Abstract
RNA silencing (RNAi) induced by virus-derived double-stranded RNA (dsRNA), which is in a sense regarded as a pathogen-associated molecular pattern (PAMP) of viruses, is a general plant defense mechanism. To counteract this defense, plant viruses express RNA silencing suppressors (RSSs), many of which bind to dsRNA and attenuate RNAi. We showed that the tobacco calmodulin-like protein, rgs-CaM, counterattacked viral RSSs by binding to their dsRNA-binding domains and sequestering them from inhibiting RNAi. Autophagy-like protein degradation seemed to operate to degrade RSSs with the sacrifice of rgs-CaM. These RSSs could thus be regarded as secondary viral PAMPs. This study uncovered a unique defense system in which an rgs-CaM–mediated countermeasure against viral RSSs enhanced host antiviral RNAi in tobacco.
Keywords: innate immunity, arms race, ubiquitin-26S proteasome
Recent studies show that plants have evolved networks of defense mechanisms to coordinately act against pathogens (1, 2). For example, plants have two types of innate immunities against bacteria. One is induced by a receptor-like kinase on the plasma membrane for recognizing pathogen-associated molecular patterns (PAMPs), such as flagellin and elongation factor Tu, thus preventing infection (PAMPs-triggered immunity) (3). In response, pathogenic bacteria deliver to host cells a number of virulence effectors that attenuate the PAMP-triggered immunity (4, 5). The second immunity involves nucleotide-binding leucine-rich repeat (NB-LRR) proteins that recognize virulence effectors to induce hypersensitive reaction, programmed cell death, and the generation of reactive oxygen species (effector-triggered immunity). Thus, cooperative action between PAMP-triggered immunity and effector-triggered immunity enables plants to control both pathogenic and potentially pathogenic bacteria. These defense networks are thought to have evolved as a result of an arms race between plants and their bacterial pathogens. This interplay between the plant defense system and its suppression by pathogens has been portrayed as “a zigzag model” (1).
In plant–virus interactions, RNA silencing (also known as RNA interference, RNAi) triggered by viral double-stranded RNA (dsRNA) is a general mechanism involved in immunity against viruses (6, 7). In RNAi, viral dsRNA is cleaved into short interfering RNAs (siRNAs), which then silence viral RNA expression (8). Thus, by analogy with the zigzag model, dsRNA and RNAi could be regarded as a viral PAMP and PAMP-triggered immunity, respectively. Most viruses counter the RNAi defense by expressing RNA silencing suppressor (RSS) genes, which encode proteins that typically bind to siRNAs or dsRNAs (9). Binding of RSSs with viral dsRNAs decreases the availability of siRNAs for the silencing machinery. Therefore, RSSs may be regarded as viral effectors that facilitate viral infection and viral replication in plants. As suggested by the zigzag model, plants may have developed a countermeasure against viral RSSs, perhaps involving an NB-LRR protein. However, so far, NB-LRR proteins have not been found to detect viral RSS proteins, with a few exceptions (10).
Recent studies showed that transgenic tobacco plants expressing an RSS called HC-Pro, derived from tobacco etch virus (TEV) or potato virus Y, showed enhanced resistance against some viruses and other pathogens (11, 12). This surprising discovery led us to postulate that transgenic HC-Pro might elicit an immune response and that a host factor might interact with HC-Pro to elicit the response. We considered that one such candidate protein might be the tobacco calmodulin-like protein rgs-CaM, because it was previously found to interact with TEV HC-Pro (13). Although rgs-CaM was previously reported to be an endogenous RSS, here we have explored a different function for rgs-CaM in RNAi-based antiviral defense.
Results and Discussion
Binding of rgs-CaM to RSS Proteins.
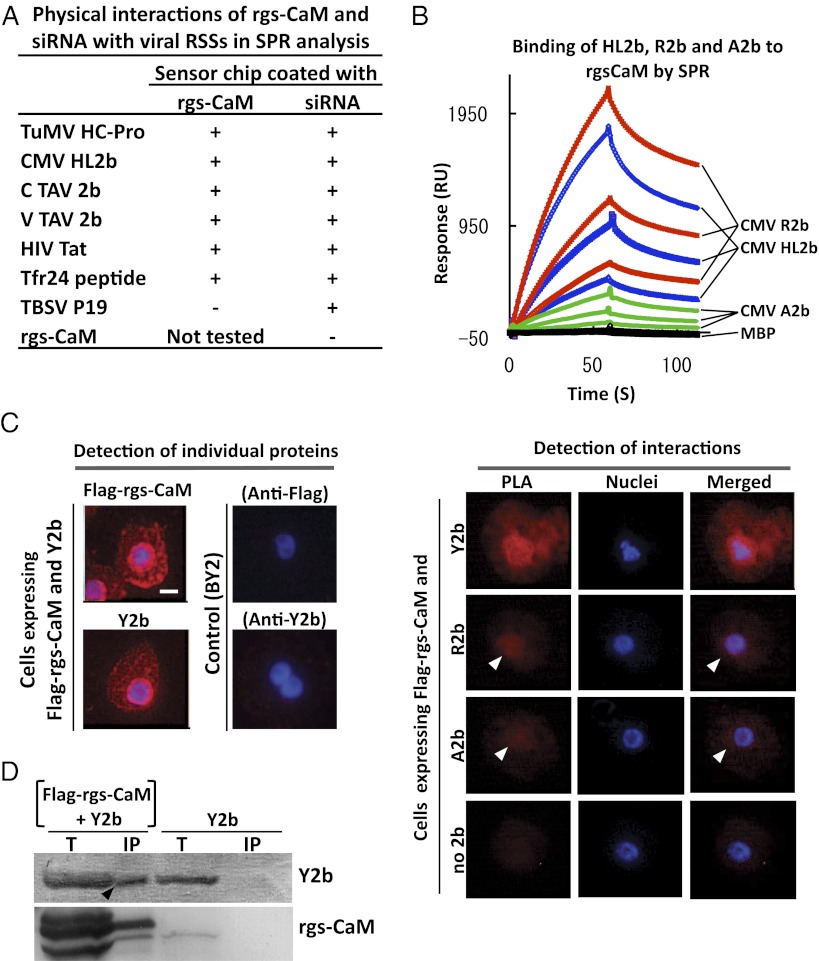
