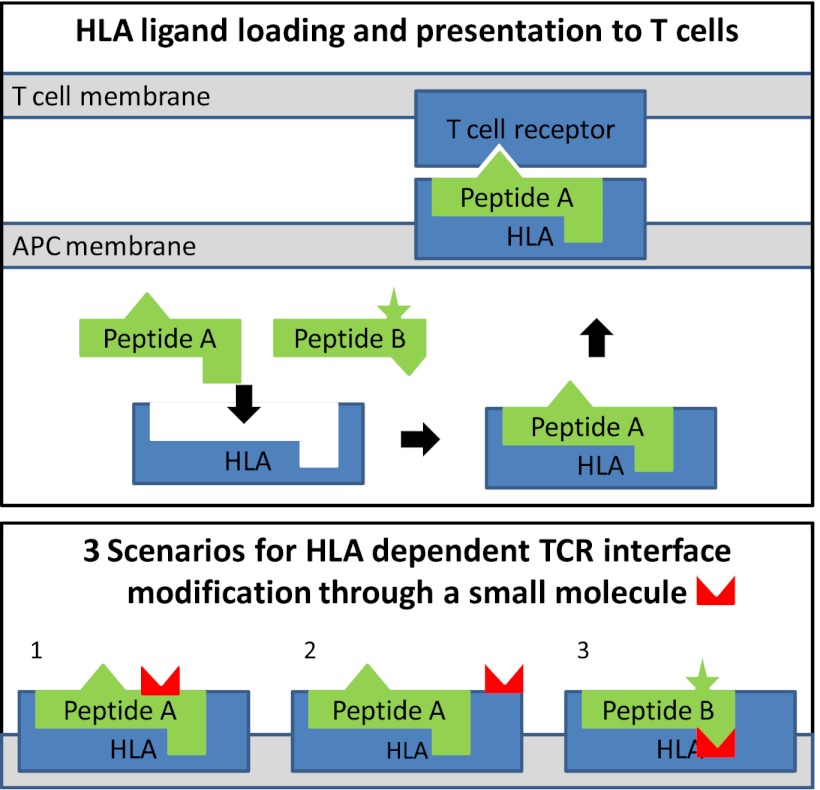
Fig. 1.
Schematic presentation of HLA antigen presentation and HLA-linked mechanisms of adverse reactions. (Upper) T-cell receptors monitor the universe of antigens to which an individual is exposed by surveying the ligands presented on an antigen-presenting cell membrane in the context of HLA molecules. The HLA ligands are typically peptides loaded onto the HLA molecule inside the antigen-presenting cells and subsequently exposed on the surface. Different allelic variants of HLA molecules have different binding specificities, resulting in a specific profile of presented ligands. In the example shown, peptide A, but not peptide B, can bind to the HLA molecule. Self-peptides presented to T cells in this manner do not trigger an immune response, because T cells that are self-reactive are negatively selected during thymic development. However, when T cells encounter an unknown ligand (eg, a virus-derived peptide), an immune response is triggered. (Lower) There are three scenarios for HLA-dependent drug-induced modifications that affect the TCR interface: (1) A ligand that is uniquely presented by the HLA allele is modified by the drug; (2) the HLA molecule itself is modified in a region exposed to the TCR; and (3) the binding specificity of the HLA molecule is altered by the presence of the drug, resulting in presentation of novel ligands such as peptide B.
Results
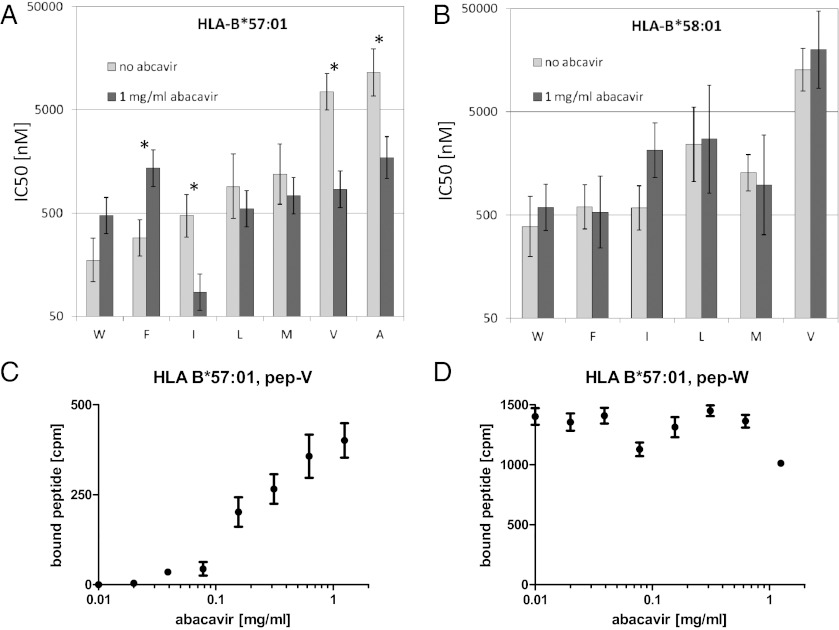
To determine whether abacavir can impact the peptide-binding specificity of HLA-B*57:01, we tested its binding affinity using positional scanning combinatorial peptide libraries (13) in the presence and absence of the drug. Each library consisted of 9-mer peptides that share the same residue at one position but were otherwise random in sequence. In the absence of abacavir, our binding measurements reproduced the known motif of HLA-B*57:01 for C-terminal peptide residues, namely a preference for large hydrophobic residues such as tryptophan and phenylalanine and a disfavoring of small hydrophobic residues like alanine and valine (Fig. 2A). We were specifically interested in residues showing an increased affinity in the presence of abacavir, which could potentially lead to presentation of de novo peptides. The most dramatic gains were seen for peptides with C-terminal valine (8.8-fold increase), alanine (6.7-fold increase), and isoleucine (5.5-fold increase). The only other residue with a fivefold or greater increase was leucine at position 7 (SI Appendix, Table S1). In contrast, the control MHC molecule HLA-B*58:01 showed no increases in affinity exceeding threefold for any residue at any position (Fig. 2B and SI Appendix, Table S2).
