Abstract
MicroRNAs are important regulators of various developmental and physiological processes. However, their roles in the CD8+ T-cell response are not well understood. Using an acute viral infection model, we show that microRNAs of the miR-17-92 cluster are strongly induced after T-cell activation, down-regulated after clonal expansion, and further silenced during memory development. miR-17-92 promotes cell-cycle progression of effector CD8+ T cells, and its expression is critical to the rapid expansion of these cells. However, excessive miR-17-92 expression enhances mammalian target of rapamycin (mTOR) signaling and strongly skews the differentiation toward short-lived terminal effector cells. Failure to down-regulate miR-17-92 leads to a gradual loss of memory cells and defective central memory cell development. Therefore, our results reveal a temporal expression pattern of miR-17-92 by antigen-specific CD8+ T cells during viral infection, the precise control of which is critical to the effector expansion and memory differentiation of CD8+ T cells.
CD8+ T cells play a pivotal role in the control of numerous intracellular infections and malignancies. Upon antigen encounter, a program triggers the few antigen-specific naïve precursor cells to undergo extensive proliferation and differentiate into effector cells, which are able to produce cytokines and cytolytic proteins (1–3). In an acute infection, antigen clearance is followed by a contraction phase during which the majority of effector CD8+ T cells undergo apoptosis. However, a small fraction of effector cells manage to survive through this phase and gradually differentiate into memory cells, which are capable of long-term self-renewal and rapid response to antigen reencounter (2, 3).
It is now well established in several different infection models that effector CD8+ T cells comprise a heterogeneous population consisting of at least two subsets: (i) CD127(IL-7Rα)high killer cell lectin-like receptor G1 (KLRG1)low memory precursor cells, which are more likely to survive the contraction phase and differentiate into memory cells and (ii) CD127lowKLRG1high terminal effectors, which are short-lived, more terminally differentiated, and lack the capacity for antigen-independent homeostatic proliferation (4–6). Additional features such as high expression of CD27 as well as rapid reexpression of CD62L can also be used to distinguish memory precursors from terminal effectors (2, 6). Memory cells are also considered to be heterogeneous, consisting of central memory T cells (TCM cells) and effector memory T cells (TEM cells) (7). Central memory T cells, which express higher levels of lymph node homing receptors (e.g., CD62L and CCR7) and have better homeostatic turnover, gradually dominate in the lymphoid organs, whereas effector memory T cells preferentially reside in the peripheral organs (8). High expression of CD27, secretion of IL-2 upon restimulation, and greater proliferation potential upon antigen reencounter are also hallmarks of central memory T cells.
T-cell differentiation is regulated by an orchestration of T-cell receptor (TCR), costimulatory, and cytokine signals and is further stabilized by lineage-specific transcription factors in response to these signals (1, 2, 9). It was recently shown that microRNA (miRNA) is also a major regulator of the T-cell immune response (10). miRNAs are small noncoding RNAs consisting of ∼22 nt that bind to the 3′ UTR of the target mRNA and suppress the expression of the encoded protein by blocking translation as well as promoting degradation of the transcript (11). Experiments using mice deficient in enzymes critical to miRNA biogenesis have demonstrated an indispensable role of miRNAs in T-cell development (12). A recent study has shown that Dicer, an enzyme involved in miRNA synthesis, is indispensable for CD8+ T-cell responses (13). However, less is known about the specific miRNAs regulating effector and memory CD8+ T-cell differentiation in the context of viral infection.
In this study, we profiled the miRNA expression of naïve, effector, and memory CD8+ T cells by using the mouse model of lymphocytic choriomeningitis virus (LCMV) infection and demonstrated that multiple miRNAs in the miR-17-92 cluster and its paralogs are highly expressed in proliferating effector cells. We then showed that miR-17-92 is critical to maintain a proliferative and terminally differentiated effector state and that down-regulation of the cluster after viral clearance is necessary for CD8+ T cells to transit into the quiescent memory phenotype.
Results
miRNAs Are Crucial for CD8+ Effector T-Cell Expansion During Acute Viral Infection.