Because rgs-CaM was reported to interact with TEV HC-Pro (13), we tested for in vitro protein–protein interactions between purified rgs-CaM and RSS proteins using surface plasmon resonance (SPR) analysis (14). We found that rgs-CaM interacted with the HC-Pro protein encoded by the potyvirus, turnip mosaic virus (TuMV) (Fig. 1A and Fig. S1A). We also found that rgs-CaM interacted with the 2b class of RSS proteins from cucumber mosaic virus (CMV) and tomato aspermy virus (TAV) (Fig. 1 A and B). The 2b proteins are structurally unrelated to HC-Pro. We then looked for an interaction in tobacco BY-2 protoplasts between rgs-CaM and either clover yellow vein virus (ClYVV), HC-Pro, or CMV-Y Y2b by in situ proximity ligation assay (PLA) (15, 16). PLA measures protein–protein interaction within cells by a fluorescent signal that is detected by microscopy. As shown in Fig. 1C and Fig. S2, a strong interaction was observed between rgs-CaM and either ClYVV HC-Pro or CMV-Y Y2b. Other 2b proteins (R2b and A2b) also appeared to interact with rgs-CaM because relatively weak PLA signals (indicated by arrowheads) were detected. The signal in cells transfected only with the plasmid expressing rgs-CaM was extremely faint (Fig. 1C). Furthermore, immunoprecipitation of rgs-CaM from cell extracts specifically coprecipitated Y2b protein (Fig. 1D). These results overall indicate that RSS proteins can associate with rgs-CaM in tobacco cells.
Fig. 1.
Interaction of rgs-CaM with viral RSSs. (A) Purified RSS proteins were tested for binding to immobilized rgs-CaM protein and siRNA by SPR analysis. Plus sign, interaction detected; minus sign, no interaction detected. (Raw data are in Fig. S1A.) (B) CMV-R2b, -A2b, or -HL2b (at 150, 75, and 37.5 μg/mL) fused with maltose-binding protein (MBP) or MBP alone was tested for binding to immobilized rgs-CaM by SPR. (C) Interactions between transiently expressed Flag–rgs-CaM and Y2b, R2b, or A2b were detected in the proximity ligation assay (PLA) as fluorescent signals in BY2 cells. Each of these proteins was individually detected by single recognition PLA with anti-Flag (Flag–rgs-CaM) and anti-2b (Y2b) antibodies (Left). In cells expressing Flag-rgs-CaM and Y2b, a PLA signal means interactions between the two (Right, PLA). Weaker PLA signals (white arrowheads) were also detected for rgs-CaM combined with other 2bs (R2b and A2b). Hoechst 33342-stained nuclei (Nuclei) and merged images (Merged) are also shown. (Scale bar, 10 μm.) (D) Flag-tagged rgs-CaM (Flag–rgs-CaM) and its associated proteins were precipitated by anti-Flag antibody in crude extracts from tobacco infected with CMV/Y2b and either the PVX vector expressing Flag–rgs-CaM or the empty vector at 16 d postinoculation (dpi). Total (T) and precipitated proteins (IP) were fractionated by SDS/PAGE, and rgs-CaM and 2b were detected using specific antibodies. The 2b band in the IP fraction with PVX/Flag–rgs-CaM (arrowhead) indicates rgs-CaM interaction with 2b.
The diversity of the sequences of these various viral RSS proteins piqued our curiosity as to how rgs-CaM protein could associate with all of them. The 2b protein and other RSSs have been reported to bind to siRNA duplexes (17–20). Using SPR analysis, we confirmed that HC-Pro and 2b RSSs bind to siRNAs in vitro (Fig. 1A and Fig. S1A). It was possible that rgs-CaM associates with RSSs by specifically recognizing their small dsRNA-binding domains. To test this hypothesis, we examined whether rgs-CaM could associate with a dsRNA-binding protein that was not a plant viral RSS. We chose Tat, an HIV protein that binds to the Tar stem loop RNA located in the HIV RNA genome. A 24-amino acid, arginine-rich region (Tfr24) derived from Tat is necessary and sufficient to bind not only to Tar RNA (21, 22) but also siRNA (23). To confirm these findings, we showed that Tat and Tfr24 were able to bind an siRNA duplex in a SPR assay (Fig. 1A and Fig. S1A). We then found that rgs-CaM and Tat (Tfr24) associated with each other in vitro, suggesting that rgs-CaM binds to proteins that recognize short dsRNA (Fig. 1A and Fig. S1A).
We excluded the possibility that these in vitro interactions were either indirect or nonspecific. First, we determined that rgs-CaM itself did not interact with siRNA (Fig. 1A), arguing against the bridging of the RSS interactions by contaminating RNA. Second, we tested whether rgs-CaM interacts with the P19 RSS from tomato bushy stunt virus (TBSV). Because P19 binds to siRNA duplexes by making contact with the siRNA termini (24, 25), it recognizes dsRNA by a different mechanism from other RSSs, whose binding to dsRNA rely upon positively charged residues (26, 27). We observed no interaction between rgs-CaM and P19 by SPR (Fig. 1A and Fig. S1A), further supporting our hypothesis that rgs-CaM specifically binds to certain types of RSS proteins.
Binding of rgs-CaM to the Positively Charged Domains in RSSs.
