Abstract
The full regulatory potential of the ferric uptake regulator (Fur) family of proteins remains undefined despite over 20 years of study. We report herein an integrated approach that combines both genome-wide technologies and structural studies to define the role of Fur in Campylobacter jejuni (Cj). CjFur ChIP-chip assays identified 95 genomic loci bound by CjFur associated with functions as diverse as iron acquisition, flagellar biogenesis, and non-iron ion transport. Comparative analysis with transcriptomic data revealed that CjFur regulation extends beyond solely repression and also includes both gene activation and iron-independent regulation. Computational analysis revealed the presence of an elongated holo-Fur repression motif along with a divergent holo-Fur activation motif. This diversity of CjFur DNA-binding elements is supported by the crystal structure of CjFur, which revealed a unique conformation of its DNA-binding domain and the absence of metal in the regulatory site. Strikingly, our results indicate that the apo-CjFur structure retains the canonical V-shaped dimer reminiscent of previously characterized holo-Fur proteins enabling DNA interaction. This conformation stems from a structurally unique hinge domain that is poised to further contribute to CjFur’s regulatory functions by modulating the orientation of the DNA-binding domain upon binding of iron. The unique features of the CjFur crystal structure rationalize the binding sequence diversity that was uncovered during ChIP-chip analysis and defines apo-Fur regulation.
Keywords: gene regulation, metal regulation, transcription factor, DNA-protein interactions, structural characterization
Iron is critical to many fundamental biological processes, including DNA synthesis, respiration, and the tricarboxylic acid cycle. Unfortunately, although iron is essential for life, it can also be toxic under physiological conditions. Its ability to undergo facile oxidation–reduction can catalyze the production of noxious radical species by Fenton or Haber–Weiss chemistry (1). Most Gram-negative bacteria maintain a remarkably precise control over cytoplasmic iron levels through the transcriptional regulator Fur (ferric uptake regulator) (1, 2). In the classical Fur regulation paradigm, Fur binds ferrous ions and the dimeric Fe2+–Fur complex (holo-Fur) recognizes target sequences upstream of iron-regulated genes and represses their transcription (2). In addition to regulating genes involved in iron homeostasis, Fur has been shown to play an important role in the modulation of bacterial virulence, acid, nitrosative, and oxidative resistances, and redox metabolism (2–9).
In some bacterial species, Fur has been reported to directly activate gene expression, establishing a significant departure from the classical model of Fur regulation (6, 7, 10). In Helicobacter pylori (Hp), apo-HpFur represses transcription of the bacterioferritin-like gene pfr and the superoxide dismutase gene sodB, resulting in transcriptional activation of these genes in the presence of iron (10). Also, work on Bradyrhizobium japonicum Fur protein (BjFur) and the Fur homolog BosR in Borrelia burgdorferi have demonstrated that certain Fur-family proteins can recognize multiple consensus binding sequences (11, 12). Recent transcriptomic analyses have revealed that Campylobacter jejuni Fur (CjFur) regulates, either positively or negatively, more than 60 genes encoding proteins involved in iron acquisition, oxidative stress defense, flagellar biogenesis, and energy metabolism (4, 13). However, these studies failed to discriminate direct from indirect gene-regulatory mechanisms.
The structural basis underlying the regulation of gene expression by Fur proteins has been extensively documented (14–22). Fur proteins consistently fold into two domains consisting of an N-terminal DNA-binding domain (DBD) linked by a hinge region to a C-terminal dimerization domain (DD). In contrast to the common fold of these proteins, the coordination, number, and geometry of the metal binding sites within Fur-family proteins diverge (2, 14, 15, 20, 22, 23). Despite these studies, the structural determinants controlling the divergent mode of regulation of gene expression by this family of proteins have remained unresolved.
We herein provide the full extent of the C. jejuni Fur regulon using a chromatin immunoprecipitation and microarray analysis approach (ChIP-chip). Our results establish that CjFur regulates 95 transcriptional units and report the presence of apo- and holo-CjFur gene repression and activation in C. jejuni. Correspondingly, crystallographic studies reveal that apo-CjFur adopts the canonical V-shaped dimer characteristic of holo-Fur proteins, with two zinc ions per protomer. However, comparative analysis of apo-CjFur with other known Fur proteins reveals that apo-CjFur’s DBD is rotated by 180° compared with other known Fur structures, a structural rearrangement stemming from a reorientation of the apo-CjFur hinge region. Overall, our results highlight the structural diversity of the Fur family of proteins and rationalize the consensus binding sequences revealed by our ChIP-chip and transcriptomics data.