We used Dicer conditional knockout mice to examine the role of miRNAs in regulating the effector CD8+ T-cell response to an acute viral infection. To avoid defective thymic T-cell development caused by Dicer deficiency, we crossed mice bearing floxed Dicer alleles (Dicer loxP/loxP) to a transgenic strain expressing Cre recombinase driven by a truncated human granzyme B promoter (GzB-cre) (14), which is only active among mature T cells activated by TCR signal. In this study, Dicer loxP/loxP;GzB-cre+ (Dicer−/−) mice were compared with their littermate controls (Dicer loxP/loxP;GzB-cre− or Dicer loxP/wt;GzB-cre+). Although Dicer−/− mice had normal T-cell compartments before infection (Fig. S1A), they mounted a severely dampened CD8+ T-cell response compared with littermate controls (Fig. S1B) on day 8 postinfection (p.i.) with LCMV Armstrong strain (Arm). The overall numbers of LCMV-specific CD8+ T cells for the two main epitopes, DbGP33–41 and DbNP396–404, were ∼20-fold lower in the spleens of Dicer−/− mice than in those of the littermate controls, as determined by both tetramer staining (Fig. S1C) and intracellular IFN-γ staining (Fig. S1D). Accordingly, the frequency and total number of CD44high effector CD8+ T cells were also significantly reduced in the knockout mice (Fig. S1 E and F). Moreover, the defective expansion of Dicer−/− effector CD8+ T cells was accompanied with impaired viral clearance (Fig. S1G). Therefore, our results in the LCMV infection model are consistent with the previous observation that Dicer is essential for CD8+ T-cell responses during Listeria monocytogenes and vesicular stomatitis virus infections (13).
miR-17-92 Cluster and Its Paralogs Are Up-Regulated in Expanding Effector CD8+ T Cells.
We next sought to identify the miRNAs whose loss of function could account for the defective CD8+ T-cell expansion observed in Dicer−/− mice by profiling miRNA expression in LCMV-specific CD8+ T cells at different stages of the immune response. We used the P14 TCR transgenic system (TCR specific to DbGP33–41 of LCMV) (5) and sorted naïve, day 5 effector, day 8 effector, and memory (day >60) P14 cells. Effector CD8+ T cells are rapidly proliferating on day 5 p.i., and their number reaches the peak on day 8 p.i., when proliferation largely stops (Fig. S2A). Therefore, miRNAs more highly expressed in day 5 effectors than in the other three populations are more likely to play a role in clonal expansion.
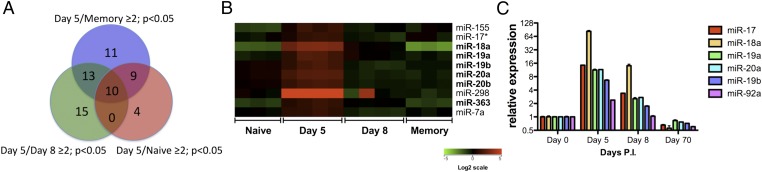
Unsupervised hierarchical analysis successfully segregated the four groups representing four different stages of LCMV-specific CD8+ T-cell differentiation (Fig. S2B). A one-way ANOVA analysis identified 160 miRNAs that were differentially regulated among the four populations (Dataset S1). miRNAs that were up-regulated in day 5 effectors by more than twofold (P < 0.05) relative to naïve cells are shown in Fig. S2C. Remarkably, multiple miRNAs in the miR-17-92 cluster and its paralogs, namely miR-106a-363 and miR-106b-25, were up-regulated on day 5 p.i. but down-regulated during the differentiation from effector to memory T cells. We identified 10 miRNAs up-regulated (fold change ≥ 2; P < 0.05) in day 5 effectors relative to all three other populations (Fig. 1A). Strikingly, six of those miRNAs belong to the miR-17-92 or miR-106a-363 cluster, suggesting a potential role of these miRNAs in the expansion phase (Fig. 1B). The expression kinetics of individual members in the miR-17-92 cluster were confirmed by quantitative RT-PCR (QRT-PCR) (Fig. 1C).
Fig. 1.
miRNAs in the miR-17-92 cluster and its paralogs are up-regulated in the proliferating effector CD8+ T cells. (A) Venn diagram of genes up-regulated by more than twofold (P < 0.05) in the day 5 effector P14 CD8+ T cells relative to the naïve (red), day 8 effector (green), or memory P14 (blue) cells. (B) Heat map of the expression of miRNAs that were up-regulated more than twofold (P < 0.05) in the day 5 effector P14 CD8+ T cells relative to naïve, day 8 effector, and memory P14 cells. miRNAs belonging to the miR-17-92 or miR-106a-363 cluster are in bold. (C) QRT-PCR analysis of the expression of individual members in the miR-17-92 cluster in naïve, day 5 p.i., day 8 p.i., and memory P14 cells. Bars represent the fold changes relative to naive. Sno-142 was used as the loading control.
miR-17-92 Deficiency Impairs Effector CD8+ T-Cell Proliferation.