The arginine-rich region of Tfr24 is essential for binding to Tar RNA (22). Similarly, CMV 2b also has an arginine-rich region that is required for siRNA binding (19). This region appears to be sufficient for dsRNA-binding because a peptide containing the region was found to bind to siRNAs in vitro. A single arginine-to-cysteine substitution at position 46 in this region results in the 2b protein having reduced affinity for binding siRNAs (19). We reasoned that if rgs-CaM binds to the arginine-rich domain of 2b, then the R46C mutant form of 2b (called A2b) will have impaired binding to rgs-CaM. Indeed, the binding affinity of A2b for rgs-CaM and siRNA was significantly lower than that of the wild-type R2b protein (Fig. 1 B and C and Fig. S1C).
If rgs-CaM directly binds to the arginine-rich region, then what structure confers the binding affinity? Homology models predicted that the rgs-CaM protein had a negatively charged cleft separating its two EF hand domains for calcium binding (Fig. S3). Simulated docking of the 2b protein structure confirmed that 2b would easily fit within the negatively charged cleft. rgs-CaM is a calmodulin-like protein, and so we wondered whether other calmodulin and calmodulin-like proteins have binding affinities for the 2b RSS. To test this possibility, we selected NtCaM13, a calmodulin protein identified in tobacco (28). NtCaM13 is reported to be highly expressed during the hypersensitive response induced by tobacco mosaic virus (29). When we tested NtCaM13 for binding to CMV 2b proteins by SPR, we observed interactions with wild-type 2b proteins but not with the mutant A2b (Fig. S1B). Human calmodulin binds to the RNA binding domain of Tat (30). Overall, these results suggest that calmodulin and calmodulin-like proteins might interact with dsRNA-binding proteins via their basic binding domains. Calmodulin is known to be a hub protein, which can bind to hundreds of proteins via its disordered binding domain with a high surface charge (31). Our homology modelings suggest that rgs-CaM has a similar negatively charged binding domain for a positively charged target (Fig. S3), which enables rgs-CaM to interact with diverse viral RSSs.
Effects of rgs-CaM on Viral RSS Activity.
What effect does rgs-CaM binding have on RSS activities? If rgs-CaM binds to the same RSS domain that mediates siRNA binding, we predicted that rgs-CaM would act as a competitive inhibitor of siRNA binding. Preincubation of rgs-CaM with TAV 2b protein inhibited 2b from binding to siRNA (Fig. S1D). However, siRNA- and rgs-CaM binding were not identical because siRNA preincubation had a much greater effect on the ability of TAV 2b to bind to siRNAs than did rgs-CaM preincubation.
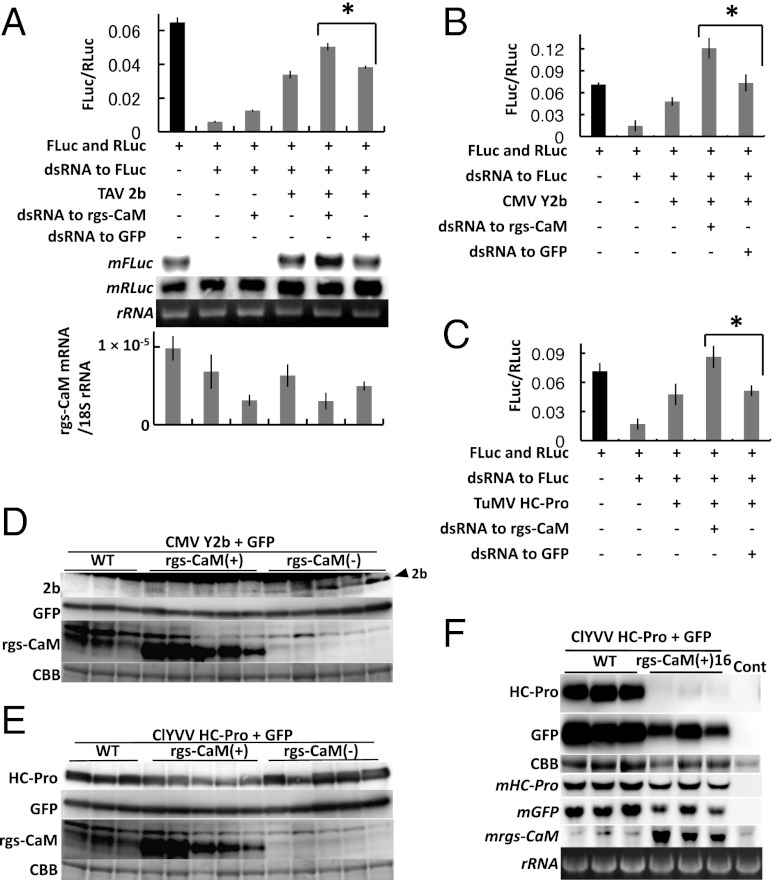
The siRNA-binding activity of RSSs is important for their antisilencing activities (14, 19, 32). Therefore, by interfering with siRNA binding, rgs-CaM might attenuate RSS antisilencing activity. To test this idea, we measured RSS antisilencing activity in protoplasts of Nicotiana benthamiana using a dual-luciferase reporter assay (14). Depletion of endogenous rgs-CaM in protoplasts by dsRNA-mediated RNAi resulted in a significant increase in the antisilencing activities of 2b and HC-Pro (Fig. 2 A–C). It was possible that some of the effects on luciferase silencing were simply due to the addition of more dsRNA into the cells. If the RNAi machinery is limiting, then rgs-CaM dsRNA could compete with luciferase dsRNA for silencing. We found that the addition of rgs-CaM dsRNA itself tended to inhibit luciferase silencing by 10% (Fig. 2A). Normalization for this contribution to antisilencing did not affect the statistical significance we observed on antisilencing with 2b and HC-Pro (Fig. 2 A–C). We also tried to evaluate the effect of overexpressed rgs-CaM on RSS activities. However, we were not able to increase the rgs-CaM transcript levels when we introduced the rgs-CaM expression construct into protoplasts.
