Abstract
Ribonucleotide reductase (RNR) catalyzes reduction of the four different ribonucleotides to their corresponding deoxyribonucleotides and is the rate-limiting enzyme in DNA synthesis. RNR is a well-established target for the antiproliferative drugs Gemzar and Hydrea, for antisense therapy, and in combination chemotherapies. Surprisingly, few novel drugs that target RNR have emerged, partly because RNR activity assays are laboratory-intense and exclude high-throughput methodologies. Here, we present a previously undescribed PCR-based assay for RNR activity measurements in microplate format. We validated the approach by screening a diverse library of 1,364 compounds for inhibitors of class I RNR from the opportunistic pathogen Pseudomonas aeruginosa, and we identified 27 inhibitors with IC50 values from ∼200 nM to 30 μM. Interestingly, a majority of the identified inhibitors have been found inactive in human cell lines as well as in anticancer and in vivo tumor tests as reported by the PubChem BioAssay database. Four of the RNR inhibitors inhibited growth of P. aeruginosa, and two were also found to affect the transcription of RNR genes and to decrease the cellular deoxyribonucleotide pools. This unique PCR-based assay works with any RNR enzyme and any substrate nucleotide, and thus opens the door to high-throughput screening for RNR inhibitors in drug discovery.
Keywords: National Cancer Institute diversity set II, nucleotide metabolism, ribonucleotide reductase assay, high-throughput assay
Ribonucleotide reductase (RNR) is an essential enzyme for de novo synthesis of DNA building blocks via reduction of the 2′-hydroxyl of ribonucleotides (1, 2). With its key role in the DNA synthesis pathway, RNR is an absolute requirement for cellular proliferation and a prerequisite for life. RNR is found in all free-living organisms as well as in some dsDNA viruses. The only known exceptions to this ubiquitous presence are a few parasites and obligate intracellular endosymbionts (3) that rely on the host cell for production of DNA precursors. Thus, RNR is a potential antimicrobial drug target in a wide variety of organisms.
RNR enzymes exist in three different classes, each with different cofactor requirements. Eukaryotes generally possess only one class, whereas bacteria possess any combination of RNR classes (3). Different oligomeric states of RNRs are known, and a common active form of the major variants of RNR is a dimer of dimers (α2β2) (4). RNR exhibits a radical-based catalytical mechanism that involves redox cycling of cysteine residues, specific metal ion dependencies, essential subunit interactions, and sophisticated allosteric regulation (1). Thus, in addition to conventional competitive inhibition, pharmaceuticals may interfere with subunit interactions, binding to allosteric effector sites, metal chelation, radical formation and transfer, or inhibition of cysteine disulfide exchange. These sites of potential intervention offer a plethora of possibilities for the design of pathogen-specific antibiotics. Because sequence identities between bacterial and human RNRs are typically well below 50%, the chances of designing species-specific inhibitors are improved even more.
Current drugs targeted at RNR are used in anticancer therapy [e.g., the radical scavenger Hydrea (hydroxyurea) and the nucleoside analog Gemzar (gemcitabine, 2′,2′-difluoro-2′-deoxycytidine)] that convert to a suicidal substrate analog in vivo. Inhibitors that explore other chemical features of RNR (e.g., iron chelators and peptides, peptide analogs mimicking subunit interactions) (5, 6) have been developed as an approach to antileishmanial drugs (7), as well as the basis for novel antibiotics against Mycobacterium tuberculosis (8) and antivirals against herpes simplex virus (9–11). To date, none of these efforts has led to development of an approved antimicrobial or antiviral drug.
There is a limited chemical variation of RNR-targeted drugs and inhibitors. A reason for this is that available enzyme activity assays have not allowed an unbiased search for novel RNR inhibitors (i.e., high-throughput screening (HTS)]. Current methodologies are all markedly labor-intensive because of the fact that ribonucleotides and deoxyribonucleotides are difficult to resolve experimentally (12–15). This severely limits the number of samples that can be processed per day. Therefore, the development of RNR inhibitors has been restricted to obvious chemical properties inherent in RNR enzymology, mostly by nucleotide analogy and radical chemistry. An efficient RNR activity assay that allows inhibitor screening in microplate format would have the potential to identify a range of novel inhibitors against this promising and ubiquitous drug target.