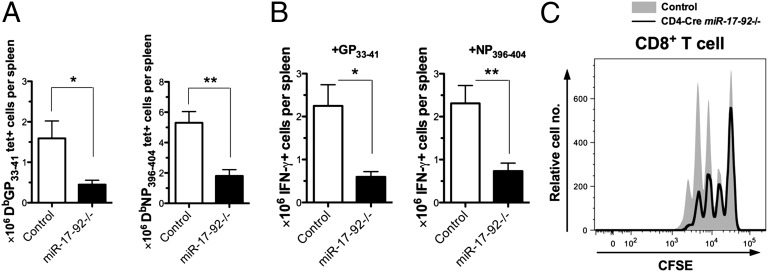
The observation that expression of miRNAs in the miR-17-92 cluster positively correlates with the proliferation of effector CD8+ T cells prompted us to speculate that miR-17-92 may promote CD8+ T-cell expansion during the immune response. We bred miR-17-92 loxP/loxP mutants (15) to GzB-cre transgenic mice to generate miR-17-92 loxP/loxP;GzB-cre (miR-17-92−/−) mice. Although a previous study showed that conventional miR-17-92 knockout mice had normal T-cell development (15), we still confirmed in our system that the miR-17-92−/− mice showed no obvious defect in the T-cell compartment (Fig. S3A). We infected miR-17-92−/− mice and their littermate controls (miR-17-92 loxP/loxP or miR-17-92 loxP/loxP;GzB-cre) with LCMV Arm. The frequency and number of effector CD8+ T cells were examined on day 8 p.i. As predicted, both DbGP33–41 and DbNP396–404 tetramer+ CD8+ T-cell frequencies in the spleens of miR-17-92−/− mice were lower than those of the littermate controls (Fig. S3B). The total numbers of DbGP33–41- or DbNP396–404-specific cells, determined by either tetramer staining or intracellular IFN-γ staining after peptide stimulation, were three- to fourfold lower in miR-17-92−/− mice than in the littermate controls (Fig. 2 A and B). Also, fewer activated CD44high CD8+ T cells were found in the knockout mice (Fig. S3 C and D). miR-17-92−/− mice had slightly higher frequencies of CD127highKLRG1low (memory precursor) effector CD8+ T cells, although not statistically significant, than their littermate controls on day 8 p.i. (Fig. S3 E and F). The trend toward higher frequencies of CD127highKLRG1low LCMV-specific CD8+ T cells was also observed in the knockout mice on day 91 p.i. (Fig. S3 G and H).
Fig. 2.
miR-17-92 deficiency reduces effector CD8+ T-cell response by inhibiting proliferation. miR-17-92−/− mice and littermate controls were infected with LCMV Arm and killed on day 8 p.i. (A and B) Numbers of tetramer+ cells per spleen (A) and numbers of IFN-γ+ cells per spleen after 5-h stimulation with GP33–41 or NP396–404 (B) were determined. Results are representative of at least three independent experiments with at least five mice per group. Student’s t test was used. *P < 0.05, **P < 0.01. (C) Carboxyfluorescein succinimidyl ester (CFSE) dilution of purified CD4-cre miR-17-92−/− and WT CD8+ T cells after culture with plate-bound anti-CD3 and soluble anti-CD28 for 48 h. Results are representative of at least two experiments with n ≥ 6.
The diminished expansion of effector CD8+ T cells observed in the miR-17-92−/− mice is likely to be caused by impaired proliferation. After stimulated for 48 h with anti-CD3 and anti-CD28 antibodies in vitro, CD8+ T cells from miR-17-92 loxP/loxP;CD4-cre (CD4-cre miR-17-92−/−) mice proliferated less than those from their littermate controls (miR-17-92 loxP/loxP or miR-17-92 loxP/loxP;CD4-cre) (Fig. 2C) despite similar expression of TCR, CD3ε, and CD28 (Fig. S3I). Our results indicate that miR-17-92 is necessary for optimal proliferation of CD8+ effector T cells.
Overexpression of miR-17-92 Promotes Effector CD8+ T-Cell Expansion.