Fig. 2.
Effects of rgs-CaM–RSS interactions on the activity and stability of RSS proteins. (A–C) RSS activity of TAV 2b (A), CMV Y2b (B), and TuMV P1/HC-Pro (C) were measured by a dual luciferase assay in protoplasts of N. benthamiana. When endogenous rgs-CaM was silenced by a dsRNA cognate to rgs-CaM mRNA, RSS activities of all RSS proteins increased significantly, and even in the absence of RSS, RNAi activity was reduced (A; *P < 0.01, paired Student t test). Firefly and Renilla luciferase mRNAs (mFLuc and mRLuc, Middle) were also detected in RNA extracts from the same samples by Northern blotting (A, Middle), and results were consistent with the luciferase assay (A, Upper graph). The accumulation of endogenous rgs-CaM mRNA was monitored by real-time PCR to confirm its silencing (A, Lower graph). The x axis is the same as in Upper graph. Bars indicate SD. (D and E) CMV Y2b (D) or ClYVV P1/HC-Pro (E) and green fluorescent protein (GFP) were transiently expressed by agroinfiltration in transgenic T0 tobacco plants that either overexpressed rgs-CaM [rgs-CaM(+)] or silenced endogenous rgs-CaM [rgs-CaM(−)], and in nontransgenic plants (WT). Accumulated CMV 2b, ClYVV HC-Pro, GFP, and rgs-CaM proteins were detected by Western blotting. Arrowhead marks the 2b protein. Below each panel set, a Coomassie brilliant blue (CBB)-stained gel is shown as a loading control. Accumulation of the RSS proteins relative to GFP is shown in bar graphs in Fig. S4. (F) The same experiment shown in E was carried out using line 16 of T1 transgenic plants expressing rgs-CaM [rgs-CaM(+)16]. HC-Pro, GFP, and rgs-CaM mRNAs (mHC-Pro, mGFP, and mrgs-CaM) were additionally detected in total RNA extracts by Northern blotting. Lane control (Cont): Extracts from nontransgenic plant. Ribosomal RNA (rRNA) stained with ethidium bromide is shown as a loading control. We here note that the GFP mRNA levels were increased by the coinfiltrated HC-Pro in WT.
We then analyzed transgenic tobacco plants in which endogenous rgs-CaM protein was depleted [rgs-CaM(−)] or overexpressed [rgs-CaM(+)] (Fig. 2 D–F). The rgs-CaM(−) plants were constructed by transforming them with an expression vector containing a tandem antisense–sense fragment of the rgs-CaM cDNA under control of the 35S promoter of cauliflower mosaic virus to induce RNAi against rgs-CaM. The rgs-CaM(+) plants were constructed by transforming them with an expression vector containing the rgs-CaM ORF under the 35S promoter. When the Y2b RSS protein was transiently expressed by agroinfiltration into rgs-CaM(−) plants, Y2b accumulation increased compared with control (Fig. 2D). Y2b was not detected when agroinfiltrated into rgs-CaM(+) plants (Fig. 2D and Fig. S4). We also introduced ClYVV HC-Pro into these plants. HC-Pro was reduced in rgs-CaM(+) plants but there was no significant increase in HC-Pro in rgs-CaM(−) plants (Fig. 2E and Fig. S4).
The effect of rgs-CaM on antisilencing by RSSs in vivo could be strictly mediated by its competition with RSS–siRNA binding. However, the results of Fig. 2D implied that rgs-CaM inhibits RSS antisilencing by additional mechanisms. Redundant inhibition would be a more robust strategy for quenching RSSs because multiple modes for antisilencing have been implicated in some RSSs (33–35). We looked at the effect of rgs-CaM on RSS protein abundance in vivo. Western and Northern blots revealed that rgs-CaM inhibited the abundance of HC-Pro protein but had no effect on HC-Pro mRNA (Fig. 2F). This result indicated that rgs-CaM either stimulates HC-Pro protein degradation or limits its translation, or both.
Involvement of Autophagy-Like Protein Degradation in Control of rgs-CaM and RSS Expression.
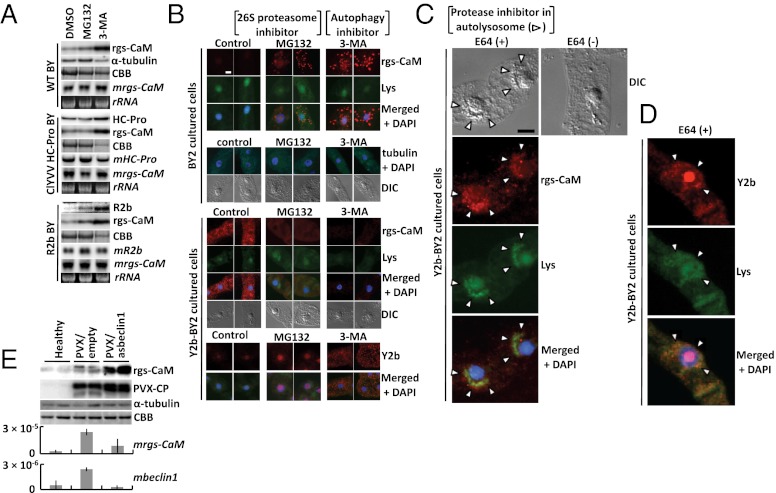
We examined the degradation mechanism of rgs-CaM and RSS proteins. Preliminary analysis had found that rgs-CaM protein expressed by the potato virus X (PVX) vector was sensitive to 26S proteasome inhibitors in tobacco (Fig. S5). We initially thought that endogenous rgs-CaM and even RSSs might also be sensitive to 26S proteasome inhibitors, but they had little effect on the accumulation of endogenous rgs-CaM and RSSs (Fig. 3 A and B). Instead, endogenous rgs-CaM protein was sensitive to 3-methyladenine (3-MA), an inhibitor of another proteolytic pathway, autophagy (36–38). A similar effect of 3-MA was observed on viral RSSs that were expressed in transgenic tobacco plants and cultured BY2 cells (Fig. 3 A and B).