Results
Identification of in Vivo Genome-Wide CjFur-Regulatory Targets.
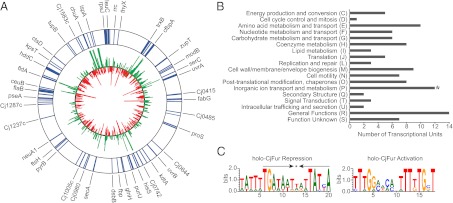
Transcriptomic analysis of a Δfur mutant in the presence or absence of iron established that CjFur may activate and repress gene expression in both holo and apo forms (4). Given that transcriptomic approaches cannot distinguish direct from indirect regulation, we sought to identify CjFur-binding regions on a genomic scale using ChIP-chip experiments. We identified 95 high-confidence CjFur-regulated transcriptional units (Table S1); however, due to the high gene density in the C. jejuni genome, we could not distinguish between intergenic and intragenic binding. Fig. 1A shows a schematic representation of the CjFur binding sites along the C. jejuni chromosome together with C. jejuni’s response to iron limitation at midlog phase. Our results are in agreement with previously known CjFur-regulated genes, and ChIP-chip results were validated by quantitative (q)PCR analysis for several identified Fur-binding regions (Fig. S1) (4, 13). Fur ChIP-enriched genes were categorized into clusters of orthologous groups (COG) categories according to their functional annotation (Fig. 1B). As expected, the COG category for inorganic ion transport and metabolism was found to be statistically overrepresented. This functional category comprises genes encoding iron transporters, including the enterobactin (ceuB), heme (chuAB), and ferric-lactoferrin (ctuA/chaN) transporters.
Fig. 1.
(A) Genomic map of CjFur-enriched transcriptional units overlaid with C. jejuni’s transcriptional response to iron limitation. The outer ring lists all of the transcriptional units that were enriched under Fur ChIP (≥1.5 enrichment, P ≤ 10−4). Blue denotes the gene that was found to be enriched under Fur ChIP. The inner ring displays the transcriptional response of each gene to iron limitation, with green denoting iron-repressed genes and red denoting iron-activated genes (≥1.5 fold change, P ≤ 10−4). The figure was made using Circos version 0.54 (34). (B) COG functional groups present in CjFur ChIP-enriched transcriptional units. CjFur ChIP-enriched genes encompass a diverse range of COG functional categories, indicating that CjFur plays regulatory roles beyond iron metabolism. The COG functional category inorganic ion transport and metabolism was found to be statistically overrepresented (*). (C) Consensus sequences for CjFur binding sites. (Left) Holo-CjFur repression consensus sequence, with its palindromic sequence highlighted with arrows. (Right) Holo-CjFur activation consensus sequence. Consensus sequences were made using WebLogo 3 (http://weblogo.berkeley.edu).
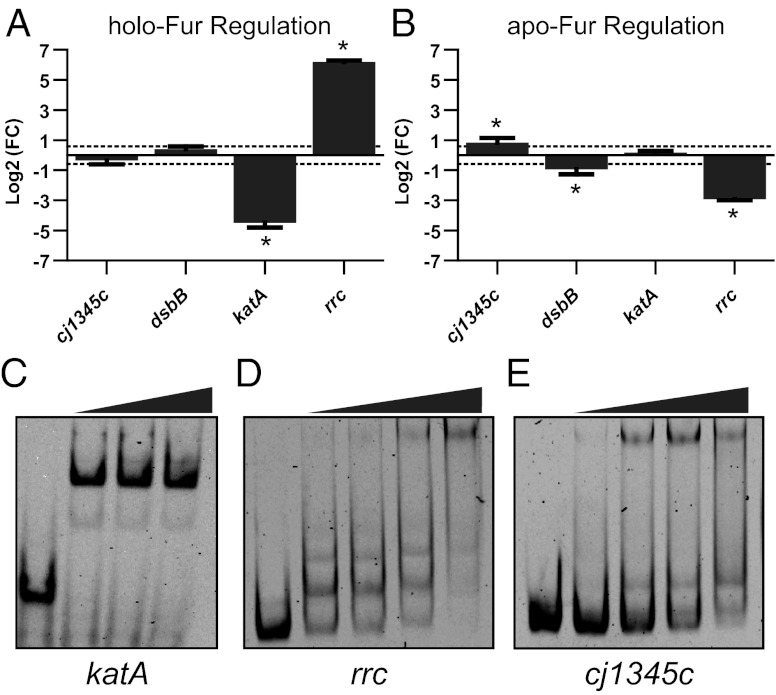
Identified CjFur-enriched genes were also compared with the CjFur regulon and iron stimulon that were previously obtained by comparing the transcriptional profiles of the wild type with a fur deletion mutant and by studying the transcriptional response of C. jejuni to iron limitation (4, 13) (Table S1). The CjFur regulon identified using ChIP-chip is significantly different from those previously proposed using transcriptomic approaches (4, 13). This is most likely due to the fact that transcriptional approaches cannot differentiate between direct and indirect regulation (and will identify both), cannot identify genes transcriptionally silent under the tested conditions, and would not detect genes exhibiting small changes in expression. In contrast, ChIP-chip will identify all direct targets that are bound by Fur under the tested conditions independently of fold change. Only 17 of our identified 95 ChIP targets were previously found to be members of the CjFur regulon. Whereas the majority of these genes were found to be under holo-CjFur repression (∼53%), our analysis also revealed holo-CjFur activation (four genes), apo-CjFur repression (four genes), and apo-CjFur activation (two genes). It should be noted that in some cases our transcriptomic data may not be able to definitively differentiate between some of the different forms of CjFur regulation, as several genes display both apo and holo forms of CjFur regulation. Therefore, we used RT-qPCR and gel-shift assays with apo-Fur protein to confirm the CjFur-dependent regulation of several CjFur ChIP targets (Fig. 2). Importantly, the apo-Fur protein extract was confirmed to be free of iron by inductively coupled plasma mass spectrometry (ICPMS) analysis (Table S2). As shown Fig. 2, apo-Fur exhibits a strong DNA-binding activity with fragments derived from the upstream regions of the genes rrc and cj1345c. In addition, it has recently been reported that dsbB is apo-Fur–repressed in C. jejuni (24), and this gene was both identified as a target in our ChIP results and also found to be apo-Fur–regulated by RT-qPCR (Fig. 2). There were 78 genes that were identified as Fur targets in our ChIP assay that were not previously thought to be Fur-regulated. Only 30 of these genes are iron-regulated (Table S1). The fact that ∼50% of the identified CjFur genes are not iron-regulated indicates that many of CjFur’s regulatory roles are iron-independent. Moreover, as the transcript level of most CjFur ChIP targets (∼82%) was not significantly affected in a Δfur mutant, this indicates the presence of complex regulatory circuits comprising additional transcription factors.
Fig. 2.
(A and B) Genes differentially expressed under iron-replete (A) or iron-limiting (B) conditions in a Δfur mutant. Positive fold changes indicate activation, whereas negative fold changes indicate repression. The dotted lines represent the 1.5-fold cutoff value. *P < 0.05. (C–E) Gel-shift assays of holo-Fur (C) and apo-Fur (D and E) demonstrating direct Fur binding to the promoter regions of genes found to be differentially expressed by RT-qPCR. Holo-Fur concentrations were 0, 5, 50, and 100 nM, whereas apo-Fur concentrations were 0, 50, 100, 200, and 1,000 nM. The promoter region for dsbB has been previously shown to be bound by apo-Fur in vitro (24).
Bioinformatic Analysis of Potential Fur Binding Sites.
The 5′ noncoding regions of the CjFur targets were further analyzed for the presence of conserved sequences that would represent CjFur binding sites. The dataset comprised all CjFur targets and subsets of transcriptional units categorized based on the modes of CjFur regulation (holo/apo, activation/repression). Despite our extensive testing of different subgroups and alternate transcriptional start sites, we failed to identify a universal consensus sequence for all of the CjFur targets and, similarly, we were unable to identify a motif for genes regulated by apo-CjFur. After these analyses, we then sought to identify enriched motifs in the genes regulated by CjFur in the presence of iron. In sharp contrast with apo-CjFur–regulated genes, we identified consensus motifs for holo-CjFur–activated and –repressed genes. The consensus sequence of the holo-Fur–repressed CjFur targets (Fig. 1C) is very similar to the CjFur consensus sequence previously reported (4). This consensus sequence is typical of classical Fur boxes and contains an internal palindromic 7-1-7 sequence. Interestingly, our analysis also detected the presence of a consensus sequence for holo-Fur–activated genes (Fig. 1C). The identified consensus sequence is significantly different from previously identified Fur boxes. It is nonpalindromic, and contains two direct repeat sequences of TTTGG that differ markedly from the two inverted repeats in the Fur box (TGATAAT).
Crystal Structure of CjFur.