Fig. 2.
The presence of abacavir alters the binding specificity of HLA-B*57:01. (A and B) Combinatorial peptide libraries were tested for binding to HLA-B*57:01 (A) and HLA-B*58:01 (B) in competitive binding assays as described previously (13, 39). Results for libraries with different C-terminal residues are shown for those residues with affinities of 5,000 nM or better, a minimal threshold for binding. Error bars indicate 95% confidence intervals for the mean, and residues marked with an asterisk had significantly different IC50 values in the presence vs. absence of abacavir (P < 0.001, two-tailed Student t test comparing log IC50 values). The most pronounced affinity increases for HLA-B*57:01 in the presence of 1 mg/mL of abacavir were found for peptides with a valine at the C terminus, which increased by more than eightfold, followed by alanine and isoleucine, which increased by fivefold. In contrast, the maximum affinity increase for any peptide library binding to HLA-B*58:01 was less than threefold. (C and D) Individual peptides HSITYLLPV (pep-V) and HSITYLLPW (pep-W) were radiolabeled and tested for binding to HLA-B*57:01 and B*58:01 in increasing doses of abacavir. After washing, no pep-V binding to HLA-B*57:01 was detectable in the absence of abacavir, but strong binding was detected in the presence of abacavir. No significant effect of abacavir was observed for pep-W binding to HLA-B*57:01.
Based on these results, we synthesized individual peptides with the sequence HSITYLLPV (pep-V) and HSITYLLPW (pep-W). Residues 1–8 of these peptides were chosen based on their high affinity in combinatorial library scans in the presence of abacavir. The C terminus was chosen so that pep-V was expected to be unable to bind efficiently in the absence of abacavir, but would do so in its presence. As a control, pep-W with a C-terminal tryptophan was expected to be able to bind HLA-B*57:01 readily in the presence or absence of abacavir. The peptides were radiolabeled and tested for binding in increasing concentrations of abacavir. Peptide binding assays demonstrated that pep-V requires abacavir in a dose-dependent manner to bind a detectable amount of peptide (Fig. 2C), whereas pep-W bound well regardless of the presence or absence of abacavir (Fig. 2D). Similar results were obtained using two independent peptide–HLA-B*57:01 interaction assays measuring either binding (14) or stability (15) (SI Appendix, Fig. S1) and an additional control allele, HLA-B*57:03 (SI Appendix, Table S3). In summary, we found that specific peptides such as pep-V have a significantly increased affinity for HLA-B*57:01 in the presence of abacavir, that this effect is abolished when the C-terminal P9 residue is switched to a tryptophan (pep-W), and that this effect is not observed for control HLA alleles.
Structural analysis was used to further dissect the mechanism by which abacavir may facilitate the binding of pep-V to HLA-B*57:01. Computational solvent mapping (16), molecular docking (17), and molecular dynamics simulations (18) of 30 ns in length identified a potential binding site for abacavir localized to the F pocket in the vicinity of residue Ser-116, which has been shown to be required for abacavir T-cell recognition (2) (SI Appendix, Fig. S2). In contrast, a similar protocol did not identify stable complexes of MHC, abacavir, and peptide with either pep-W or abacavir-insensitive HLA-B*57:03.