Fig. 3.
Control of rgs-CaM and RSS expression by proteolytic activities. (A) Western blots of transgenic BY tobacco leaves expressing ClYVV HC-Pro or CMV R2b and of nontransgenic tobacco leaves after 16-h treatment with inhibitors of host proteolytic pathways [20 μM clastolactacystin–lactone (Sigma-Aldrich) and 40 μM MG132 (MG132; Sigma-Aldrich) to inhibit 26S proteasome, 5 mM 3-methyladenine (3-MA; Sigma-Aldrich) to block autophagy or DMSO as control]; RSSs, α-tubulin and endogenous rgs-CaM were detected using urea as described in SI Materials and Methods and Fig. S8. Their mRNAs (mHC-Pro, mR2b, and mrgs-CaM) were also detected by Northern blotting of RNAs extracted from the same leaves. CBB-stained and ethidium-bromide-stained gels are shown as loading controls. (B–D) After treatment of BY2 cultured cells with inhibitors of host proteolytic pathways [20 μM MG132 to inhibit 26S proteasomes, 5 mM 3-MA and 10 μM E64 (Sigma-Aldrich) to block autophagy or DMSO as control] overnight, wild type (B) and Y2b-expressing (B–D) BY2 cultured cells were stained with LysoTracker (Lys; Life Technologies) and DAPI. Endogenous rgs-CaM (B and C), CMV Y2b (B and D), and α- and β-tubulin (tubulin) (B) were detected using specific antibodies. Differential interference contrast (DIC) images are also shown. (Scale bar, 10 μm.) (E) Western blots of inoculated leaves of tobacco infected with the PVX vector expressing antisense of beclin1 (PVX/asbeclin1) or not (PVX/empty) detected rgs-CaM, PVX CP, and α-tubulin at 14 dpi. The CBB stained gel was shown as loading control. The rgs-CaM and beclin1 mRNA levels were also investigated by real-time PCR. The relative accumulation of these mRNA to 18S ribosomal RNA was shown with bars of SD.
These data suggested that rgs-CaM and RSSs were degraded by autophagosomes. If so, then were the proteins localized in these organelles? We were unable to see colocalization of rgs-CaM and Y2b in the presence or absence of inhibitors of the proteolytic pathways (Fig. 3B). However, we observed that rgs-CaM and Y2b colocalized with LysoTracker-stained bodies in the perinuclear region of BY2 cells (Fig. 3 C and D). LysoTracker specifically marks autolysosomes. This was only observed when cells were subjected to sucrose starvation and treated with the cysteine protease inhibitor, E64. E64 treatment following sucrose starvation was reported to stimulate the accumulation of active autolysosomes at the nuclear periphery of BY2 cells (38, 39). Our observations therefore indicate that both rgs-CaM and Y2b are recruited into autolysosomes. Moreover, we could detect the colocalization of Y2b with rgs-CaM in autolysosomes only when protein degradation was blocked by E64 (Fig. 3 C and D), suggesting that the rgs-CaM and Y2b were likely degraded by autophagy-like protein degradation (ALPD) immediately after they form a complex.
The involvement of ALPD was genetically confirmed in tobacco by using PVX-induced gene silencing to inhibit an essential gene for autophagy called beclin1 (autophagy-related gene 6, atg6) (40) (Fig. 3E). Infection of the PVX empty vector itself induced the expression of beclin1 and rgs-CaM genes (Fig. S6E), consistent with a previous study (40). Infection increased accumulation of rgs-CaM mRNA, but the protein level of rgs-CaM changed little. The PVX vector carrying the antisense sequence of beclin1 completely suppressed beclin1 expression and increased the level of the rgs-CaM protein (Fig. 3E). These results suggest that rgs-CaM transcription is induced by PVX infection, but most of the induced rgs-CaM protein is quickly degraded by ALPD unless the PVX infection is accompanied by a suppression of autophagy. Autophagy is thought to be a nonspecific, bulk degradation process. However, recent studies have shown that protein degradation by autophagy is selective for specific organelles, harmful protein aggregates, and even foreign pathogens, including bacteria and viruses (xenophagy: autophagy of foreign matter). The ubiquitination of these substrates has been identified as the necessary signal for selective degradation by auto- and xenophagy (36, 41).
Reactions of rgs-CaM Against Viral Infection.