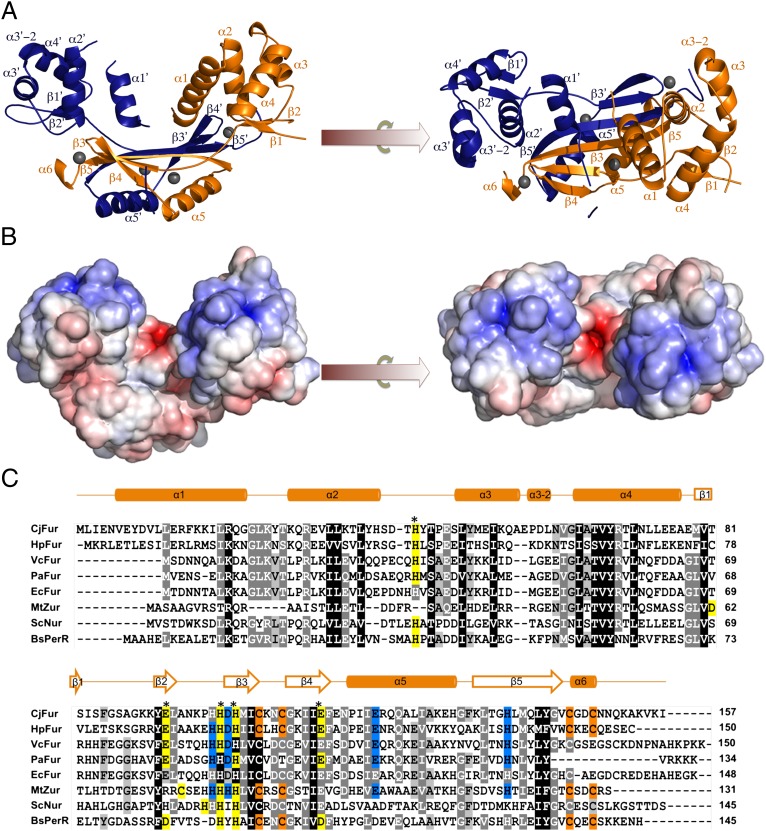
To understand the biochemical determinants underlying the different modes of Fur regulation, the structure of CjFur was determined at 2.1 Å resolution using single-wavelength anomalous dispersion experiments (Table S3). The structure consists of two protomers (A and B) that form the asymmetric unit and the functional dimer (Fig. 3 and Fig. S2). Protomer A consists of residues 4–83 and 90–149, whereas protomer B comprises residues 2–16 and 26–154. CjFur contains two modular domains forming an N-terminal DNA-binding domain and a C-terminal dimerization domain. The DBD of CjFur is composed of five consecutive α-helices (α1, α2, α3, α3−2, and α4) followed by a two-stranded antiparallel β-sheet (β1−β2). The tip of β2 is connected to the DD by an 8-residue hinge region. The DD folds as a mixed-α/β domain in which β3-β4-β5 form a twisted β-sheet intersected, between β4 and β5, by the α5-helix. The structure ends with a short α-helix (α6) that coordinates one zinc ion.
Fig. 3.
(A) Crystal structure of CjFur. Orthogonal views of the CjFur crystal structure in which protomers A and B are rendered in orange and blue, respectively. β-Sheets and α-helices are labeled accordingly, and zinc atoms are depicted as gray spheres. (B) Electrostatic surface potential of the CjFur crystal structure. Electrostatic potentials are contoured from +10 kbTe−1 (blue) to −10 kbTe−1 (red) (kb = Bolzmann's constant, T = temperature in Kelvin and e = charge of an electron). (C) Sequence alignment of Fur and Fur-like proteins. Sequence alignment of Fur proteins [C. jejuni (Cj), H. pylori (Hp), V. cholerae (Vc), P. aeruginosa (Pa), E. coli (Ec)] and of the Fur-like Zur from Mycobacterium tuberculosis (MtZur), Nur from Streptomyces coelicolor (ScNur), and PerR from B. subtilis (BsPerR). Sequences were aligned using the ClustalW option in MEGA5 (35). CjFur secondary-structure elements are shown above the alignment. Asterisks indicate the residues involved in the predicted CjFur S2 site. S1 residues are shaded in orange, predicted S2 residues are in yellow, and S3 residues are in blue. Positions with 100% amino acid conservation are indicated by dark gray, 100–80% by medium gray, and 80–60% by light gray. kb, Bolzmann's constant; T, temperature in Kelvin; e, charge of an electron.