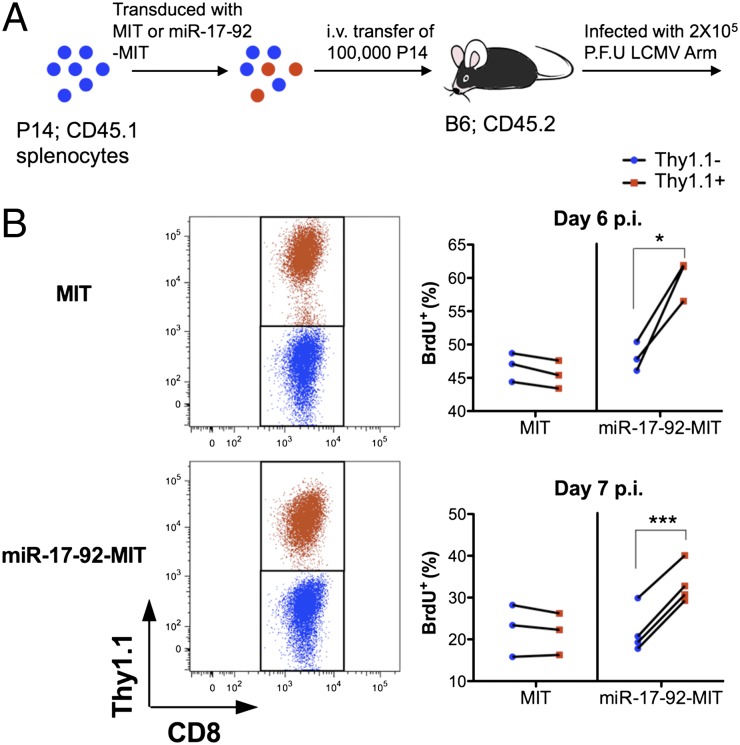
To test whether miR-17-92 has an effect on cell-cycle progression of effector CD8+ T cells, we overexpressed miR-17-92 in P14 CD8+ T cells. P14 cells were infected by retrovirus packaged with MSCV-IRES-Thy1.1 (MIT) vector with or without a miR-17-92 insert. After infection, all P14 cells, both transduced (Thy1.1+) and nontransduced (Thy1.1−), were transferred into C57BL/6 recipients, which were subsequently infected with LCMV (Fig. 3A). This procedure allows us to determine the effect of a vector by directly comparing transduced to nontransduced cells within the same mouse. Thus, any environmental factors can be ruled out, and better sensitivity is achieved. BrdU was i.p. injected into mice on day 6 or 7 after LCMV infection; 6 h later, mice were killed, and cells were checked for BrdU incorporation. Although the transduction with MIT empty vector showed little effect on the BrdU+ frequency, the transduction with miR-17-92-MIT clearly increased BrdU incorporation in the P14 cells (Fig. 3B). Therefore, increasing the expression of miR-17-92 promotes cell-cycle progression of effector CD8+ T cells.
Fig. 3.
Overexpression of miR-17-92 promotes cell-cycle progression of effector CD8+ T cells. (A) Splenocytes from Thy1.1− P14 mice were transduced with MIT or miR-17-92-MIT and transferred to C57BL/6 mice, which were subsequently infected with LCMV Arm and pulse-labeled with BrdU on day 6 or 7 p.i. (B Left) Gating of transduced (Thy1.1+; red) and nontransduced (Thy1.1−; blue) P14 cells. (Right) Frequencies of BrdU+ cells within transduced and nontransduced P14 T cells in the spleens of each group (MIT or miR-17-92-MIT) on days 6 and 7 p.i. Each line represents data from one individual mouse. Results are representative of at least two experiments with n ≥ 3. Paired Student’s t test was used. *P < 0.05, **P < 0.01, ***P < 0.001.
To test whether miR-17-92 overexpression enhances clonal expansion, we transduced purified P14 CD8+ cells with MSCV-Puro-IRES-GFP (MSCV-PIG) vector with or without a miR-17-92 insert, cultured the cells with IL-2 for 2–3 d, sorted for GFP+-transduced cells, and adoptively transferred the GFP+ T cells to C57BL/6 recipients (Fig. S4 B and C). The chimeras were subsequently infected with LCMV and killed on day 5 p.i. As shown in Fig. S4 D and E, the P14 cells transduced with the miR-17-92 overexpression vector accumulated approximately threefold more on day 5 p.i. than the P14 cells transduced with the empty MSCV-PIG.
Overexpression of miR-17-92 Skews Effector CD8+ T Cells to CD127 (IL-7Rα)lowKLRG1high Terminal Effectors.