Here, we present a PCR-based method [patent pending (16)] for activity determination of RNR that is suitable for screening of compound libraries in microplate format. The method relies on quantification via PCR of the amount of a dNTP formed by RNR. Only three dNTPs are added in excess to the PCR mixture, and the fourth limiting dNTP is supplied via the RNR reaction mixture. For RNR enzymes using ribonucleoside diphosphates as substrates, the PCR-required dNTP is obtained from the RNR reaction via an incubation step with nucleoside diphosphate kinase (NDPK). The amount of DNA formed in the PCR is related to the amount of the limiting dNTP, and it can be quantified by various means (e.g., via fluorescence intensity of DNA binding dyes or radioactivity-based detection). To exemplify the usefulness of the methodology, we have screened the diversity set II compound library (http://dtp.cancer.gov) of the National Cancer Institute (NCI) for inhibitors of RNR from Pseudomonas aeruginosa, an opportunistic pathogen and major cause of nosocomial infections (17, 18). Among 1,364 diverse compounds, we identified 27 RNR inhibitors with IC50 values ranging from ∼200 nM to 30 μM. These RNR inhibitors were tested for inhibition of growth of P. aeruginosa PAO1, and four compounds exhibited potencies in the same range as or better than carbenicillin, tetracycline, and hydroxyurea. Among the RNR inhibitors with antibacterial activity, two were found to lower cellular dNTP levels and to affect RNR gene expression, which are observations compatible with RNR being targeted in vivo.
Results
PCR-Based Assay for Identification of RNR Inhibitors.
PCR experiments with limiting amounts of dCTP indicated that DNA formation was approximately linear up to 12 μM limiting dCTP and that NDPK conversion of dCDP to dCTP was sufficiently effective to give comparable PCR results and linearity (Fig. 1). Assay performance was also verified with different incubation times and different amounts of RNR in the reactions (Fig. S1). Assay conditions were adapted for SYBR green-based detection and CDP as substrate for RNR (Fig. 1). All four RNR products (dCTP, dUTP, dATP, and dGTP) and dTTP could be used as limiting dNTP, with dCTP and dTTP giving the highest sensitivities (Fig. S2).
Fig. 1.
PCR-based quantification of RNR enzyme activity. (Upper Left) PCR-based assay with three dNTPs in excess and limiting amounts [1 (a), 2 (b), 4 (c), 6 (d), 12 (e), and 15 (f) μM] of either dCTP (Right) or dCDP (Left) in which NDPK was used to catalyze conversion of dCDPs into dCTPs shows that quantifications are linear up to 12 μM dCTP. The concentrations of dATP, dGTP, and dTTP were 200 μM. DNA (ca. 180 bp) from PCRs was separated on an ethidium bromide-containing (0.5 μg/mL) agarose gel (2%). (Upper Right) NDPK-treated dCDP gives a similar response in the PCR-based assay as corresponding amounts of dCTP. An aliquot (10 μL) from each PCR was mixed with SYBR green dissolved in TAE buffer (190 μL), and the fluorescence intensity was recorded. (Lower Left) Distribution of relative P. aeruginosa RNR enzyme activity in 1,364 assays each containing 100 μM of a compound from the NCI’s compound library (diversity set II). Approximately 110 compounds inhibited RNR enzyme activity to >50%, and 28 compounds inhibited it to >90% (red bars). (Lower Right) Representative 96-well plate shows 80 samples (blue bars, 0–90% inhibition; red bars, >90% inhibition), eight fully inhibited controls (purple bars), and eight noninhibited controls (yellow bars).
When the assay was used to screen for RNR inhibitors, conditions were calibrated to create maximal separation between positive and negative control samples. Thus, measures were taken to ensure that samples with the highest product formation (positive controls) gave a response close to the upper detection limit of the assay. This rendered the response of samples with an activity higher than the positive controls to be partly flattened and favored detection of inhibitors. The final screening conditions gave good separation between positive and negative control samples, as judged by a Z-factor of 0.86 (Fig. S3).
Identification of 27 Potent Inhibitors of P. aeruginosa RNR.