After elucidating the CjFur structure, we sought to analyze the structural differences between CjFur and other structurally characterized Fur and Fur-like homologs. Using lsqkab (25), the HpFur, Vibrio cholerae (Vc)Fur, Pseudomonas aeruginosa (Pa)Fur, and CjFur structures were overlaid and the rmsd was calculated (26). We noted that, with the exception of CjFur, all Fur structures aligned reasonably well, with an rmsd of ∼1.8 Å for all atoms (Fig. S3). Other Fur proteins consistently orient their α1-helix outside of the V-shaped cleft and their two-stranded antiparallel β-sheet inside the dimer. In contrast, alignment of CjFur with any Fur proteins (rmsd of ∼15 Å for all atoms) revealed drastic differences in the position of the DBD secondary-structure elements. These differences include a 180° rotation of CjFur’s DBD, which positions the β1−β2 β-sheet on the exterior of the structure and the α1-helix within the V-shaped dimer (Fig. 3A). Close inspection of the overlay of CjFur with HpFur (CjFur’s closest homolog) revealed that CjFur’s DBD repositioning stems from the conformation of the CjFur hinge region. Indeed, in CjFur, the hinge region is elongated, whereas the structurally equivalent region of HpFur is bent in such a way that it adopts a loose turn. Overall, these observations suggest that the CjFur hinge region plays an important role in controlling the orientation of the DBD.
Metal Binding Sites of CjFur.
After establishing that CjFur adopts a peculiar structural conformation, we hypothesized that metal coordination would also diverge from other Fur homologs. To confirm this hypothesis, anomalous Fourier difference maps were calculated and the resulting electron density was analyzed. For consistency, we have used the nomenclature recently used for designating metal binding sites in HpFur (14). The CjFur structure contains two occupied Zn2+ binding sites (referred therein as S1 and S3) per protomer. The S1 site contains a Zn2+ ion that is tetracoordinated by two pairs of cysteine residues (C105/108 and C145/148) (Fig. S4A) and is found in the DD of CjFur. This zinc binding site is known to be important for maintaining the structural integrity of the protein and dimerization in HpFur (27). Given that the S1 site is also found in HpFur and Bacillus subtilis (Bs)PerR and that both proteins exhibit an additional C-terminal α-helix (Fig. 3C and Fig. S4A), these results suggest that Fur proteins harboring an additional C-terminal α-helix coordinate a structural Zn2+ ion in the S1 site.
The second metal binding site, S3, lies between β5 and α5 and is in close proximity to the C-terminal end of the hinge that links the DBD to the DD. In the S3 site, the Zn2+ ion is hexacoordinated by residues D101, E120, and H137 and two water molecules. The first water molecule (referred therein as W1) is located 2.2 Å from the Zn2+ ion and makes a 2.6-Å hydrogen bond with the side-chain amide group of N123. The second water molecule (W2) engages in two hydrogen bonds with the carbonyl group and carboxylate moiety of H99 and E115, respectively (Fig. S4C). This type of coordination is drastically different from the HpFur and VcFur S3 sites, which tetracoordinate the Zn2+ ion and lack metal-coordinating water molecules. Although the number of coordination in CjFur is analogous to PaFur, there are differences in the mode of coordination. In PaFur, W2 is absent and replaced by the imidazole side chain of H86. In contrast, CjFur is able to coordinate W2 due to the aforementioned 180° rotation of CjFur’s DBD, which places the backbone carbonyl group of H99 in an orientation permissive for a W2-mediated hydrogen bond with the Zn2+ ion. In addition, E115, which engages in the second W2-mediated hydrogen bond, is unique to CjFur.
Recent reports have suggested that the S2 metal binding site is the iron-sensing site in several Fur proteins (2). The crystal structures of VcFur, HpFur, and PaFur revealed that all these proteins coordinate metal ions at S2 using different geometries and modes of coordination (14, 15, 22). Close analysis of the calculated Fourier maps in proximity to the CjFur S2 site failed to detect electron density, suggesting that the CjFur S2 site is unoccupied. Consistent with this observation, only three metal-coordinating residues (H100, H102, and E113) could be located at the putative iron binding S2 site (Fig. S4B). Moreover, ICPMS analysis confirmed the absence of contaminating iron in our protein preparation (Table S2). The absence of metal in the S2 site can be explained by the rotation of CjFur’s DBD, which positions two of the putative Fe2+-coordinating residues, namely E93 and H99, in a nonpermissive orientation for engaging in metal coordination. Because there is a lack of metal in the S2-regulatory site, the CjFur crystal structure is in the apo form. However, our apo-CjFur crystal structure still adopts the canonical V-shaped conformation that is characteristic of other holo-Fur crystal structures.
