Abstract
For the most part, studies of grass genome structure have been limited to the generation of whole-genome genetic maps or the fine structure and sequence analysis of single genes or gene clusters. We have investigated large contiguous segments of the genomes of maize, sorghum, and rice, primarily focusing on intergenic spaces. Our data indicate that much (>50%) of the maize genome is composed of interspersed repetitive DNAs, primarily nested retrotransposons that insert between genes. These retroelements are less abundant in smaller genome plants, including rice and sorghum. Although 5- to 200-kb blocks of methylated, presumably heterochromatic, retrotransposons flank most maize genes, rice and sorghum genes are often adjacent. Similar genes are commonly found in the same relative chromosomal locations and orientations in each of these three species, although there are numerous exceptions to this collinearity (i.e., rearrangements) that can be detected at the levels of both the recombinational map and cloned DNA. Evolutionarily conserved sequences are largely confined to genes and their regulatory elements. Our results indicate that a knowledge of grass genome structure will be a useful tool for gene discovery and isolation, but the general rules and biological significance of grass genome organization remain to be determined. Moreover, the nature and frequency of exceptions to the general patterns of grass genome structure and collinearity are still largely unknown and will require extensive further investigation.
Keywords: chromosomal evolution, gene discovery, genome rearrangement, genetic maps, microcollinearity
Very little is known about the structure of the nuclear genomes of higher plants, although comprehensive investigations are now underway into the sequence composition of the unusually small [about 110-megabase pair (mbp)] genome of Arabidopsis thaliana. Most plant nuclei contain more than five times as much DNA as that of Arabidopsis, ranging up to the over 110,000 mbp of Fritillaria assyriaca (1). Part of this genome size variation is caused by differences in ploidy, but the majority is caused by differences in the amounts of repetitive DNA (2). Some of these repetitive sequences are found in tandemly repeated satellites, like the chromosomal knobs of maize (3), but most are represented by interspersed repeats that vary in copy number from tens to hundreds to thousands per haploid nucleus (4). The nature and organization of these repeats, and their functional or structural relationship to genes, are not well understood.
Low-density genetic maps, including those for several cereal species (5–7), have shown that conserved DNA markers (primarily genes) often are found in the same linear order in different plant species. This discovery of the collinearity of “orthologous” genes (i.e., those with a direct evolutionarily relationship) in cereals has allowed a wholly new perspective on how genes and information can be used synergistically in the study and improvement of all grasses (8–10). Although many exceptions to collinearity exist in these comparative genetic maps (5, 7, 10), collinearity supplies an exceptional and unique tool for both gene discovery and gene isolation. Moreover, collinearity provides an opportunity to discover how evolution has created new morphologies, physiologies, pathways, and species from the same set of starting genetic material.
Recent advances in the technologies needed to generate near-saturated recombinational maps, including fine structure maps of gene clusters and large (>100 kb) cloned segments of chromosomes, will yield our first detailed insights into the evolved structures of complex plant genomes. In addition, DNA sequencing technologies have improved to a level where contiguous genomic sequences, covering multiple genes and intergenic spaces, can be generated rapidly and at reasonable cost. We have decided to use these technologies to investigate the complex genomes of maize, sorghum, and rice. These three grass species were chosen partly because of their agronomic importance and genetic history, but mainly because of their differences in genome size (430 mbp for rice, 750 mbp for sorghum, and 2500 mbp for maize) (11) and their known phylogenetic relatedness (12). Our data indicate that a comparative analysis of grass genomes will be tremendously useful, but that conclusions regarding the most efficient route to this synergistic knowledge acquisition now require additional experiments on local genome structure and evolution.
MATERIALS AND METHODS
Materials.
The nature and sources of yeast artificial chromosome clones from maize and bacterial artificial chromosome (BAC) clones from sorghum and rice have been described (13–15). Additional BACs containing Arabidopsis genomic DNA were obtained from the Arabidopsis Biological Resource Center at Ohio State University.
Gel Blot Hybridization, Restriction Mapping, and DNA Sequencing.
Genomic and cloned DNA isolation, gel blot analysis, and hybridization were all as described previously (14, 16). Restriction enzymes were used according to the manufacturer’s recommendations. Sequencing and sequence analysis were as described (16, 17).
RESULTS
The Structure of a Maize Chromosomal Segment.