We screened the NCI’s diversity set II (1,364 compounds) with the unique assay, and 110 compounds were found to inhibit class I RNR from P. aeruginosa by >50% (Fig. 1). We selected 28 compounds exhibiting >90% inhibition for dose–response analysis using the conventional assay (14, 15) with radiolabeled CDP, chromatographic purification of formed dCDP, and subsequent quantification using liquid scintillation counting. In addition to assessment of inhibitor potency, this served to confirm the hits with a complementary assay. All derived dose–response curves allowed acceptable model-to-data fit and determination of IC50 values. Interestingly, two of the selected strong inhibitors were duplicates in the NCI diversity set; thus, the screen identified 27 compounds with confirmed inhibition of RNR activity.
IC50 values for the 27 active compounds ranged from 0.2 to 34 μM (Fig. 2 and Figs. S4–S7), which corresponds to Ki values of ∼46 nM to 8 μM, for the experimental conditions applied and assumed competitive inhibition [Ki = IC50/(1 + [S]/Km)]. Notably, the enzyme concentration in the assay was high (0.25 μM and 1 μM for the respective RNR subunits), which suggests that the most potent inhibitors may be tight binders with Ki values even lower than the apparent ones reported. In addition, the relatively high enzyme concentration in relation to the potency of the inhibitors indicates that inhibition likely involves interference with the catalytical machinery of RNR rather than trivial mechanisms, such as aggregation leading to promiscuous inhibition.
Fig. 2.
Four RNR inhibitors that also have bactericidal effect on growth of P. aeruginosa (Table 1): toluidine blue (NSC36758, ○), streptonigrin (NSC45383, ●), NSC361666 (□), NSC228155 (■), and hydroxyurea (△). All four inhibitors were significantly more potent than hydroxyurea. Toluidine blue exhibited >70% inhibition even at 13 nM (discussed in main text).
Four Main Groups of RNR Inhibitors.
On a structural basis and with respect to functionality and possible mode of action, the inhibitors could be divided into a few groups. Three groups contained compounds with functionalities known to be redox-active and that exhibited some similarity to compounds known to affect RNR activity. These three groups were defined by 5 anthraquinone-like (Fig. S4), 10 naphthoquinone-like (Fig. S5), and 4 phenol-containing (Fig. S6) substances. Eight (one-third) of the confirmed inhibitors were more diverse and did not possess functionalities obviously related to known RNR inhibitors, and they were defined as a separate group of diverse compounds (Fig. S7).
The most potent inhibitors were found within the anthraquinone-like group and exhibited IC50 values below 1 μM (Fig. S4). In order of potency as an RNR inhibitor, the group consists of the dye toluidine blue (NSC36758), a derivative of the redox indicator methylene blue (NSC40273), the resorufin analog questiomycin A (NSC94945), crystal violet 47 (NSC23123), and an isoalloxazine derivative (NSC3064). Toluidine blue exhibited an apparent IC50 value well below the enzyme concentration of the assay, and the lowest concentration tested (0.2 μM) still gave pronounced inhibition (Fig. 2). Compounds of the naphthoquinone-like group contain naphthoquinone or hydronaphthoquinone moieties. These compounds typically exhibited IC50 values from 1 to 10 μM. One subgroup is defined by three differently substituted hydronaphthoquinones (Fig. S5; NSC278631, NSC128281, and NSC111552). Of these, NSC278631 (IC50 = 2.4 μM) with the largest substituent exhibited a fourfold lowered IC50 value, thus suggesting a possible structure–activity relationship. Conversely, the subgroup defined by four differently substituted naphthoquinones all exhibited similar IC50 values around 1.2 μM (Fig. S5; NSC645330, NSC641396, NSC641395, and NSC661221). Compounds in the phenol-containing group (Fig. S6) had IC50 values in the same range as compounds of the naphthoquinone-like group. For the diverse group of compounds, IC50 values ranged from 1.1 to 26 μM (Fig. S7).
Four RNR Inhibitors with Antibacterial Activity Against P. aeruginosa.