Discussion
Our study defines the apo-Fur structure and the Fur regulon in C. jejuni. The structural analysis also reveals a possible mechanism for apo-CjFur regulation, whereas our ChIP-chip results significantly expand known CjFur target gene loci, unveiling regulatory roles beyond iron homeostasis. The central regulatory role of CjFur is highlighted by the identification of over 95 Fur binding sites in proximity to genes encoding proteins involved in diverse biological pathways ranging from metal homeostasis (including iron, zinc, tungsten, and molybdenum) to flagellar and membrane biogenesis, energy production and conversion, and stress responses.
Consistent with the classical role of Fur as a holo-Fur repressor of iron-acquisition genes (2), the CjFur targets comprise all of the genes known to be involved in iron acquisition (e.g., ferric-enterobactin, heme and lactoferrin transporters). Remarkably, whereas half of the CjFur binding sites are associated with iron regulation, only a fourth of the CjFur targets were previously reported to be deregulated in a fur mutant. This pattern is not unprecedented, and was also observed for the HpFur and PaFur regulons (28, 29).
Genes that were found to be CjFur targets but not differentially expressed in a Δfur mutant include genes involved in flagellar biogenesis and genes involved in zinc and molybdate/tungsten transport. Members involved in flagellar biogenesis include the major C. jejuni flagellins (flaAB) and numerous glycosylation proteins (e.g., pseA and pseF). By analogy, the HpFur regulon also contains several genes involved in flagellar biogenesis (28). This absence of differential expression of the flagellum genes in the fur mutant likely reflects the complex regulatory transcriptional cascade for flagellar genes in C. jejuni. Indeed, transcriptional control over flagellar biogenesis is quite extensive in C. jejuni, involving two σ factors (σ28 and σ54), the FlgSR two-component system, and the FlhF regulator (30). The CjFur regulon also includes the zinc, molybdate, and tungstate transporters ZupT, ModB, and TupB, respectively (31, 32). The direct regulation of these transporters by CjFur reveals additional roles for CjFur in transition-metal homeostasis beyond iron regulation.
Interestingly, although we identified four modes of CjFur regulation, holo- and apo-CjFur repression and activation, only two distinct CjFur DNA-binding motifs could be identified. The motif identified for holo-CjFur–repressed genes matches the consensus sequence previously identified, whereas the motif identified in holo-CjFur–activated genes bears little similarity to the holo-Fur–repressed motif (4). The fact that CjFur recognizes two distinct consensus sequences is not unprecedented, as recent work on the Fur homolog BosR in B. burgdorferi demonstrated that this Fur-like protein could recognize three distinct consensus sequences (11). Similarly, multiple unique DNA recognition sites were also reported for the B. japonicum Fur protein (12). Overall, our observations highlight the expanding repertoire of DNA sequences recognized by the Fur family of proteins.
Although CjFur should share similar structural features with Fur proteins from other bacteria, none of the previously crystallized proteins has been demonstrated to bind multiple consensus sequences, and only HpFur is known to regulate gene expression in its apo form. Moreover, the crystal structure of HpFur failed to provide structural insights into apo-HpFur function, and previous work has suggested that the structure of CjFur differed from other Fur-family proteins (33). Because our results indicate that CjFur recognizes divergent DNA consensus sequences and regulates gene expression independently of iron, we reasoned that CjFur must possess unique structural features that have not yet been described for the Fur family of proteins. Accordingly, comparative analyses of our CjFur crystal structure with other Fur and Fur-like proteins reveal several similarities and differences. Comparable to all Fur proteins, CjFur contains two protomers that form the canonical V-shaped dimer characteristic of holo-Fur proteins. Similarly, analysis of CjFur metal binding sites reveals that the S1 site in CjFur contains a zinc ion that is tetracoordinated by two pairs of cysteine residues, forming a C4 zinc-finger motif, an important structural determinant for dimerization in HpFur and also found in BsPerR (27). The S3 site is also occupied by a zinc ion hexacoordinated by residues including E120, D101, and H137 and two water molecules. Although the position of the S3 site is analogous to that of other Fur proteins, it differs significantly from the tetracoordination state observed in HpFur. The final metal binding site is known as the S2 site, and is the regulatory metal binding site in HpFur, VcFur, and PaFur. Strikingly, this S2 site is unoccupied in CjFur, indicating that the structure of CjFur represents the apo form of the protein. However, unlike the previously obtained apo-BsPerR structure (21), which has lost its V-shaped conformation upon reorientation of its DBD in a near-planar arrangement (Fig. S4), apo-CjFur maintains the canonical V-shaped conformation reminiscent of other holo-Fur proteins. These observations likely stem from several interdomain contacts between the CjFur DBD and DD. First, residues located at the N-terminal end of α1, which include N5, V6, and GE7, make several van der Waals contacts, hydrophobic interactions, and hydrogen bonds with the CjFur DD. Second, residues found in the C-terminal end of α2, which include Y38 and H39, make extensive interactions with residues encompassing the β3−β4 hairpin of the CjFur DD. Third, the residues succeeding the C4 zinc finger of the DD fold back onto the DBD and engage in several hydrophobic contacts with a loop connecting α2 and α3 of the DBD. Altogether, this extensive network of interactions is strikingly different from apo-BsPerR, in which no contacts between the DBD and DD domains are observed. Altogether, these interactions provide a rationale underlying the formation of apo-CjFur’s V-shaped conformation. Finally, this conformation places several basic residues on the tip of the V-shaped structure (Fig. 3B) in a position amenable for engaging in electrostatic interactions with DNA. Thus, our structure provides a snapshot view of an apo-Fur protein.