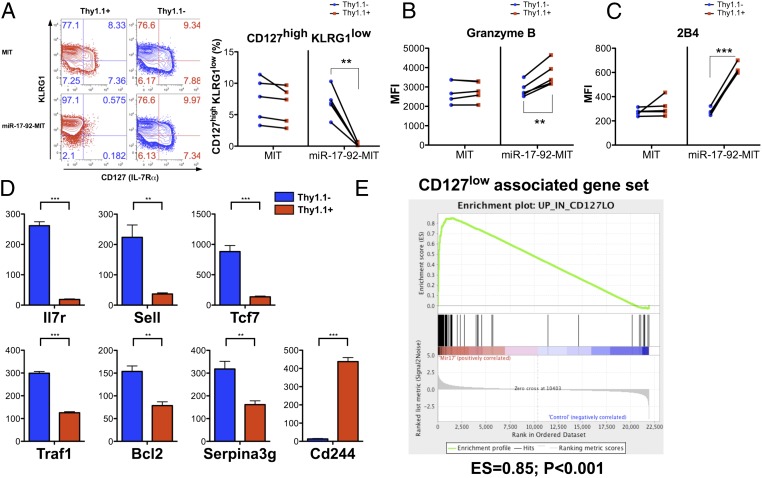
As described above, the expression of miRNAs in the miR-17-92 cluster peaks when the CD8+ T cells are rapidly proliferating, decreases by day 8 p.i. when the proliferation nearly stops, and further decreases during memory development (Fig. 1 B and C). Therefore, we reasoned that maintaining the high expression of miR-17-92 by overexpression might affect the differentiation of LCMV-specific effector and memory CD8+ T cells. We first examined the impact of miR-17-92 overexpression on effector CD8+ T-cell differentiation by comparing the day 8 P14 cells transduced with miR-17-92-MIT to the nontransduced P14 cells in the same mice as well as the empty MIT-transduced P14 cells. The nontransduced P14 cells in both the MIT and miR-17-92-MIT groups, as well as the empty MIT-transduced P14 cells, showed similar expression patterns of CD127 and KLRG1 (Fig. 4A and Fig. S5 A and B). In striking contrast, the P14 cells transduced with miR-17-92-MIT were almost exclusively CD127lowKLRG1high, a pattern associated with short-lived terminal effector cells (4, 6). Moreover, miR-17-92-MIT–transduced P14 cells also expressed lower levels of CD62L (L-selectin), CD27, and Bcl2, but higher levels of granzyme B, all consistent with a more terminally differentiated effector phenotype (Fig. 4B and Fig. S5 C–E). Interestingly, miR-17-92-MIT–transduced P14 cells also showed heightened expression of 2B4 (Fig. 4C and Fig. S5F), which is highly expressed in exhausted CD8+ T cells (16). In addition, consistent with a previous report that miR-19 directly targets TNF-α mRNA (17), the TNF-α production after restimulation by GP33–41 was lower with miR-17-92 overexpression (Fig. S5G).
Fig. 4.
Overexpressing miR-17-92 compromises the differentiation of memory precursor effector CD8+ T cells. Chimeras transferred with MIT- or miR-17-92-MIT–transduced P14 cells were generated and infected as described in Fig. 3. Phenotypic analysis of transduced (Thy1.1+) or nontransduced (Thy1.1−) P14 cells in the spleens on day 8 p.i. was performed. (A) Representative plots and statistics of CD127 and KLRG1 expression on transduced (red) and nontransduced (blue) P14 CD8+ T cells from each group (MIT or miR-17-92-MIT). Paired Student’s t test was performed. Each line represents the frequencies of CD127highKLRG1low cells in the transduced and nontransduced P14 from one individual mouse. (B and C) The same statistical analysis was performed on the expression of granzyme B, and 2B4. Experiments were repeated at least three times with n ≥ 3. (D) Relative gene expression values of Il7r (CD127), Sell (CD62L), Tcf7, Traf1, Bcl2, serpina3g, and Cd244 (2B4) in the miR-17-92-MIT–transduced (Thy1.1+; red) or nontransduced (Thy1.1−; blue) P14 cells. Student’s t test was used. (E) Gene signature of CD127low effector CD8+ T cells is overrepresented in miR-17-92-MIT–transduced P14 on day 8 p.i., as determined by GSEA. *P < 0.05, **P < 0.01, ***P < 0.001.
We examined the mRNA profiles of miR-17-92-MIT–transduced and nontransduced P14 cells by microarray analysis. Student’s t test identified 350 probes down-regulated by more than 1.5-fold (P < 0.05) and 546 probes up-regulated by more than 1.5-fold (P < 0.05) in the transduced P14 cells relative to the nontransduced P14 cells (Dataset S2). Consistent with our FACS data, Il7r, Sell (CD62L), and Bcl2 were lower and Cd244 (2B4) was higher at the transcript level in the cells transduced with miR-17-92-MIT (Fig. 4D). Notably, Tcf7, a transcription factor essential for central memory T-cell development (18), as well as Traf1 and serpina3g, which facilitate memory CD8+ T-cell survival by suppressing Bim or cathepsin B, respectively (19, 20), were also down-regulated when miR-17-92 was overexpressed. To determine whether the overall gene expression pattern of miR-17-92–overexpressing cells resembles that of terminal effector cells, we compared our data with published microarray data of terminal effector cells and memory precursors (4). Gene-set enrichment analysis (GSEA) showed that the gene signature of CD127low effectors was overrepresented in the miR-17-92-MIT–transduced P14 cells whereas the gene signature of CD127high effectors was underrepresented in these cells (Fig. 4E and Fig. S5H).