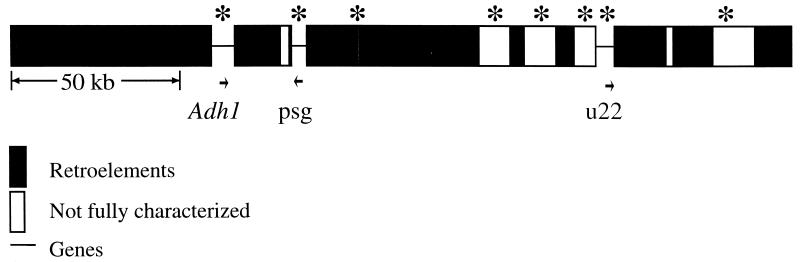
We have used DNA sequencing and retroelement structural analysis to characterize the sequence of a 240-kb region of maize DNA flanking the Adh1-F locus. Our data have identified three genes in this region, as determined by homology to cDNA sequences in the standard databases (Fig. 1). There are five additional putative genes in this region, identified by cross-hybridization to the same regions in sorghum (Fig. 1, asterisks) (15) and/or by homology to a cDNA sequence but without the confirming evidence of an intron found in the genomic sequence that is lacking from the cDNA (data not shown). Most of the remaining DNA is composed of uninterrupted boxes of repetitive DNAs, primarily consisting of retrotransposons inserted within other retrotransposons (Fig. 1) (17). We also have seen similar repeat blocks adjacent to all of the other maize genes that we have examined (18).
Figure 1.
Physical map of the Adh1-F region of maize. A 240-kbp region flanking the maize Adh1-F allele has been partially sequenced. Narrow lines indicate the locations of known or predicted genes, and the boxes indicate either confirmed retroelements (filled boxes) or sequences that have not yet been fully characterized (open boxes). Asterisks depict the sites of restriction fragments that cross-hybridized with the orthologous region of sorghum (15). Arrows below the line indicate putative or known transcripts and their orientations. Only one of these transcripts is associated with a gene that has an official designation, Adh1, whereas the others have only our operational names.
Repetitive DNAs make up over 70% of this region of the maize genome (13, 17), which is a number in line with the total percentage of repetitive DNA in the maize genome (4). The most abundant repetitive DNAs in the Adh1-F region are six retrotransposons with copy numbers ranging from 2,000 to 30,000. Combined, these elements make up over 50% of the maize nuclear genome. Lower-copy-number retrotransposons (from a few copies per nucleus up to a thousand) account for at least another 10% of maize nuclear DNA (ref. 17 and unpublished observations). Database screens and in situ hybridization indicate that the most highly repetitive retrotransposons are scattered throughout the maize genome and flank most maize genes (17, 19). These intergene retrotransposons are largely absent within the ribosomal DNA repeats of the nuclear organizer and are somewhat under-represented in centromeric regions (19). However, beyond these notable exceptions, these retrotransposons appear to flank most or all maize nuclear genes (17–19). Hence, we feel that the Adh1-F region of maize is fairly typical of the maize genome.
Microcollinearity Between the Orthologous Adh1 Regions of Maize and Sorghum.
Maize and sorghum are both members of the tribe Andropogonae and have evolved independently for about 15–20 million years (12). We used a maize Adh1 probe to isolate a BAC from sorghum that contained a homologue to maize Adh1 (15). The sorghum BAC was found to also cross-hybridize to several locations on the orthologous maize yeast artificial chromosome (Fig. 1, asterisks), including to all of the other known genes in the maize region but not to any of the known retrotransposons. The linear order of cross-hybridizing fragments between maize and sorghum also were found to be identical (15). Hence, gene composition and order (i.e., microcollinearity) appear to be conserved in this area. Moreover, we found that this cross-hybridizational criterion was the most effective way to localize the gene islands in this great sea of maize repetitive DNAs (15). The Adh1 and u22 homologs were only about 50 kbp apart in sorghum, whereas they are separated by more than 120 kbp in maize (15).
The Structure of a Rice Chromosomal Segment.
Extensive investigation of structure and recombination in the Sh2/A1 region of maize (20, 21) provided a point of comparison for the orthologous genes of smaller genome grasses, like rice and sorghum. We selected BAC clones containing rice genomic DNA by hybridization to the maize Sh2 gene (14). These clones were found to contain orthologs of A1, and these two loci and the intergene region were fully sequenced (22).