The 27 compounds that were active inhibitors of P. aeruginosa RNR by the enzymatic dose–response test were further analyzed for antibacterial activity against P. aeruginosa PAO1. Four compounds [NSC36758 (toluidine blue), NSC45383 (streptonigrin), NSC361666, and NSC228155; Fig. 2] were active in disk diffusion tests, and minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values were in the micromolar range for three of these (Table 1). In relative terms, these three compounds were as effective P. aeruginosa bactericidals as the common antibiotics tetracycline and carbenicillin. Compared with the cytostatic RNR inhibitor hydroxyurea (IC50 = 150 μM; Fig. 2), all four compounds exhibited higher antibacterial activities, and streptonigrin and NSC228155 were about two to three orders of magnitude more potent than hydroxyurea on the bacterial level (Table 1). Whereas both streptonigrin and its methyl ester derivative (NSC45384) showed similar inhibition at the enzymatic level (Fig. S5), only streptonigrin was active against P. aeruginosa (Table 1). Interestingly, the four compounds with obvious antibacterial activity each belonged to a separate functional group of RNR inhibitors. This demonstrates that our HTS approach for identification of RNR inhibitors can identify potential leads of various chemical types.
Table 1.
Antibacterial effect of unique RNR inhibitors compared with a known RNR antiproliferative drug and two known antibiotic drugs
| Compound | DDT*, mm | MIC†, μM | MBC‡, μM | IC50§, μM |
| NSC36758 (toluidine blue) | 17 | 400 | 400 | <0.2 |
| NSC45383 (streptonigrin) | 18 | 50 | 200 | 7.4 |
| NSC361666 | 8 | >400¶ | n.d.‖ | 7.4 |
| NSC228155 | 11 | 50 | 400 | 26 |
| Hydroxyurea | ND | 5,700 | 180,000 | 150 |
| Tetracycline | 16 | 15 | 450 | N/A |
| Carbenicillin | 20 | 150 | 625 | N/A |
N/A, not applicable; ND, not detected.
*Disk diffusion test (38).
†MIC (LB media) (39).
‡MBC (LA plates) (39).
¶Distinct but not complete growth inhibition was observed at the highest concentration tested (400 μM).
‖Not determined (n.d.) because complete growth inhibition was not observed at highest concentration tested (400 μM).
Two Inhibitors Increase RNR Expression and Lower Cellular dNTP Levels.
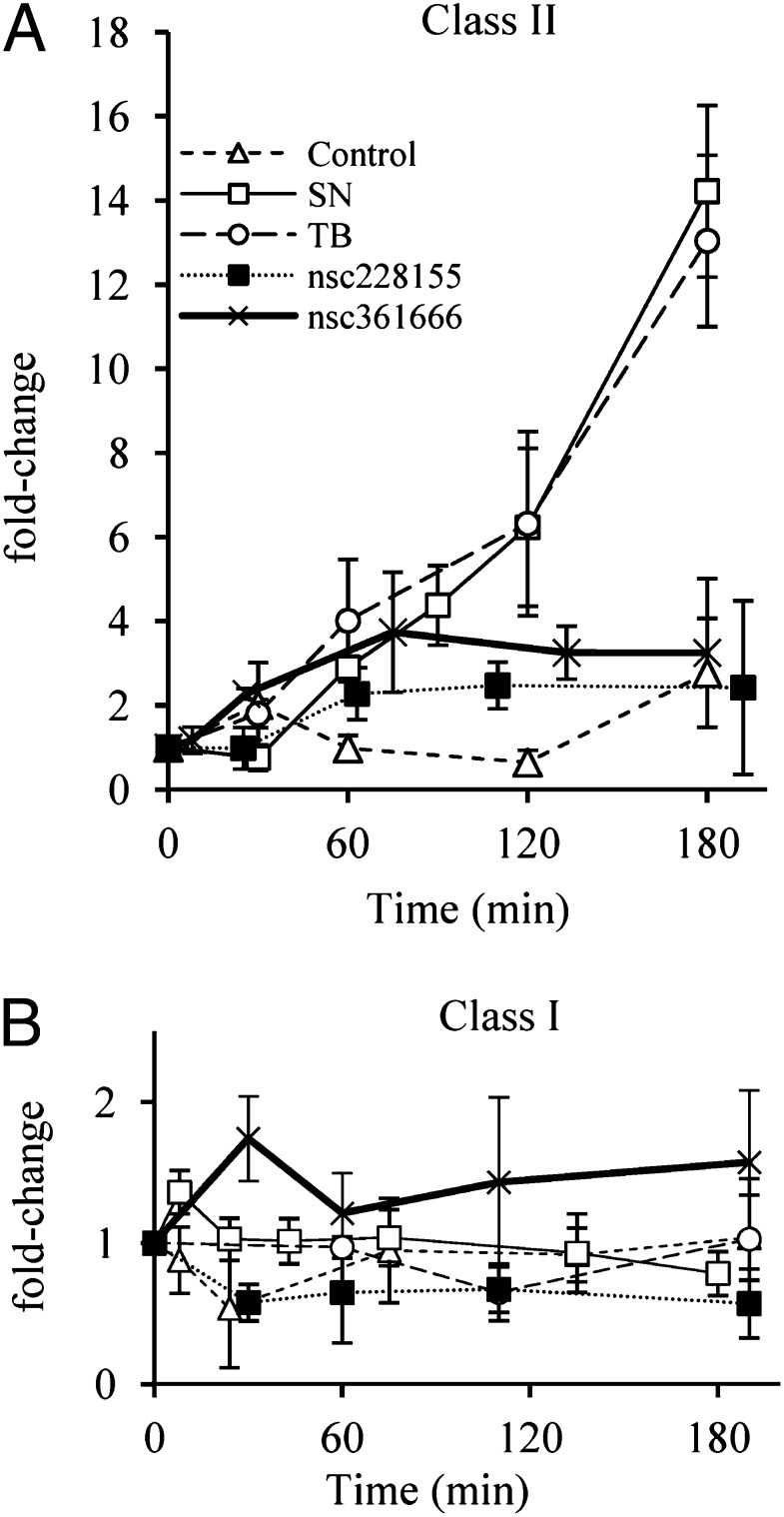