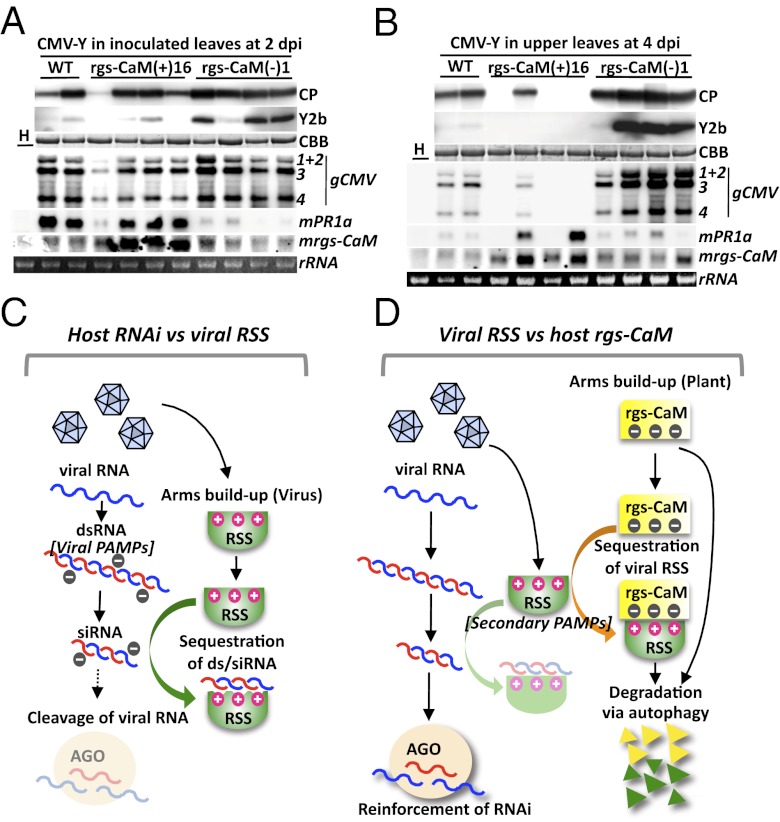
Expression of endogenous rgs-CaM protein was induced in a tobacco plant that also expressed TEV HC-Pro (13). We confirmed that other RSSs and various viral infections could also induce rgs-CaM expression at both the mRNA and protein levels (Fig. S6). rgs-CaM protein was induced in response to Y2b expression and was distributed throughout BY2 cells (Fig. 3B). We hypothesized that rgs-CaM protein induction had a role in antiviral defense and inhibited virus infection. To test this hypothesis, tobacco plants were infected with CMV. Plants overexpressing rgs-CaM were less susceptible to viral infection, whereas those in which rgs-CaM was knocked down were more susceptible (Fig. 4 A and B and Fig. S7). This finding supports the model that rgs-CaM counteracts RSSs in viral infections and has an impact on the outcome of virus infection (Fig. 4 C and D).
Fig. 4.
Resistance and response to CMV-Y infection in tobacco plants and schematic model of antiviral defense in tobacco. (A and B) T1 transgenic tobacco plants overexpressing rgs-CaM [rgs-CaM(+)16] or with knocked down rgs-CaM [rgs-CaM(−)1] and nontransgenic (WT) tobacco plants were inoculated with CMV-Y. Capsid (CP) and Y2b proteins, viral genome RNAs (gCMV), and pathogenesis-related protein 1a and rgs-CaM mRNAs (mPR1a, mrgs-CaM) were detected by Western and Northern blotting as in Fig. 2F. Lane H: healthy WT plants. (C) Presently accepted model of the viral counterdefense against the host’s RNAi-based defense. The viral RNA genome is configured into intermolecular and/or intramolecular dsRNA, which induces antiviral RNAi in the plants. To counteract this host defense, the virus expresses RSSs, many of which can bind to siRNA and/or dsRNA. Viral RSSs, in turn, may sequester viral siRNAs and long dsRNAs from inducing RNAi and eventually enhance viral fitness in the host. (D) The proposed model of the tobacco countermeasure against viral RSSs is depicted, as uncovered in the present study. Tobacco expresses the calmodulin-like protein rgs-CaM, which has an affinity for positively charged siRNA/dsRNA-binding surfaces of viral RSSs. The rgs-CaM prevents RSS from binding to dsRNAs/siRNAs and affects RSS protein stability by autophagy, resulting in a more potent RNAi defense against viral infection.
We postulate that tobacco plants use rgs-CaM as a defense factor that binds viral RSSs through its affinity for positively charged siRNA/dsRNA-binding domains. Binding by rgs-CaM reduces the ability of viral RSSs to bind and titrate viral siRNAs away from the plant’s RNAi machinery, and eventually degrades viral RSSs in cooperation with ALPD. As a consequence, the viral counterdefense system using RSSs is weakened, allowing the plant’s RNAi system to mount a more vigorous antiviral response. Thus, rgs-CaM cooperates with the plant’s RNAi system, but at a level one step removed, as a counter countermeasure to viral infection. The role of rgs-CaM therefore relies upon the virus countering a different plant defense. Autophagy may be a general strategy for an innate immune system against viruses in plants and animals, as Li et al. (42) recently demonstrated that human cells use autophagy to defend against invasion of tobacco mosaic virus RNA.
Anandalakshmi et al. (13) previously reported that rgs-CaM is an endogenous host RSS. Our results are not in agreement with this conclusion because we were not able to detect any RSS activity of rgs-CaM in the protoplast system (Fig. 2A). We also failed to detect any RSS activity of rgs-CaM in conventional agroinfiltration of N. benthamiana leaves (Fig. S9) or in transgenic tobacco plants that overexpress or lack rgs-CaM (Fig. S10). However, heterologous expression of rgs-CaM in Drosophila S2 cells partially inhibited RNAi activity (Fig. S11), suggesting that under some circumstances, rgs-CaM might have some RSS-like activity. For example, when overexpressed, rgs-CaM could conceivably interfere with the siRNA-binding activities of a factor such as R2D2, a dsRNA-binding protein in the Drosophila RNAi pathway that bridges the initiation and effector steps of the pathway. Perhaps rgs-CaM expression is strictly induced in cells in response to stress conditions. When tobacco is not infected, rgs-CaM might be unstable by virtue of proteolytic activities, especially ALPD (Fig. 3 A–C). Conversely, rgs-CaM protein levels might be strongly increased by virus infection (or RSS expression) (Fig. 3B and Fig. S6). We think that rgs-CaM might be potentially harmful to the host silencing machinery controlling cellular RNA levels, and thus it is kept at a low level until needed. Therefore, rgs-CaM seems to act, in coordination with ALPD, to homeostatically govern RNAi-based antiviral defense.