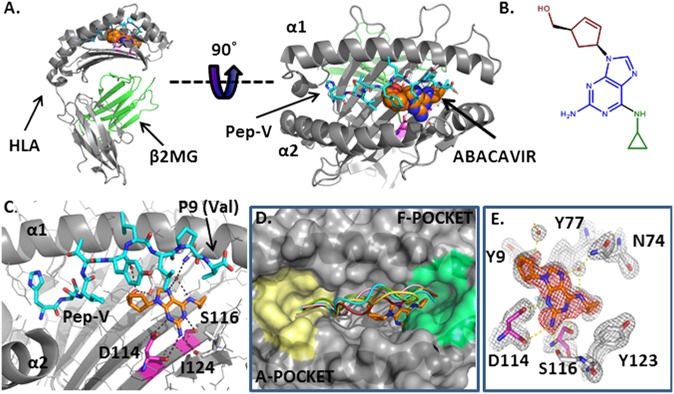
To directly test our hypothesis that abacavir binds within the antigen-binding cleft, we solved the X-ray crystal structure of HLA-B*57:01 bound to pep-V in the presence of abacavir. We believe that choosing pep-V instead of a ligand with a large side chain at the C terminus, such as those used in previous crystal structures (2), was crucial to obtaining crystals that could resolve the location of abacavir. The structure was refined to an R value of 18% and an Rfree value of 22% using X-ray diffraction data to 2.0 Å (PDB ID: 3UPR); details are provided in Materials and Methods, and complete refinement statistics are presented in SI Appendix, Table S4. Abacavir is bound to a largely hydrophobic pocket in the antigen-binding cleft forming van der Waals contacts with both HLA-B*57:01 (Tyr9, Tyr-74, Ile-95, Val97, Tyr99, Tyr123, Ile-124, and Trp147) and pep-V (Ile3, Leu7, and Val9) (Fig. 3A and SI Appendix, Fig. S3).
Fig. 3.
Crystal structure of the abacavir–peptide–MHC complex solved to a resolution limit of 2.0 Å reveals intermolecular contacts within the antigen-binding cleft. (A) Cartoon diagram of HLA-B*57:01 in gray. The peptide HSITYLLPV is shown in cyan carbons. Abacavir is shown as spheres, orange for carbon, blue for nitrogen and red for oxygen. (B) Chemical structure of abacavir, with the cyclopropyl moiety shown in green, the purine core in blue, and the hydroxymethyl cyclopentene moiety in red. (C) Abacavir forms H bond interactions (black dashes) with both the peptide and HLA-B*57:01. The residues that distinguish the abacavir-sensitive allele HLA-B*57:01 from abacavir-insensitive HLA-B*57:03 are shown in magenta for carbon, blue for nitrogen, and red for oxygen. (D) Abacavir binding in the F pocket does not alter the peptide conformation compared with other peptide/HLA-B complexes. A cartoon representation of peptide in the crystal structure complexed to abacavir and HLA-B*57:01 is shown in cyan (HSITYLLPV; PDB ID: 3UPR). A 9-mer self peptide (LSSPVTKSF) complexed to HLA-B*57:01 (PDB ID: 2RFX (2) is shown in red, the 8-mer peptide epitope HIV1 Nef 75–82 (VPLRPMTY) bound to HLA-B*35:01 (PDB ID: 1A1N) (40) is shown in pink, a 9-mer EBV peptide (FLRGRAYGL) complexed to HLA-B8 (PDB ID: 1MI5) (41) is shown in green, and the 11-mer EBV peptide HPVGEADYFEY complexed to HLA-B*35:01 (PDB ID: 3MV9) (42) is shown in yellow. The molecular surface of HLA-B*57:01 from 3UPR is shown in gray. The F pocket residues (9) are colored green, and the A pocket is yellow. (E) Experimental electron density corresponding to abacavir in an Fo-Fc difference map contoured at 3.5σ (red mesh) after molecular replacement. Gray mesh depicts the final 2Fo-Fc electron density map of abacavir in the antigen-binding cleft of HLA-B*57:01 (contour level, 1.5σ). H bond interactions between abacavir and HLA-B*57:01 are shown as yellow dashed lines. The residues that distinguish the abacavir-sensitive allele HLA-B*57:01 from abacavir-insensitive HLA-B*57:03 are shown in magenta for carbon, blue for nitrogen, and red for oxygen.
To determine whether the conformation of pep-V is altered by abacavir binding compared with conventionally presented peptides, we compared our structure with four published structures of peptides bound to HLA-B molecules. The main chain conformation of pep-V is similar to that of other peptides bound to HLA-B (Fig. 3D). The amino and carboxy termini of pep-V and the other HLA-B–bound peptides shown in Fig. 3D are buried in the A and F pockets, respectively, forming conventional contacts with highly conserved residues. The central portion of the pep-V main chain is within the range of variability demonstrated by other peptide–HLA-B complexes. The pep-V main chain does not protrude in a central bulge as does the longer 11-mer peptide bound to HLA-B*35:01. These data suggest that pep-V in the presence of abacavir is bound in a regular antigen conformation, allowing for conventional recognition by TCRs rather than the need for hapten or superantigen recognition modes (19, 20).