miR-17-92 Enhances Mammalian Target of Rapamycin (mTOR) Signaling in Effector CD8+ T Cells.
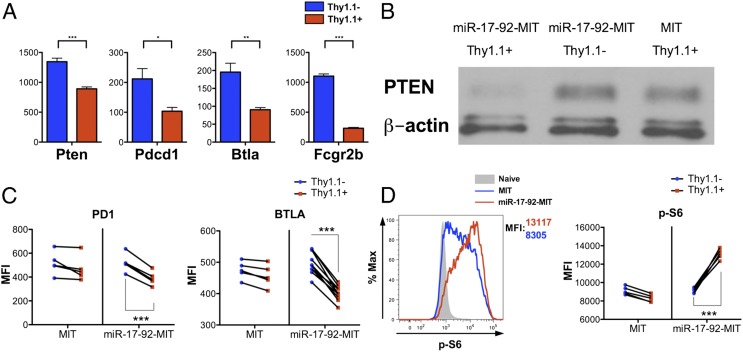
Our previous work demonstrated that reducing mTOR signaling in effector CD8+ T cells favors their differentiation into memory precursors and increases the generation of central memory T cells (21). Interestingly, our microarray data showed that the transcripts of multiple negative regulators of the PI3K–Akt–mTOR axis, namely phosphatase and tensin homolog (Pten), programmed cell death 1 (Pdcd1; PD1), B- and T-lymphocyte associated (Btla), and Fc fragment of IgG, low affinity IIb, receptor (Fcgr2b), were significantly lower in the miR-17-92-MIT–transduced P14 cells (Fig. 5A). The results of Western blots and FACS confirmed that the protein levels of PTEN, PD1, and BTLA were lowered by miR-17-92 overexpression on day 4.5 p.i. (Fig. 5 B and C), when mTOR signaling is high. Notably, the 3′ UTR of Pten mRNA contains target sites for five of the six miRNAs in the cluster (miR-17, miR-19a, miR-19b, miR-20a, and miR-92a), suggesting that miR-17-92 can directly suppress PTEN expression by interacting with its mRNA (22, 23). To determine mTOR pathway activity in the P14 cells, we stained for the phosphorylated ribosome protein S6 (Ser235/236) on day 4.5 p.i. As shown in Fig. 5D, miR-17-92 overexpression increased the phosphorylation of S6, indicating heightened mTOR signaling. Therefore, miR-17-92 relieves the suppression on the PI3K–Akt–mTOR axis and enhances mTOR activity. Strengthened mTOR signaling by miR-17-92 overexpression may explain the absence of memory precursor cells and indicates a potential defect in memory differentiation.
Fig. 5.
miR-17-92 enhances mTOR signaling by suppressing multiple negative regulators up-stream of mTOR. (A) Relative expression values of Pten, Pdcd1 (PD1), Btla, and Fcgr2b in miR-17-92-MIT–transduced and nontransduced P14 cells on day 8 p.i., as determined by microarray. (B and C) Protein levels of PTEN (B) as well as PD1 and BTLA (C) on day 4.5 p.i. were measured by Western blotting or FACS, respectively. β-Actin was used as loading control for Western blots. (D) Representative histogram of S6 phosphorylation in MIT- or miR-17-92-MIT–transduced P14 cells on day 4.5 p.i. (Left) and statistical analysis of the effect of MIT or miR-17-92-MIT transduction on S6 phosphorylation (Right). The results shown, except the microarray data, were representative of at least two independent experiments with n ≥ 4. Paired t test (C and D) and unpaired t test (A) were used. *P < 0.05, **P < 0.01, ***P < 0.001.
Down-Regulation of miR-17-92 Is Necessary for Optimal Memory CD8+ T-Cell Development.