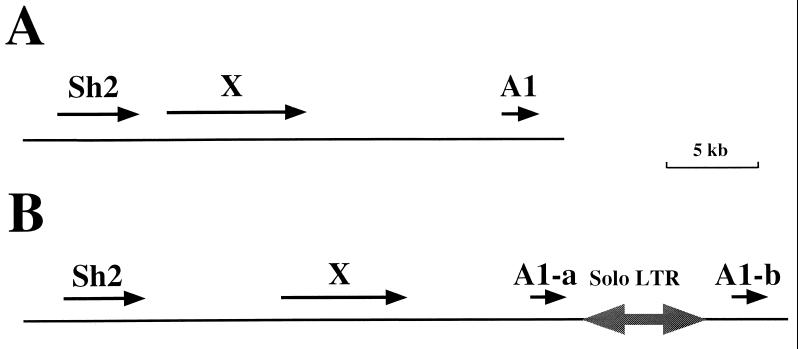
The sequence data indicated three genes in a 28,717-bp region (Fig. 2A). All three genes are apparently transcribed in the same orientation, including a novel gene (“gene X”) that encodes coil-coil protein binding and zinc finger motifs suggestive of a transcription factor (22). In contrast to our results from investigating the flanking sequences of maize genes (13, 17, 18), retrotransposon homologies were not observed in this region. Most of the intergenic space in the sequenced chromosomal segment, amounting to about one-third of the total length, lacked any distinctive features except the presence of a few miniature inverted repeat transposable elements (23) and a 1,432-bp direct repeat just upstream of the A1 homologue (22).
Figure 2.
Physical maps of the Sh2/A1-homologous regions of rice and sorghum. Maps are based on the completed sequences of these regions from rice (A) (22) and sorghum (B) (24). Arrows above the line indicate the size and orientation of known or predicted transcripts. We have data to unambiguously support the expression of one of these genes, gene X of rice, which exhibits 100% homology with the sequenced portion of a rice cDNA (22). The other genes are thought to be expressed primarily because their predicted exons are well-conserved relative to the maize genes (22, 24). The two-headed arrow depicts the one identified retroelement in these two orthologous regions, a solo long terminal repeat of Leviathan (24, 25).
The Similar Structure of Rice and Sorghum Sh2/A1-Homologous Regions.
To determine the nature and degree to which the structure of the Sh2/A1-homologous region has varied over a relatively long evolutionary time, we decided to clone and sequence the orthologous region from sorghum. By molecular clock criteria, sorghum and rice have undergone about 50 million years of independent evolutionary descent (12).
A maize Sh2 probe was used to isolate sorghum BAC clones and, as in rice, several of these clones also contained A1 homology (14). A 42,446-bp region of contiguous DNA was sequenced, and four genes were found in the region (Fig. 2B) (24). These genes were homologs of Sh2 and gene X, plus two tandem homologs of the A1 gene. As in rice, these four genes all were transcribed in the same orientation and were not separated by large blocks of retrotransposons or other repetitive DNAs. The sole exception was the presence of the solo long terminal repeat of the Leviathan retrotransposons (25), located between the two A1 homologs (24). Several miniature inverted repeat transposable elements were present in this region (but different ones from those seen in the rice region), at different locations, but about 25% of the region was composed of intergene spaces with no distinctive sequence characteristics.
Interestingly, the gene X homologue in sorghum does not contain the zinc finger motif. The sequences of the sorghum and rice gene X orthologs otherwise are quite well conserved, however, suggesting that they are both active genes. The difference in the presence of a DNA and/or protein binding domain between the two related genes suggests that they may have evolved quite different functions (24).
In previous experiments, we had observed that only three regions cross-hybridized between the rice and sorghum BAC clones (14), at the locations we now know to be occupied by the Sh2 homologs, gene X, and the A1 homologs. Our sequencing results confirmed this observation that only the genes, and some of their putative transcriptional regulatory signals, have been conserved in this region (24).
Microcollinearity Not Seen in the Adh1-Homologous Region of Rice or in Sh2/A1-Homologous Regions of Arabidopsis.
A maize Adh1 probe was used to isolate BACs containing the orthologous locus from rice. These BACs did not contain any other sequences that cross-hybridized with the other genes (or any other sequence except Adh1) in the maize Adh1 region. In addition, a rice BAC containing a homologue of the maize u22 gene was selected and found to not contain a homologue of the maize Adh1 gene. Hence, these data suggest that the orthologous rice and maize Adh1 regions are not collinear.
Similarly, Arabidopsis expressed sequence tags (i.e., cDNAs) with high homology to the maize A1 and Sh2 loci were used to isolate several BACs containing these homologs. None of these clones contained homology to both A1 and Sh2 on the same BAC (data not shown). Hence, these data suggest a lack of microcollinearity between Arabidopsis and maize for these related genes.
In both of the above cases, it was not possible to determine for certain whether the actual clones analyzed were true orthologs or just members of the same gene family (paralogs). With both Adh1 and Sh2, however, only one strong cross-hybridizing band is seen in each species, and this was the clone that was analyzed.
DISCUSSION
Grass Genome Structure.