Only two residues, Asp114 and Ser116, distinguish HLA-B*57:01 from the abacavir-insensitive allele HLA-B*57:03. Abacavir interacts directly with these residues (Fig. 3 C and E). The Oδ1 atom of Asp114 is within H bonding distance of the main purine group N2 and N3 atoms of abacavir. The hydroxyl group of Ser116 forms an H bond with the 2-amino group on the purine ring of abacavir. The exchange of Ser116 to Tyr116 seen in HLA-B*57:03 is expected to disrupt these interactions. Indeed, it was shown that this single residue exchange is sufficient to abrogate abacavir-associated recognition by CD8+ T cells (2). The HLA-B*58:01 allelic variant is also very similar to HLA-B*57:01. These molecules have identical amino acids at positions 114 and 116 but different amino acids at five other positions. These include Val97, part of the hydrophobic pocket in HLA-B*57:01 that forms van der Waals contacts with abacavir (SI Appendix, Fig. S3), which is replaced with a charged Arg97 in HLA-B*58:01, abrogating these interactions. In addition, the structure reveals that the side chain of Val9 of pep-V is within van der Waals contact distance of the cyclopropyl moiety of abacavir. Finally, the contact made between the Leu7 residue of the peptide and abacavir explains why the MHC binding assays showed that this residue had the highest increase in affinity in the presence of abacavir apart from C-terminal residues. In summary, our findings give a structural explanation for why distinct repertoires of peptides with short hydrophobic P9 side chains are bound by HLA-B*57:01 in the presence of abacavir, whereas other HLA alleles are unaffected.
To explore the biological relevance of these findings, we determined whether live cells treated with abacavir present a different set of self-peptides on HLA-B*57:01 molecules compared with untreated cells. Our binding assays predicted that HLA-B*57:01 in the presence of abacavir would favor presentation of peptides with a small C-terminal residue, such as valine and isoleucine, rather than the tryptophan and phenylanine normally preferred by HLA-B*57:01 molecules. To answer this question, we eluted peptides from an HLA-B*57:01 single allele-transfected 721.221 cell line (21) treated with and without abacavir. Eluted peptides were analyzed by nanoflow-HPLC coupled to an orbitrap mass spectrometer equipped with front-end electron transfer dissociation (22, 23). We identified 539 and 682 peptide sequences from the drug-treated and untreated samples, respectively, 287 of which were found in both samples (SI Appendix, Table S5). No peptides with valine at the C terminus were identified in untreated cells, but 15 peptides with valine at the C terminus were identified in the presence of abacavir. Three of these peptides were present at levels (>100 copies/cell) that place them among the top 5% of all peptides in the drug-treated sample. Table 1 compares the frequency of C-terminal residues in peptides identified uniquely in either the abacavir-treated or untreated samples. In the presence of abacavir, there was significant enrichment not only for peptides with valine at the C terminus, but also for isoleucine. In contrast, there were significantly fewer peptides with tryptophan and phenylalanine at the C terminus. The results for these residues match exactly the predictions made by the binding assays. In contrast to valine, no peptides with C-terminal alanine were discovered in the presence of abacavir, even though both showed a similar increase in affinity. However, the absence of peptides with an alanine at the C terminus can be explained by the antigen processing machinery, including proteasomal cleavage and TAP transport, which restricts the peptide repertoire available for binding to MHC and disfavors peptides with C-terminal alanine (24, 25). In summary, we find that the self-peptide repertoire presented by HLA-B*57:01–positive cells in the presence of abacavir is significantly altered in a manner consistent with results obtained from the molecular MHC binding assays.
Table 1.
Distribution of C-terminal residues in peptides uniquely presented by abacavir treated and untreated cells
| Abacavir |
|||
| C-terminal residue | Untreated | Treated | P value* |
| W | 218 | ↓ 95 | 2E-05 |
| F | 89 | ↓ 31 | 1E-03 |
| Y | 42 | 33 | 0.38 |
| L | 25 | 25 | 0.10 |
| I | 14 | ↑ 45 | 1E-09 |
| M | 6 | 7 | 0.39 |
| V | 0 | ↑15 | 5E-07 |
*Two-tailed Fisher exact test. All C-terminal residues for which two or more peptides were identified are listed.