In addition to adopting the V-shaped conformation, further comparative analyses of apo-CjFur with known holo-Fur structures identified notable differences in the orientation of its DNA-binding domain (14–17, 19, 22). Indeed, whereas all holo-Fur proteins align relatively well with each other (rmsd of ∼1.8 Å), apo-CjFur displays striking structural differences in the orientation of its DBD. This conformational difference stems from a notable rearrangement of the CjFur hinge region, which positions the α1-helices inside the V-shaped dimer. Thus, the CjFur hinge region is poised to play an important role in modulating the orientation of the DBD, and likely contributes to the rotation of the DBD upon iron binding. These results are in line with a recent study showing that metallation of HpFur S2 triggers a conformation change, which results in the formation of an active holo-Fur protein (14). These observations may also suggest that residues composing the CjFur hinge region allow for an increased degree of freedom and thereby the positioning of the DBD in multiple orientations. However, given that the CjFur S2 site lacks a metal ion but the apo-CjFur structure maintains a V-shaped conformation, we postulate that additional mechanisms confer to CjFur the ability to bind to divergent DNA sequences and maintain a V-shaped conformation in the absence of metal in the S2 site. This added complexity would give CjFur the ability to selectively regulate gene expression depending on other environmental factors along with iron.
In conclusion, our results imply that the DNA-binding properties of CjFur will diverge depending both on the occupancy of its regulatory S2 site and the orientation of its unconventional hinge region. In a broader context, these results support the observed four modes of Fur regulation, apo- and holo-Fur activation and repression. Finally, our study has not only provided a genome-wide view of Fur binding in C. jejuni but also provides a view of an apo-Fur structure.
Materials and Methods
C. jejuni and Escherichia coli strains were grown under standard conditions with antibiotic supplementation as needed. CjFur was purified and used to generate anti-CjFur antibodies and for crystallization trials. ChIP-chip experiments were completed and the results were analyzed as previously described (4, 34). CjFur ChIP enrichment was confirmed using qPCR of known Fur targets. Consensus sequence analysis of CjFur ChIP targets using MEME (http://meme.nbcr.net) was done as described previously (4). The CjFur crystal structure was solved using single-wavelength anomalous diffraction datasets generated from the Life Sciences Collaborative Access Team beamline at the Advanced Photon Source in Chicago. For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Fredric Poly for the construction of the pASK-Fur expression vector. We also thank the Quantitative Bioelemental Imaging Center, Northwestern University, for ICPMS analysis. This work was supported by the Canadian Institutes of Health Research (CIHR) (J.-F.C. and A.S.) and a CIHR-Banting graduate scholarship (to J.B.). J.-F.C. acknowledges a Canada Research Chair in structural biology and epigenetics. An Early Research Award by the Ministry of Research and Innovation of Ontario supported this research (to J.-F.C.).
Footnotes
The authors declare no conflict of interest.
Database deposition: Crystallographic data, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4ETS).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118321109/-/DCSupplemental.