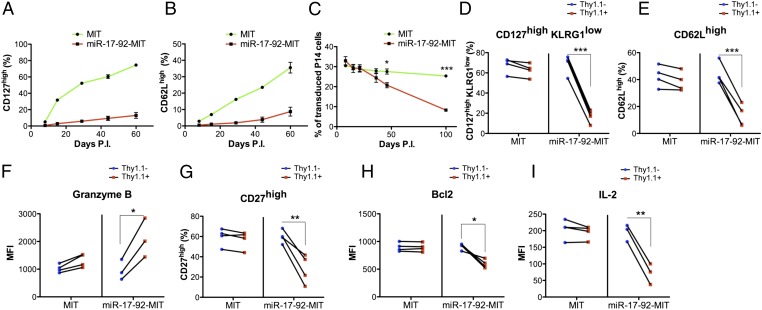
Normally, during the CD8+ T-cell response to acute LCMV infection, the proportions of CD127high and CD62Lhigh cells gradually increase after day 8 during the contraction and memory development. Meanwhile, the expression of miR-17-92 decreases to a level similar to that in naïve T cells (Fig. 1 B and C). However, when ectopically overexpressing miR-17-92, the reexpression of CD127 and CD62L in the P14 cells was strongly delayed, suggesting a defective memory differentiation program (Fig. 6 A and B). As a result, although the frequency of the empty MIT-transduced cells within the donor P14 cell pool was largely unchanged over time, the miR-17-92-MIT–transduced P14 cells were outcompeted by the nontransduced cells in the same mice (Fig. 6C). The remaining miR-17-92-MIT–transduced cells displayed a phenotype closer to what seen in effector T cells or effector memory T cells: The majority of the cells were CD127lowKLRG1high with limited expression of CD62L and high levels of granzyme B (Fig. 6 D–F and Fig. S6 A–E). The impaired central memory T-cell development was further supported by the observation that the miR-17-92-MIT–transduced memory P14 cells expressed lower CD27 and Bcl2 and produced less IL-2 upon restimulation than the control P14 cells did (Fig. 6 G–I and Fig. S6 F–H).
Fig. 6.
Overexpression of miR-17-92 impairs the development of LCMV-specific memory CD8+ T cells. (A–C) Frequencies of CD127high (A) and CD62Lhigh (B) cells within transduced P14 cells as well as the portion of transduced cell (Thy1.1+) within donor P14 cells in the peripheral blood mononuclear cells (PBMC) of each group (MIT or miR-17-92-MIT) (C) were tracked longitudinally starting from day 8 p.i. (D–I) Phenotypic analysis of transduced (Thy1.1+) or nontransduced (Thy1.1−) P14 cells in the spleens on day 66 p.i. was performed. Experiments were repeated at least three times with n ≥ 3. Unpaired t test (A–C) and paired t test (D–I) were used. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In this study, we determined the miRNA profiles of LCMV-specific CD8+ T cells during and after an acute viral infection and demonstrated that a group of miRNAs, predominantly members of the miR-17-92 cluster or its paralogs, are more expressed in rapidly proliferating effectors than in naïve, memory, or nonproliferating effector cells. The miR-17-92 cluster encodes precursors for six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a) and has two paralogs (miR-106b-25 and miR-106a-363) generated by ancient genomic duplication (24). miR-17-92 is frequently involved in genomic translocation and amplification and is overexpressed in various hematopoietic malignancies and solid tumors (24). Studies on CD4+ T cells showed that overexpressing the miR-17-92 cluster overrides the need for costimulatory signals (22) and that several miRNAs in the cluster can enhance proliferation and inhibit activation-induced cell death of T-helper cells after in vitro antigen stimulation (22, 25, 26). Accordingly, using the in vivo LCMV acute infection model, we showed that knocking out miR-17-92 with GzB-cre reduced the number of LCMV-specific CD8+ T cells and thus demonstrated that the loss of miR-17-92 at least partially accounts for the phenotype observed in Dicer knockout mice. In addition to the loss-of-function experiments, we found that overexpressing miR-17-92 promotes the expansion of effector cells. Altogether, our data reveal a proproliferative role of miR-17-92 in effector CD8+ T cells. Although the effector CD8+ T-cell response to LCMV infection is largely independent of CD4+ T-cell help (27), it is worth pointing out that GzB-cre can also induce recombination in effector CD4+ T cells that express granzyme B and potentially delete miR-17-92 in this subset of CD4+ T cells (14). Whether this process has any effect on the differentiation of memory CD8+ T cells needs to be determined in future studies.