Our data indicate very different sequence compositions in gene-containing regions of the maize, sorghum, and rice genomes. In agreement with its large genome size (about 2,400 mbp) (1, 4, 15), maize was found to have the most repetitive DNA and the greatest distance between genes. In the Adh1-F region, there is about 30–80 kbp per gene. The arrangement of the repetitive DNAs, interspersed repetitive sequences that were mostly intergene retrotransposons (17), is exactly in the interleaved pattern with short stretches of unmethylated and low-copy-number genic sequences that was predicted by both renaturation (“Cot”) analysis (5) and by the pulsed-field gel analysis of genomic DNA digested with 5-methyl cytosine-sensitive restriction enzymes (18).
In contrast, Sh2/A1-homologous regions of both sorghum and rice were found to be much more gene rich, averaging one gene per every 9–12 kbp. The Sh2 and A1 homologs are about 22 kbp apart in sorghum and 21 kbp apart in rice (22, 24), whereas these two genes are separated by about 140 kbp in maize (20). This result is in agreement with the smaller genome sizes of sorghum (750 mbp) and rice (430 mbp) (1, 11). Notably missing from the sorghum and rice regions analyzed were the numerous retrotransposons seen in maize. Studies from many laboratories (reviewed in ref. 25) indicate that retrotransposons are present in all investigated plant species, but that high-copy-number retrotransposons (>1,000 copies/genome) are found only in plants with large genome sizes. Hence, it seems likely that the great variation in plant genome size, otherwise known as the C-value paradox, is caused by differences in the presence and amplification of these retroelements.
One could propose a simple model for genome organization in plants, by using the maize Adh1-F example as a paradigm for large genome species and the rice and sorghum Sh2/A1-homologous regions as exemplars for small genome species. In maize, we feel that the Adh1-F region is fairly standard, because we have seem similar structure at all of the other unlinked regions we have investigated (18). However, in other plants, we have very little data on which to base such models, and these data are almost completely lacking from large genome species of gymnosperms or dicotyledonous angiosperms.
Moreover, even in rice and sorghum, we are extrapolating from a single genomic location. Our analyses of methylated DNA blocks in sorghum (26) provided similar results to those in maize (18), suggesting that equally large blocks of heterochromatic and methylated retrotransposons also exist in this species. Maize, sorghum, and rice may not differ in the nature or size of these repetitive blocks, but only in the frequency with which they are present between any pair of genes. More experimentation is needed to characterize genome organization in rice and sorghum before any comprehensive conclusions can be drawn.
Grass Genomic Collinearity and its Exceptions.
Most investigations of map collinearity in the cereals and other plants have used the hybridization of low-copy-number DNA markers that are usually genes. Both collinearity and microcollinearity, when observed, appear to be limited to genes.
Although comparative maps of the grasses show large regions of collinearity, chromosomal rearrangements involving entire arms or segments of arms are not uncommon (5, 7, 27). Particularly confusing for comparative mapping studies are “distantly tandem” duplications that place the same markers or linear array of markers at two well-separated locations on the same chromosome arm (28). These large rearrangements are not necessarily problematic, as chromosome walking to clone a gene or mapping to identify an ortholog would be hindered only if the investigated area crossed one of the breakpoints of the rearrangement.
However, comparisons of recombinational maps between closely related plant species often yield 20–40% of the markers that do not fall into any obvious collinearity, or even synteny, relationship (10). These markers are usually left off of the comparative maps to simplify the presentation. Reports of collinearity and orthology between mapped genes (including DNA markers and morphological traits) also are biased toward the successes. Failures are simply not reported, partly because negative results rarely receive attention and partly because a lack of observed collinearity could be caused by technical error or to an inappropriate comparison between paralogs rather than orthologs.
Genomic Collinearity as a Tool for Crop Improvement.
Genetic map comparisons make it clear that gene composition and order, otherwise known as map collinearity, are common among plants, particularly between the cereals (5–10). In some cases, this collinearity may extend even between monocotyledonous and dicotyledonous plants (27). However, many exceptions have been ignored and local rearrangement (i.e., microcollinearity) has not been extensively examined. We have seen a lack of collinearity between maize and rice clones in Adh1-homologous regions. Microcollinearity between Arabidopsis and any monocotyledonous plant may be rare. At this stage, it is premature to conclude that the frequency of small scale rearrangements that would be missed in most comparative genetic maps will be as low as the frequency of the large rearrangements that have been observed (5, 7, 27).