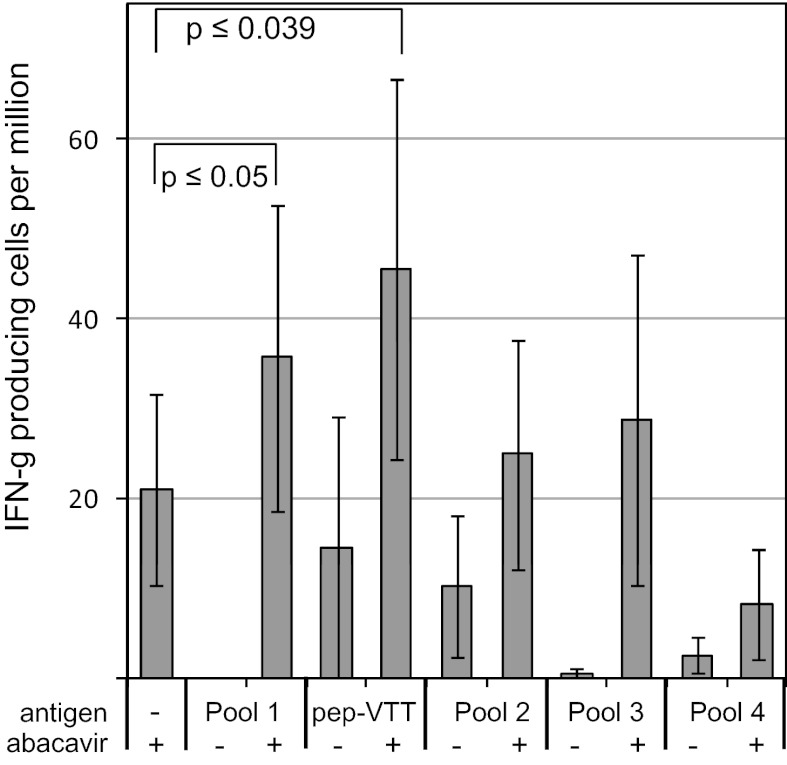
Finally, we set out to determine whether we could detect T cells in hypersensitive patients who recognize HLA-B*57:01–restricted peptides that are presented only in the presence of abacavir. Of note, we did not expect to see high-frequency T-cell responses against any individual peptide, given that the altered ligand mechanism suggests that the response is directed against a very large number of different peptides. We screened peripheral blood mononuclear cells (PBMCs) from five HLA-B*57:01–positive donors with a clinical history of abacavir hypersensitivity for recognition of peptides with valine at the C terminus that were identified after elution from abacavir-treated HLA-B*57:01 cells. The PBMCs were incubated for 15 min with a high concentration of 10 μg/mL of endogenous peptides in the presence of 100 μg/mL of abacavir, followed by washing to optimize loading of HLA-B*57:01 with the specific exogenously added peptide(s), reduce the abacavir entering cells, and enable presentation of other endogenous ligands. After a first round of screening four pools of three or four peptides in ELISPOT assays using PBMCs from two donors with sufficient samples available, the peptides from the pool with the highest response (pool 1) were tested individually. Peptide VTTDIQVKV showed the greatest response in this screen (SI Appendix, Fig. S4). Subsequently, we tested the four pools and the individual peptide VTTDIQVKV in the remaining three donors. A significantly greater response was detected when cells were pulsed with peptide VTTDIQVKV and abacavir compared with the response against cells pulsed with either abacavir alone or peptide alone (Fig. 4). These data demonstrate that memory T-cell responses in abacavir-hypersensitive donors are directed against a self-peptide that requires abacavir to efficiently bind HLA-B*57:01.
Fig. 4.
T cells from hypersensitive donors respond to specific self-peptides in an abacavir- dependent fashion. PBMCs from five HLA-B*57:01-positive donors with a clinical history of abacavir hypersensitivity were pulsed for 15 min with peptide antigens in the presence or absence of abacavir or with abacavir alone, washed, and then tested by an IFN-γ ELISpot assay (Materials and Methods). The figure shows the calculated mean (± SEM) IFN-γ spots per million input PBMCs. Statistically significant responses compared with the response induced by the abacavir pulse alone were obtained for peptide pool 1 and the individual peptide VTTDIQVKV (paired two-tailed Student t test on the square root of the SFC counts). Incubating PBMCs from these donors with 10 μg/mL of abacavir overnight yielded an average of 200 ± 63 SFCs per million.
Discussion
Our findings provide a mechanistic explanation for abacavir-induced adverse drug reactions. We found that abacavir can bind inside the peptide-binding groove of HLA-B*57:01, thereby enabling the presentation of peptide ligands that normally cannot bind in substantial amounts. Because T cells are generally tolerant only to MHC-restricted peptide ligands presented during T-cell development in the thymus (26), presentation of an altered repertoire of class I MHC binding peptides will be perceived as being foreign and trigger CD8+ T-cell responses. These responses have been shown to be a hallmark of patients with abacavir hypersensitivity (2). Although abacavir is metabolized inside cells (27, 28), our finding that HLA-B*57:01 is affected by the abacavir parent drug suggests that the parent drug itself is present during peptide loading in the endoplasmic reticulum (ER). Indeed, others have detected unaltered intracellular abacavir, which rapidly colocalizes with HLA-B57 in the ER (27, 29). Abacavir might well be delivered to the ER directly from the extracellular medium without traversing the cytosol, as was described for HLA class I binding peptides (30). In this context, it also should be noted that only a small fraction of peptide–MHC loading events affected by abacavir can cause a physiological effect, given that T cells have a very high sensitivity to detect nonself peptides, which is essential to their function, and remarkably few peptide–MHC complexes are sufficient to trigger a T-cell response (31, 32).