Role of rgs-CaM as an Agent for Viral Recognition in the Plant–Virus Arms Race.
The existence of a counter countermeasure to viral infection intuitively makes sense from the point of view of a biological arms race; rgs-CaM might be part of an ongoing struggle by plants to keep viral infections in check. Binding of dsRNA/siRNA is thought to be a general strategy for viruses to suppress RNAi in plants (17, 18). By recognizing and blocking the dsRNA-binding sites on RSSs, which act as viral PAMPs (3), rgs-CaM may be able to recognize most RNA viruses in the initial stage of infection. However, to escape from such PAMP recognition by rgs-CaM, some viruses may have evolved different strategies. We have no idea whether viruses that had used dsRNA/siRNA-binding strategy at one time then developed new RSSs as a consequence of an arms race to escape from rgs-CaM. However, some viral RSSs do not use dsRNA binding to suppress RNAi (43–45). Considering that rgs-CaM can bind to diverse viral RSSs and reinforce host RNAi defense by sequestering RSSs, we should regard such viral RSSs as “secondary viral PAMPs” for cellular recognition of viruses (Fig. 4 C and D).
It is well known that plants have a two-step bacterial immune system consisting of PAMP-triggered immunity and subsequent effector-triggered immunity. When bacterial effectors can overcome PAMP-triggered immunity, plants recognize the effectors as avirulence factors to induce R-gene–mediated resistance usually accompanied by hypersensitive reaction. Viral RSSs may be considered as either secondary PAMPs or effectors. If precisely defined, viral RSSs may be effectors because they suppress PAMP-triggered immunity. However, unlike fungal or bacterial effectors, viral RSSs do not necessarily activate R-gene–mediated resistance. From this point of view, we tentatively use the phrase, secondary PAMPs for viral RSSs.
Materials and Methods
rgs-CaM, RSS, and NtCaM13 Genes and Viruses.
The cDNA sequence of the rgs-CaM gene, identical to that reported previously (13), was cloned from Nicotiana tabacum cv. Xanthi-nc. We used sequence information of the N. benthamiana homologous gene (TC16502), which was developed on the basis of EST information from the Gene Index Project database (Computational Biology and Functional Genomics Laboratory, Dana Farber Cancer Institute and Harvard School of Public Health, Boston; website http://compbio.dfci.harvard.edu/tgi). Its deduced amino acid sequence is highly homologous (71.1% identity) to that of rgs-CaM, and its in vitro translated protein was bound by anti–rgs-CaM antibodies.
CMV RSS 2b genes, HL2b, Y2b, A2b, and R2b were cloned from CMV strains belonging to subgroup I, HL-CMV(46), CMV-Y(47), CM95 (48), and CM95R (48), respectively. CM95 is an attenuated CMV isolate that has been used to control CMV diseases in Japan. CM95R, a spontaneous revertant generated from CM95 in the field, induces severe symptoms in inoculated plants. CMV-Y (described as CMV/Y2b in this study) is a typical strain that causes severe symptoms in infected plants. HL-CMV was isolated from an edible lily (Lillium leichtlinii cv. Hakugin). CMV-H1 was also used as an attenuated CMV lacking an RSS, 2b (49).
For the other RSSs, potyviral RSS, HC-Pro genes were from ClYVV (50) and TuMV. HC-Pros, TAV 2bs, TBSV P19, and HIV Tat were tested for binding to rgs-CaM (or tobacco calmodulin NtCaM13; ref. 28) and/or siRNA duplex using SPR analysis (14).
In Situ PLA.
At 16 h after transfection of BY2 protoplasts, treated cells were fixed on slides and reacted with primary antibodies basically according to the protocol on The Arabidopsis Information Resources (TAIR) website (http://www.arabidopsis.org/cshl-course/7-gene_expression.html, Part B, Immunofluorescence labeling of Arabidopsis protoplasts in gene expression and protein localization in Arabidopsis) with slight modifications. Slides were precoated with 0.005% (wt/vol) poly-l-lysine (Sigma). Primary antibodies were 60-fold diluted with blocking solution [2% (wt/vol) BSA]. The samples were reacted with the PLA probe (secondary antibody conjugated with oligonucleotides), and after hybridization between PLA probes, ligations, and amplifications, detection with fluorescently labeled oligonucleotides (Olink Bioscience; detection kit 613 Ex/Em: 598/613) was carried out according to the manufacturer’s protocol. This method was successfully used to detect the interaction between viral proteins of ClYVV, and of CMV 2b with Arabidopsis catalase 3 (16, 51). The expression and distribution of target proteins are detected by a single recognition PLA using one primary antibody. The in situ specific interaction between two proteins was detected by a double recognition PLA using two primary antibodies. Photomicrographs were taken using a Leica DMI6000 B microscope (Leica Microsystems). Image colors were then reassigned using AF6000 ver. 1.5 software as follows: a PLA signal, red; Hoechst 33342, blue.