References
- 1.Butcher J, Flint A, Stahl M, Stintzi A. Campylobacter Fur and PerR regulons. In: Cornelis P, Andrews SC, editors. Iron Uptake and Homeostasis in Microorganisms. Norfolk, UK: Caister Academic; 2010. pp. 168–202. [Google Scholar]
- 2.Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 3.McHugh JP, et al. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 4.Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gancz H, Censini S, Merrell DS. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect Immun. 2006;74:602–614. doi: 10.1128/IAI.74.1.602-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Silva Neto JF, Braz VS, Italiani VC, Marques MV. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic α-proteobacterium Caulobacter crescentus. Nucleic Acids Res. 2009;37:4812–4825. doi: 10.1093/nar/gkp509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson LA, et al. Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J Bacteriol. 2010;192(1):77–85. doi: 10.1128/JB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres VJ, et al. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun. 2010;78:1618–1628. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Appl Environ Microbiol. 2010;76:406–416. doi: 10.1128/AEM.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter BM, et al. A single nucleotide change affects Fur-dependent regulation of sodB in H. pylori. PLoS One. 2009;4:e5369. doi: 10.1371/journal.pone.0005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang Z, Deka RK, Norgard MV. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 2011;7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman YE, O’Brian MR. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J Biol Chem. 2003;278:38395–38401. doi: 10.1074/jbc.M306710200. [DOI] [PubMed] [Google Scholar]
- 13.Holmes K, et al. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology. 2005;151:243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- 14.Dian C, et al. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol Microbiol. 2011;79:1260–1275. doi: 10.1111/j.1365-2958.2010.07517.x. [DOI] [PubMed] [Google Scholar]
- 15.Sheikh MA, Taylor GL. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol. 2009;72:1208–1220. doi: 10.1111/j.1365-2958.2009.06718.x. [DOI] [PubMed] [Google Scholar]
- 16.An YJ, et al. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 2009;37:3442–3451. doi: 10.1093/nar/gkp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquamet L, et al. Structural characterization of the active form of PerR: Insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol. 2009;73(1):20–31. doi: 10.1111/j.1365-2958.2009.06753.x. [DOI] [PubMed] [Google Scholar]
- 18.Traoré DA, et al. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol. 2009;5(1):53–59. doi: 10.1038/nchembio.133. [DOI] [PubMed] [Google Scholar]
- 19.Lucarelli D, et al. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J Biol Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- 20.Pecqueur L, et al. Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J Biol Chem. 2006;281:21286–21295. doi: 10.1074/jbc.M601278200. [DOI] [PubMed] [Google Scholar]
- 21.Traoré DA, et al. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol Microbiol. 2006;61:1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x. [DOI] [PubMed] [Google Scholar]
- 22.Pohl E, et al. Architecture of a protein central to iron homeostasis: Crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- 23.Lewin AC, Doughty PA, Flegg L, Moore GR, Spiro S. The ferric uptake regulator of Pseudomonas aeruginosa has no essential cysteine residues and does not contain a structural zinc ion. Microbiology. 2002;148:2449–2456. doi: 10.1099/00221287-148-8-2449. [DOI] [PubMed] [Google Scholar]
- 24.Grabowska AD, et al. Campylobacter jejuni dsb gene expression is regulated by iron in a Fur-dependent manner and by a translational coupling mechanism. BMC Microbiol. 2011;11:166. doi: 10.1186/1471-2180-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Cryst. 1976;A32:922–923. [Google Scholar]
- 26.Kabsch W, Kabsch H, Eisenberg D. Packing in a new crystalline form of glutamine synthetase from Escherichia coli. J Mol Biol. 1976;100:283–291. doi: 10.1016/s0022-2836(76)80064-2. [DOI] [PubMed] [Google Scholar]
- 27.Vitale S, et al. A ZnS(4) structural zinc site in the Helicobacter pylori ferric uptake regulator. Biochemistry. 2009;48:5582–5591. doi: 10.1021/bi9004396. [DOI] [PubMed] [Google Scholar]
- 28.Danielli A, et al. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J Bacteriol. 2006;188:4654–4662. doi: 10.1128/JB.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butcher BG, et al. Characterization of the Fur regulon in Pseudomonas syringae pv. tomato DC3000. J Bacteriol. 2011;193:4598–4611. doi: 10.1128/JB.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendrixson DR. Regulation of flagellar gene expression and assembly. In: Nachamkin I, Szymanski C, Blaser MJ, editors. Campylobacter. 3rd Ed. Washington, DC: Am Soc Microbiol; 2008. pp. 545–559. [Google Scholar]
- 31.Smart JP, Cliff MJ, Kelly DJ. A role for tungsten in the biology of Campylobacter jejuni: Tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter. Mol Microbiol. 2009;74:742–757. doi: 10.1111/j.1365-2958.2009.06902.x. [DOI] [PubMed] [Google Scholar]
- 32.Grass G, et al. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol. 2005;187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles S, Carpenter BM, Gancz H, Merrell DS. Helicobacter pylori apo-Fur regulation appears unconserved across species. J Microbiol. 2010;48:378–386. doi: 10.1007/s12275-010-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danielli A, Scarlato V. Regulatory circuits in Helicobacter pylori: Network motifs and regulators involved in metal-dependent responses. FEMS Microbiol Rev. 2010;34:738–752. doi: 10.1111/j.1574-6976.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- 35.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.