Together, these biochemical and structural findings provide an explanation for why, of the more than 5,000 class I MHC alleles, only HLA-B*5701 is affected by abacavir. They also provide support for the altered peptide model in which the drug in effect creates a novel HLA-B allele presenting self-peptides to which the host has not been toleralized, analogous to the situation occurring in HLA-mismatched organ transplantation. In organ transplantation, preexisting class I restricted effector memory T-cell responses to prevalent viral infections can mediate organ rejection (33). This model of heterologous immunity might account for the clinical manifestations arising from drug-induced altered peptide presentation and explain why only 55% of HLA-B*5701–positive patients treated with abacavir develop hypersensitivity. Other possible explanations for the incomplete positive predictive value of specific HLA alleles for abacavir and other drug hypersensitivity syndromes and the varying clinical features include that the relevant peptide recognized by drug-specific T cells in the presence of drug is itself genetically polymorphic and/or only present in some patients or tissues, and that only some patients have a T-cell clonotype able to respond to the neo-antigen (34).
Over the last decade in particular, numerous HLA-associated drug toxicities have been reported (4). Further studies should explore in detail whether the mechanism for HLA-linked adverse reactions to abacavir applies to adverse reactions against other small molecules that seem to be immune-mediated and HLA-linked, such as chronic beryllium disease (35) and adverse reactions to allopurinol (36) and carbamazepine (37). Our methods and findings are particularly significant for such studies, because they can be used to identify the structural, biochemical, and functional bases of potential HLA-associated T-cell–mediated drug hypersensitivities before use of a drug in humans. This may have utility both in excluding high-risk compounds from further development and in guiding the design of compounds that do not bind high-risk HLA alleles or alter the repertoire of peptides presented. The biochemical and functional assays described here also could be used to characterize the HLA restriction and likely immunopathogenesis of cases of hypersensitivity in early clinical studies. This could facilitate the early introduction of HLA screening, as has been successfully implemented for the prevention of abacavir hypersensitivity (38), rather than having to rely exclusively on genetic association studies requiring large cohorts of affected patients. Finally, these findings have potential relevance for a broader understanding of HLA associations in the immunopathogenesis of autoimmune disease, infectious disease, and cancer. Such associations can be strong yet remain enigmatic, because the molecular recognition events underlying the associations are unclear. The discovery that a small-molecule drug can bind within the antigen-binding cleft of MHC and alter the repertoire of presented peptides suggests a mechanism of action as the causative basis for HLA associations. HLA-associated disorders may be perpetuated by drugs, small molecules of environmental origin, or self-metabolites that bind within the antigen-binding cleft and alter peptide binding.
Materials and Methods
X-Ray Crystallography.
Refolded β2-microglobulin, HLA-B*57:01, and abacavir formed crystals at 4 mg/mL in 0.17 M sodium acetate trihydrate, 0.1 M sodium cacodylate (pH 6.5), 25% PEG 8000, and 15% glycerol, which belong to the space group P21 with unit cell dimensions a = 44.8 Å, b = 130.7 Å, c = 88.3 Å, α = 90°, β = 104.6o, and γ = 90o. Two peptide–abacavir–HLA complexes in the asymmetric unit were identified by molecular replacement using coordinates for the heavy chain and β2-microglobulin from PDB 2RFX (HLA-B*57:01 bound to LSSPVTKSF). The peptide and ligands were not included in the molecular replacement model. Unambiguous electron density for the peptide and abacavir were visible in Fo-Fc difference maps and simulated annealing omit electron density maps. The structure was refined to an R value of 18% and an Rfree value of 21% using X-ray diffraction data to 2.0 Å.
The best crystals diffracted to a high-resolution limit of 1.9 Å. These crystals belonged to space group P21 and contained two HLA heterodimers (heavy chain HLA-B*57:01 and light chain β2-microglobulin) in the asymmetric unit. The phasing was done by molecular replacement. The initially calculated Fo-Fc difference map showed unambiguous electron density for both the peptide and abacavir.
Abundance Calculation for Eluted Peptides.
Peptide abundances were determined by comparing the ion current observed for 100 fmol of an internal standard, angiotensin I (DRVYIHPFHL), to that observed for individual peptides. This femtomole quantity was then converted to peptide molecules by multiplying by Avogadro’s number and to molecules (copies)/cell by dividing this number by the total number of cells used to generate the sample aliquot injected into the mass spectrometer.
T-Cell ELISpot Assays.