Supplementary Material
Acknowledgments
We thank H. Bando, H. Siomi, K. Miyoshi, M. Katahira, and S. Yamaguchi for reagents, technical advice, and encouragement. This work was supported by the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (17780032, 18108001, and 20688002) and a Grant-in-Aid (to K.S.N.) for Regional Research and Development Proposal-Based Program from Northern Advancement Center for Science and Technology of Hokkaido, Japan. This work was also supported by National Institutes of Health Grant GM068743 (to R.W.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201628109/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 4.Alfano JR, Collmer A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 5.Abramovitch RB, Martin GB. Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 8.Mlotshwa S, Pruss GJ, Vance V. Small RNAs in viral infection and host defense. Trends Plant Sci. 2008;13:375–382. doi: 10.1016/j.tplants.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Csorba T, Pantaleo V, Burgyán J. RNA silencing: An antiviral mechanism. Adv Virus Res. 2009;75:35–71. doi: 10.1016/S0065-3527(09)07502-2. [DOI] [PubMed] [Google Scholar]
- 10.Moffett P. Mechanisms of recognition in dominant R gene mediated resistance. Adv Virus Res. 2009;75:1–33. doi: 10.1016/S0065-3527(09)07501-0. [DOI] [PubMed] [Google Scholar]
- 11.Pruss GJ, et al. The potyviral suppressor of RNA silencing confers enhanced resistance to multiple pathogens. Virology. 2004;320:107–120. doi: 10.1016/j.virol.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 12.Shams-Bakhsh M, Canto T, Palukaitis P. Enhanced resistance and neutralization of defense responses by suppressors of RNA silencing. Virus Res. 2007;130:103–109. doi: 10.1016/j.virusres.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Anandalakshmi R, et al. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- 14.Shimura H, et al. A strategy for screening an inhibitor of viral silencing suppressors, which attenuates symptom development of plant viruses. FEBS Lett. 2008;582:4047–4052. doi: 10.1016/j.febslet.2008.10.046. [DOI] [PubMed] [Google Scholar]
- 15.Söderberg O, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Inaba J, Kim BM, Shimura H, Masuta C. Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol. 2011;156:2026–2036. doi: 10.1104/pp.111.180042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakatos L, et al. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mérai Z, et al. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007;48:1050–1060. doi: 10.1093/pcp/pcm074. [DOI] [PubMed] [Google Scholar]
- 20.Rashid UJ, Hoffmann J, Brutschy B, Piehler J, Chen JC. Multiple targets for suppression of RNA interference by tomato aspermy virus protein 2B. Biochemistry. 2008;47:12655–12657. doi: 10.1021/bi801281h. [DOI] [PubMed] [Google Scholar]
- 21.Weeks KM, Ampe C, Schultz SC, Steitz TA, Crothers DM. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- 22.Calnan BJ, Biancalana S, Hudson D, Frankel AD. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 23.Segura T, Hubbell JA. Synthesis and in vitro characterization of an ABC triblock copolymer for siRNA delivery. Bioconjug Chem. 2007;18:736–745. doi: 10.1021/bc060284y. [DOI] [PubMed] [Google Scholar]
- 24.Silhavy D, et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vargason JM, Szittya G, Burgyán J, Hall TMT. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 26.Chen HY, Yang J, Lin C, Yuan YA. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 2008;9:754–760. doi: 10.1038/embor.2008.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Yuan YA. A structural perspective of the protein-RNA interactions involved in virus-induced RNA silencing and its suppression. Biochim Biophys Acta. 2009;1789:642–652. doi: 10.1016/j.bbagrm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Yamakawa H, et al. Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur J Biochem. 2001;268:3916–3929. doi: 10.1046/j.1432-1327.2001.02301.x. [DOI] [PubMed] [Google Scholar]
- 29.Takabatake R, et al. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 2007;48:414–423. doi: 10.1093/pcp/pcm011. [DOI] [PubMed] [Google Scholar]
- 30.McQueen P, et al. Tat peptide-calmodulin binding studies and bioinformatics of HIV-1 protein-calmodulin interactions. Proteins. 2011;79:2233–2246. doi: 10.1002/prot.23048. [DOI] [PubMed] [Google Scholar]
- 31.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Shiboleth YM, et al. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J Virol. 2007;81:13135–13148. doi: 10.1128/JVI.01031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azevedo J, et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 38.Takatsuka C, Inoue Y, Matsuoka K, Moriyasu Y. 3-methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 2004;45:265–274. doi: 10.1093/pcp/pch031. [DOI] [PubMed] [Google Scholar]
- 39.Toyooka K, et al. Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy. 2006;2:96–106. doi: 10.4161/auto.2.2.2366. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 42.Li L, et al. The invasion of tobacco mosaic virus RNA induces endoplasmic reticulum stress-related autophagy in HeLa cells. Biosci Rep. 2012;32:171–186. doi: 10.1042/BSR20110069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pazhouhandeh M, et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giner A, Lakatos L, García-Chapa M, López-Moya JJ, Burgyán J. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 2010;6:e1000996. doi: 10.1371/journal.ppat.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda A, et al. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005;24:3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi N, et al. Genetic mapping of the compatibility between a lily isolate of Cucumber mosaic virus and a satellite RNA. J Gen Virol. 2005;86:2359–2369. doi: 10.1099/vir.0.81059-0. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki M, et al. Functional analysis of deletion mutants of cucumber mosaic virus RNA3 using an in vitro transcription system. Virology. 1991;183:106–113. doi: 10.1016/0042-6822(91)90123-s. [DOI] [PubMed] [Google Scholar]
- 48.Kosaka Y, Fukunishi T. Multiple inoculation with three attenuated viruses for the control of cucumber virus disease. Plant Dis. 1997;81:733–738. doi: 10.1094/PDIS.1997.81.7.733. [DOI] [PubMed] [Google Scholar]
- 49.Matsuo K, et al. Development of Cucumber mosaic virus as a vector modifiable for different host species to produce therapeutic proteins. Planta. 2007;225:277–286. doi: 10.1007/s00425-006-0346-5. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi Y, Takahashi T, Uyeda I. A cDNA clone to clover yellow vein potyvirus genome is highly infectious. Virus Genes. 1997;14:235–243. doi: 10.1023/a:1007940028058. [DOI] [PubMed] [Google Scholar]
- 51.Nakahara KS, et al. Involvement of the P1 cistron in overcoming eIF4E-mediated recessive resistance against Clover yellow vein virus in pea. Mol Plant Microbe Interact. 2010;23:1460–1469. doi: 10.1094/MPMI-11-09-0277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.