The four inhibitors that exhibited antibacterial activity were tested for their ability to affect expression of class I (nrdA) and II (nrdJ) RNR genes using quantitative real-time PCR. Subbactericidal concentrations of toluidine blue (100 μM) and streptonigrin (20 μM) were found to cause >10-fold increased expression of nrdJ, whereas nrdA was not significantly affected (Fig. 3). In contrast, 100 μM of either of the other two inhibitors (NSC361666 and NSC228155) did not cause any significant changes in RNR transcript levels and followed the expression profile of the DMSO-treated control sample (Fig. 3).
Fig. 3.
Transcription of RNR genes in response to inhibitor treatment. Class II RNR (nrdJ) (A) and class I RNR (nrdA) (B) P. aeruginosa cultures with an optical density of 0.15 were treated with RNR inhibitors with antibacterial activities (streptonigrin, □; toluidine blue, ○; NSC228155, ■; NSC361666, thick black line) or DMSO as a control (△), and the fold-change in mRNA expression was monitored over time by real-time PCR. Whereas streptonigrin and toluidine blue lead to >10-fold changes in transcript levels of nrdJ, changes by NSC228155 and NSC361666 are modest and close to the control. None of the inhibitors significantly affected transcription of the nrdA gene.
To investigate a coupling to inhibition of RNR activity in vivo, toluidine blue and streptonigrin were further tested for their effects on the cellular dNTP pools. As above, subbactericidal inhibitor concentrations were added to P. aeruginosa cultures, bacterial cells were harvested after 30 min of continued incubation, and dNTPs were extracted and quantified. Both streptonigrin and toluidine blue led to significantly depleted levels of dNTP pools (Fig. 4). All four dNTPs were decreased by both inhibitors, and compared with the control, streptonigrin treatment decreased the total dNTP pool by 73 ± 21% and toluidine blue decreased it by 54 ± 16%.
Fig. 4.
dNTP pool changes in response to inhibitor treatment. P. aeruginosa cultures with an optical density of 0.35 were treated with inhibitors associated with increased RNR expression [streptonigrin (SN), light gray bars; toluidine blue (TB), dark gray bars] or DMSO as a control (open bars). Cultivation was continued for 30 min after addition of inhibitor, after which cells were harvested and dNTPs were extracted and quantified by HPLC. Given concentrations refer to the total amount extracted divided by the culture volume. Both streptonigrin and toluidine blue significantly decreased the total dNTP pool compared with DMSO-treated controls as well as the four individual dNTP pools. The observed differences between individual dNTP levels (i.e., dCTP vs. dGTP vs. dTTP vs. dATP) were not statistically significant, however.