Before clinical samples were obtained, informed consent was provided by all patients, and the study was approved by both the Royal Perth Hospital and Murdoch University Ethics Committees. Cryopreserved PBMCs from abacavir-hypersensitive patients obtained at time points previously evaluated with an abacavir-specific ELISpot assay were thawed and cultured overnight in RPMI 1640 medium containing 10% FCS, 50 U/mL of penicillin, 50 μg/mL of streptomycin, and 1 mM sodium pyruvate (F10 medium; Life Technologies). The assays were controlled using a unstimulated PBMC control, a positive control for abacavir-induced endogenous ligand consisting of 10 μg/mL of abacavir added for the duration of the assay culture and an abacavir 15-min pulse and wash to control for the effects of the pulse on inducing endogenous peptide targets. Peptide ± abacavir incubation times were derived to achieve the optimum balance between detection of enhanced exogenous peptide binding and a restriction of abacavir-induced endogenous peptide presentation. To set up the assay, 5 × 105 PBMCs were then exposed to 10 μg/mL of exogenous peptides, either singly or as peptide pools containing up to four peptides, in the presence or absence of 100 μg/mL of abacavir for 15 min at 37 °C in a cell culture incubator. The cells were then immediately washed with 25 volumes of ice-cold RPMI 1640 and centrifuged for 10 min at 300 × g at 4 °C. Cells were then resuspended in 450 μL of F10 medium containing 20U/mL of recombinant human IL-2 (Peprotech). Then 2 × 105 PBMCs in duplicate were transferred into MAIPS4510 ELISpot plates coated with 2 μg/mL of anti–IFN-γ antibody (1-D1k; Mabtech) and blocked with F10 medium. Cells were cultured overnight at 37 °C in a cell culture incubator. The next day, the plates were washed with sterile PBS and incubated with anti–IFN-γ biotinylated antibody (7-B6-1; Mabtech) for 2 h, washed, incubated for 1 h with streptavidin-HRP, and then washed again. Plates were substrated for 8–12 min using TMB substrate (Mabtech). After the plates were air-dried, counts were evaluated with an AID automated microplate ELISpot reader. The following sequences of the peptides were tested: pool 1: KTIHLTLKV, RTLAEIAKV, VTTDIQVKV, and TVAPFNPTV; pool 2: HSIPVTVEV, KSNGTIIHV, and RTFHHGVRV; pool 3: ATIKLQSTV, KIYEGQVEV, RSARVTVAV, and RVAGIHKKV; pool 4: RSVALAVLA, KAAKIRVSV, KVAKVEPAV, and RTTETQVLV.
Supplementary Material
Acknowledgments
We thank Amiyah Steen, Sandy Ngo, Carrie Moore, and Victoria Tripple for technical assistance; Patrick Hogan for helpful discussions; and Janet Woodcock and Donna Mendrick for support. Funding was provided by National Institute of Health Grants AI 33993 (to D.F.H.) and HHSN 272 200900045C (to S.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3UPR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207934109/-/DCSupplemental.
References
- 1.Rauch A, et al. Prospective genetic screening decreases the incidence of abacavir hypersensitivity reactions in the Western Australian HIV cohort study. Clin Infect Dis. 2006;43:99–102. doi: 10.1086/504874. [DOI] [PubMed] [Google Scholar]
- 2.Chessman D, et al. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity. 2008;28:822–832. doi: 10.1016/j.immuni.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Uetrecht J. Idiosyncratic drug reactions: Current understanding. Annu Rev Pharmacol Toxicol. 2007;47:513–539. doi: 10.1146/annurev.pharmtox.47.120505.105150. [DOI] [PubMed] [Google Scholar]
- 4.Phillips EJ, Mallal SA. HLA and drug-induced toxicity. Curr Opin Mol Ther. 2009;11:231–242. [PubMed] [Google Scholar]
- 5.Lavergne SN, Park BK, Naisbitt DJ. The roles of drug metabolism in the pathogenesis of T-cell–mediated drug hypersensitivity. Curr Opin Allergy Clin Immunol. 2008;8:299–307. doi: 10.1097/ACI.0b013e3283079c64. [DOI] [PubMed] [Google Scholar]
- 6.Park BK, Naisbitt DJ, Gordon SF, Kitteringham NR, Pirmohamed M. Metabolic activation in drug allergies. Toxicology. 2001;158:11–23. doi: 10.1016/s0300-483x(00)00397-8. [DOI] [PubMed] [Google Scholar]
- 7.Pirmohamed M, Naisbitt DJ, Gordon F, Park BK. The danger hypothesis—potential role in idiosyncratic drug reactions. Toxicology. 2002;181-182:55–63. doi: 10.1016/s0300-483x(02)00255-x. [DOI] [PubMed] [Google Scholar]
- 8.Pichler WJ, et al. Pharmacological interaction of drugs with immune receptors: The p-i concept. Allergol Int. 2006;55:17–25. doi: 10.2332/allergolint.55.17. [DOI] [PubMed] [Google Scholar]
- 9.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharadwaj M, et al. Drug hypersensitivity and human leukocyte antigens of the major histocompatibility complex. Annu Rev Pharmacol Toxicol. 2011;52:501–531. doi: 10.1146/annurev-pharmtox-010611-134701. [DOI] [PubMed] [Google Scholar]