More experimentation is needed to determine how often collinearity holds true at the few hundred-kbp level that is significant for most molecular and genetic applications. If small rearrangements often interfere with this microcollinearity, then it may be necessary to mostly rely on closely related species for such approaches as chromosome walking. For instance, sorghum has a genome that is about 1.7 times as large as that of rice, but it has been separated from maize by about 3-fold fewer years than has rice. Hence, if microrearrangements are common, then the study of maize might be better accomplished by using sorghum as an ally rather than (or preferably, in addition to) rice.
At the very least, further investigations into genome organization and microcollinearity are needed in the grasses before one commits to a full-scale assault on any single genome. There is an enormous potential value in genomic collinearity for gene isolation, for pathway dissection, and for uncovering the basis and directions of the variation engineered by nature in plants (8–10). We have the technology to initiate this era and should use it wisely.
Acknowledgments
We thank S. Frank for excellent technical assistance. This work was supported by grants from the United States Department of Agriculture/National Research Initiative Competitive Grants Program (94–37310-0661 and 94-37300-0299) to J.L.B., a United States National Science Foundation Training Grant Fellowship to P.S., and a Purdue Research Foundation Fellowship to M.C.
ABBREVIATIONS
- BAC
bacterial artificial chromosome
- mbp
megabase pair
References
- 1.Bennett M D, Leitch I J. Ann Bot (London) 1995;76:113–176. [Google Scholar]
- 2.Flavell R B, Bennett M D, Smith J B, Smith D B. Biochem Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 3.Peacock W J, Dennis E S, Rhoades M M, Pryor A J. Proc Natl Acad Sci USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hake S, Walbot V. Chromosoma. 1980;79:251–270. [Google Scholar]
- 5.Hulbert S H, Richter T E, Axtell J D, Bennetzen J L. Proc Natl Acad Sci USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn S, Anderson J A, Sorrells M E, Tanksley S D. Mol Gen Genet. 1993;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- 7.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 8.Bennetzen J L, Freeling M. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 9.Moore G, Gale M D, Kurata N, Flavell R B. Bio/Technology. 1993;11:584–589. [Google Scholar]
- 10.Bennetzen J L, Freeling M. Genome Res. 1997;7:301–306. doi: 10.1101/gr.7.4.301. [DOI] [PubMed] [Google Scholar]
- 11.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 12.Doebley J, Durbin M, Golenberg E M, Clegg M T, Ma D P. Evolution. 1990;44:1097–1108. doi: 10.1111/j.1558-5646.1990.tb03828.x. [DOI] [PubMed] [Google Scholar]
- 13.Springer P S, Edwards K J, Bennetzen J L. Proc Natl Acad Sci USA. 1994;91:863–867. doi: 10.1073/pnas.91.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, SanMiguel P, de Oliveira A C, Woo S-S, Zhang H, Wing R A, Bennetzen J L. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avramova Z, Tikhonov A, SanMiguel P, Jin Y-K, Liu C, Woo S-S, Wing R A, Bennetzen J L. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y-K, Bennetzen J L. Plant Cell. 1994;6:1177–1186. doi: 10.1105/tpc.6.8.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 18.Bennetzen J L, Schrick K, Springer P S, Brown W E, SanMiguel P. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 19.Edwards K J, Veuskens J, Rawles H, Daly A, Bennetzen J L. Genome. 1996;39:811–817. doi: 10.1139/g96-102. [DOI] [PubMed] [Google Scholar]
- 20.Civardi L, Xia Y, Edwards K J, Schnable P S, Nikolau B J. Proc Natl Acad Sci USA. 1994;91:8268–8272. doi: 10.1073/pnas.91.17.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Hsia A-P, Zhang L, Nikolau B J, Schnable P S. Plant Cell. 1995;7:2151–2161. doi: 10.1105/tpc.7.12.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Bennetzen J L. Plant Mol Biol. 1996;32:999–1001. doi: 10.1007/BF00041383. [DOI] [PubMed] [Google Scholar]
- 23.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 24.Chen, M., SanMiguel, P. & Bennetzen, J. L. (1997) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 25.Bennetzen J L. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 26.Bennetzen J L, Liu C-N, SanMiguel P, Springer P S, Jin Y-K, Zanta C A, Avramova Z. In: Genomes of Plants and Animals. Gustafson J P, Flavell R B, editors. New York: Plenum; 1996. pp. 103–113. [Google Scholar]
- 27.Paterson A H, Lan T H, Reischmann K P, Chang C, Lin Y R, Liu S C, Burow M D, Kowalski S P, Katsar C S, DelMonte T A, Feldmann K A, Schertz K F, Wendel J F. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- 28.Sanz-Alferez S, Richter T E, Hulbert S H, Bennetzen J L. Theor Appl Genet. 1995;91:25–32. doi: 10.1007/BF00220854. [DOI] [PubMed] [Google Scholar]