Discussion
The PCR-based HTS method to screen for inhibitors of any RNR has been tested on class I RNR from the opportunistic pathogen P. aeruginosa. Among 1,364 chemically diverse compounds, ∼2% (27 compounds) were potent inhibitors and had IC50 values between 0.2 μM and 34 μM. Four of these RNR inhibitors were also found to inhibit growth of P. aeruginosa, and three compounds were at least as bactericidal against P. aeruginosa as the well-known antibiotics tetracycline and carbenicillin. Because these four antibacterial compounds were structurally different, contained different chemical functionalities, and were potent RNR inhibitors with IC50 values between 0.2 μM and 26 μM, it is conceivable that the approach presented here has a promising potential to identify novel antibiotic leads with a known intracellular target. The different chemical functionalities of the inhibitors suggest that different parts of the catalytical machinery of P. aeruginosa RNR have been targeted. Approximately 15% of the identified RNR inhibitors possessed antimicrobial activity, which is a reasonable number considering the inherent drug resistance of P. aeruginosa (19).
A phenol functionality was found in 4 of the 27 potent RNR inhibitors identified by us (Fig. S6). This functional group, as well as hydroxylamines (e.g., hydroxyurea), is known to exhibit reactivity toward RNR and to act as a radical scavenger (20, 21). Among the phenol-containing compounds, NSC361666 exhibited antibacterial activity in the disk diffusion test; however, its effect in liquid culture was weak, and we did not observe interference with RNR gene expression for this compound. Similarly, for the diverse group of compounds (Fig. S7), one (NSC228155) was found to inhibit P. aeruginosa growth but did not affect RNR gene expression. Regardless of the limited biological effects of the inhibitors in these two groups (Figs. S6 and S7), they are of interest to guide the design of novel bioactive RNR inhibitors, especially in light of the fact that most of these inhibitors are inactive in PubChem BioAssays for activity against human cell lines in culture and in vivo (Table S1). In this respect, the eight compounds (Fig. S7) with structures previously unknown to be associated with inhibition of RNR are particularly interesting and truly unique as RNR inhibitors. The most potent of these (NSC130872) is a reported inhibitor of cyclin-dependent kinase (22), and therefore a possible competitive inhibitor with respect to nucleotides.
The most potent RNR inhibitors were found within the anthraquinone-like group of substances (Fig. S4). Based on their structural features, compounds in this group can be classified as distinctly redox-active, and they most likely inhibit RNR via interference with radical formation, radical transfer, or metal oxidation state. Related redox indicators have previously been shown to interact with the radical harboring subunit of Escherichia coli RNR (23, 24). In this group, toluidine blue exhibited an extremely low IC50 value (<0.2 μM) and also exhibited antimicrobial activity. Toluidine blue is structurally very similar to methylene blue. Both are dyes with various uses in histology, for example, and are capable of two-electron redox cycling under physiological conditions (25). They induce cancer cell apoptosis and have activity against some bacteria (26, 27). The extreme IC50 of toluidine blue, below the enzyme concentration in the assay, excludes pure competitive inhibition and instead indicates that its activity is redox-related. The mechanism may involve repeated and DTT-driven inactivation of the radical harboring subunit of RNR.
The naphthoquinone-like RNR inhibitors are also known to be redox-active, and therefore have potential to interfere with this aspect of RNR catalysis (Fig. S5). Among the 10 compounds in this group, the streptonigrin exhibited antibacterial activity against P. aeruginosa that was in line with common antibiotics. Streptonigrin is produced by Streptomyces flocculus, and it has activity against several microbial pathogens (28, 29). Its cytotoxic effects are enhanced by metal ions, and its mechanism of action is proposed to involve redox cycling, formation of radicals, genotoxic effects, or interference with cell respiration (30, 31). Inhibition of RNR results in similar effects, as observed for hydroxyurea-mediated inhibition of RNR in E. coli (32). However, RNR has not been proposed as a mode of action for streptonigrin. Despite the general toxicity of streptonigrin, it is inactive in many tumor models (Table S1), suggesting that it has potential as an antibiotic lead compound.