- 11.Yang CW, et al. HLA-B*1502-bound peptides: Implications for the pathogenesis of carbamazepine-induced Stevens-Johnson syndrome. J Allergy Clin Immunol. 2007;120:870–877. doi: 10.1016/j.jaci.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Chen J, He L. Harvesting candidate genes responsible for serious adverse drug reactions from a chemical–protein interactome. PLOS Comput Biol. 2009;5:e1000441. doi: 10.1371/journal.pcbi.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidney J, et al. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harndahl M, et al. Peptide binding to HLA class I molecules: Homogenous, high-throughput screening, and affinity assays. J Biomol Screen. 2009;14:173–180. doi: 10.1177/1087057108329453. [DOI] [PubMed] [Google Scholar]
- 15.Harndahl M, Rasmussen M, Roder G, Buus S. Real-time, high-throughput measurements of peptide–MHC-I dissociation using a scintillation proximity assay. J Immunol Methods. 2011;374:5–12. doi: 10.1016/j.jim.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenke R, et al. Fragment-based identification of druggable “hot spots” of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesner RA, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 18.Case DA, et al. The Amber biomolecular simulation programs. J Computat Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR–MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudolph MG, Wilson IA. The specificity of TCR/pMHC interaction. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A, -B, -C genes using an HLA-A, -B, -C null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 22.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protoc. 2008;3:1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarling AL, et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci USA. 2006;103:14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shastri N, Schwab S, Serwold T. Producing nature’s gene-chips: The generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 25.Tenzer S, et al. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci. 2005;62:1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PubMed] [Google Scholar]
- 26.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 27.Faletto MB, et al. Unique intracellular activation of the potent anti-human immunodeficiency virus agent 1592U89. Antimicrob Agents Chemother. 1997;41:1099–1107. doi: 10.1128/aac.41.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melroy J, Nair V. The antiviral activity, mechanism of action, clinical significance and resistance of abacavir in the treatment of pediatric AIDS. Curr Pharm Des. 2005;11:3847–3852. doi: 10.2174/138161205774580642. [DOI] [PubMed] [Google Scholar]
- 29.Martin AM, et al. Immune responses to abacavir in antigen-presenting cells from hypersensitive patients. AIDS. 2007;21:1233–1244. doi: 10.1097/QAD.0b013e3280119579. [DOI] [PubMed] [Google Scholar]
- 30.Day PM, Yewdell JW, Porgador A, Germain RN, Bennink JR. Direct delivery of exogenous MHC class I molecule-binding oligopeptides to the endoplasmic reticulum of viable cells. Proc Natl Acad Sci USA. 1997;94:8064–8069. doi: 10.1073/pnas.94.15.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide–MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 32.Yewdell JW, Haeryfar SM. Understanding presentation of viral antigens to CD8+ T cells in vivo: The key to rational vaccine design. Annu Rev Immunol. 2005;23:651–682. doi: 10.1146/annurev.immunol.23.021704.115702. [DOI] [PubMed] [Google Scholar]
- 33.Amir AL, et al. Allo-HLA reactivity of virus-specific memory T cells is common. Blood. 2010;115:3146–3157. doi: 10.1182/blood-2009-07-234906. [DOI] [PubMed] [Google Scholar]
- 34.Ko TM, et al. Shared and restricted T-cell receptor use is crucial for carbamazepine-induced Stevens-Johnson syndrome. J Allergy Clin Immunol. 2011;128:1266–1276. doi: 10.1016/j.jaci.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Dai S, et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc Natl Acad Sci USA. 2010;107:7425–7430. doi: 10.1073/pnas.1001772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hung SI, et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci USA. 2005;102:4134–4139. doi: 10.1073/pnas.0409500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung WH, et al. Medical genetics: A marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 38.Mallal S, et al. PREDICT-1 Study Team HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 39.Sidney J, et al. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1803s31. Unit 18.3. [DOI] [PubMed] [Google Scholar]
- 40.Smith KJ, et al. An altered position of the α2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501. Immunity. 1996;4:203–213. doi: 10.1016/s1074-7613(00)80429-x. [DOI] [PubMed] [Google Scholar]
- 41.Kjer-Nielsen L, et al. A structural basis for the selection of dominant αβ T cell receptors in antiviral immunity. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 42.Gras S, et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J Exp Med. 2010;207:1555–1567. doi: 10.1084/jem.20100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.