Our data demonstrated that miR-17-92 expression is down-regulated when clonal expansion approaches the end and further reduced to levels seen in naïve cells during the contraction phase. Given the prosurvival role of miR-17-92 in malignancies as well as primary lymphocytes in the autoimmune model (22, 28), one might have predicted that maintaining high levels of miR-17-92 would make effector CD8+ T cells less vulnerable to apoptosis and favor the accumulation of memory cells. In sharp contrast, instead of surviving better, the P14 cells ectopically expressing miR-17-92 contracted more than the control P14 cells did. Memory development was also impaired, manifested by the loss of markers usually associated with memory or central memory cells. Phenotypic analysis of miR-17-92–overexpressing effector cells provides a logical link between the enhanced clonal expansion and increased contraction of these cells. Our data demonstrated that the enhanced cell-cycle progression driven by miR-17-92 overexpression is accompanied by a strong tendency toward terminal effector differentiation. In fact, the miR-17-92–overexpressing P14 cells on day 8 p.i. were almost exclusively CD127lowKLRG1high. The expression pattern of other markers such as CD62L and CD27 as well as the global transcription signature assessed by GSEA further support the idea that these cells resemble short-lived terminal effectors. However, knocking out miR-17-92 only slightly increased the frequencies of CD127highKLRG1low LCMV-specific CD8+ T cells at effector and memory time points. One likely explanation of the lesser impact on CD8+ T-cell differentiation caused by loss of function than gain of function of miR-17-92 is that the miR-106a-363 and miR-106b-25 may compensate for the loss of miR-17-92, given that the two share extensive targets with miR-17-92. In addition, the reduced CD8+ T-cell response in miR-17-92−/− mice may result in delayed antigen clearance or prolonged proinflammatory cytokine stimulation, which may impair the generation of memory precursor cells.
Interestingly, previous studies in our laboratory showed that extending antigen stimulation leads to more proliferation toward the tail end of clonal expansion, drives effectors toward terminal differentiation, and impedes the conversion from effector memory cells to central memory cells (6), which closely resembles our observation in miR-17-92–overexpressing CD8+ T cells. Therefore, miR-17-92 may be an intracellular signaling component that promotes proliferation and effector differentiation in response to antigen stimulation. In support of this hypothesis, NF-κB, a transcription factor downstream of TCR, was shown to bind to the human miR-17-92 promoter (29). Additional transcription factors such as STAT3 and E2Fs are also involved in the transcriptional regulation of miR-17-92 in human cell lines (30, 31), indicating that cytokine signals and proliferation itself may also regulate miR-17-92 expression.
In conclusion, we showed that miRNA expression patterns undergo dramatic changes in the course of a CD8+ T-cell response, and we identified the high expression of miR-17-92 cluster and its paralogs as a miRNA signature of proliferating effectors. We then dissected the role of miR-17-92 in clonal expansion and effector/memory differentiation. Our results may provide useful insights for the development of vaccines and therapies that target to enhance CD8+ T-cell effector function or maximize memory cell formation by modulating miR-17-92.
Materials and Methods
Standard procedures and methods such as mouse handling, plaque assay, in vitro T-cell activation, lymphocyte isolation, flow cytometry, retroviral transduction, BrdU labeling, RNA isolation, microarray analysis, QRT-PCR, and statistical analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Ventura for providing the MSCV-PIG and MSCV-PIG-miR-17-92 vectors and J. Jacob for providing GzB-cre transgenic mice. This work was supported by National Institutes of Health Grant AI030048 (to R.A.), Grant AI080192 (to R.A.), and Training Grant T32AI074492 (to J.S.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34218).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207327109/-/DCSupplemental.
References
- 1.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 2.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed R, Gray D. Immunological memory and protective immunity: Understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 4.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar S, et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 8.Wherry EJ, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Xiao C, Rajewsky K. MicroRNA control in the immune system: Basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 11.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 12.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593–597. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 15.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, et al. TNF-α is a novel target of miR-19a. Int J Oncol. 2011;38:1013–1022. doi: 10.3892/ijo.2011.924. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabbagh L, et al. A critical role for TNF receptor-associated factor 1 and Bim down-regulation in CD8 memory T cell survival. Proc Natl Acad Sci USA. 2006;103:18703–18708. doi: 10.1073/pnas.0602919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu N, et al. Serine protease inhibitor 2A is a protective factor for memory T cell development. Nat Immunol. 2004;5:919–926. doi: 10.1038/ni1107. [DOI] [PubMed] [Google Scholar]
- 21.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavrakis KJ, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiner DF, et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang S, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 28.Mu P, et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-κB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brock M, et al. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res. 2009;104:1184–1191. doi: 10.1161/CIRCRESAHA.109.197491. [DOI] [PubMed] [Google Scholar]
- 31.Sylvestre Y, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.