The cellular response by P. aeruginosa to toluidine blue and streptonigrin involved increased expression of nrdJ and decreased dNTP levels. We have previously observed a pronounced increase in the expression of the P. aeruginosa class II RNR gene and impaired dNTP synthesis caused by hydroxyurea treatment (33). The increased RNR gene expression in conjunction with depletion of dNTP levels indicates that RNR is targeted by toluidine blue and streptonigrin, thus suggesting a novel activity for these compounds that may be as relevant as their previously reported effects (26–32). Our results suggest that inhibition of growth is attributable to impaired DNA synthesis as a result of low dNTP pools. A possible feedback mechanism accounting for the induction of RNR expression in response to lowered dNTP levels might be executed by NrdR, a protein that regulates RNR expression (34, 35) and possesses an ATP cone capable of binding nucleotides, and thereby to respond to nucleotide fluctuations.
In this study, we present a simple PCR-based assay to identify novel RNR inhibitors. The assay is suitable for microplate format and can be performed with standard laboratory equipment or fully automated for HTS. Because the read-out is based on quantification of DNA, a variety of detection modes can be used (e.g., fluorescence intensity of DNA-bound dyes, scintillation-based detection of radiolabeled nucleotides). In its most simple form, only a PCR thermocycler is required and semiquantitative results can be obtained simply by photographic documentation of the fluorescence of DNA-bound dyes on UV illumination. This is in contrast to the chromatographic approaches currently used for analysis of RNR activity, whose automation requires robotic HPLC systems. The moderate detection range of the assay in its current setup can easily be extended by adjustment of PCR conditions, reagents, or the mode of detection (e.g., use of a scintillation proximity assay with streptavidin-coated beads, radiolabeled RNR substrate, and biotinylated PCR primers). Moreover, an interesting application of the assay is determination of dNTP pools.
This is a unique assay that is particularly suitable in HTS for inhibitors of any RNR, regardless of species origin or substrate preference. Using the method, we have identified several potent and previously undescribed inhibitors of P. aeruginosa RNR with distinctly different structures, and three of the identified compounds exhibited clear antibacterial activities against P. aeruginosa. Relative to hydroxyurea, an antiproliferative drug that targets RNR via radical scavenging, four compounds were more potent on both the cellular and enzymatic levels, and two were found to target P. aerugionsa RNR in vivo. Interestingly, a majority of the identified inhibitors of P. aeruginosa RNR have been found inactive in human cell lines as well as in anticancer and in vivo tumor tests as reported by the PubChem BioAssay database. RNR is a promising antimicrobial drug target that deserves attention at a time when the number of multidrug-resistant pathogens is escalating globally. To meet this challenge, we have developed a unique methodology that opens the door to exploration of RNR in HTS-based drug discovery.
Materials and Methods
Materials.
The two subunits, NrdA and NrdB, of class I RNR from P. aeruginosa were purified as described (36). Taq polymerase from Fermentas was used for PCR. NDPK, SYBR green (at a 10,000× working concentration), and standard chemicals were from Sigma–Aldrich. Tris/acetate/EDTA (TAE) buffer at 1× working solution contained 40 mM Tris-acetate pH 8.0 and 2 mM EDTA. Additional materials are described in SI Materials and Methods.
PCR with a Limiting Amount of dCTP.
Different PCR assays with three dNTPs each at a concentration of 200 μM in each sample and the fourth dNTP at variable concentrations (0–16 μM) between samples were prepared. Other components of the 50-μL PCR mixture contained 2 pg/μL DNA template, 0.5 μM DNA primers, 1.5 mM MgCl2, 0.02 U/μL Taq DNA polymerase, and PCR reaction buffer. Samples were subjected to thermocycling: 3 min at 94 °C followed by 40 cycles of 45 s at 94 °C, 50 s at 55 °C, and 60 s at 72 °C. PCR samples were analyzed by agarose gel electrophoresis and ethidium bromide staining or by fluorescence intensity measurement after mixing with SYBR green as described in SI Materials and Methods.
NDPK Conversion of dNDP to dNTP.
Samples of 25 μL contained 100 mU/μL NDPK, 4 mM ATP, 5 mM MgCl2, and variable concentrations (5, 10, 20, 30, 60, and 75 μM) of dCDP were incubated for 30 min at 37 °C, after the reactions were quenched by heating. Subsequently, 10 μL of NDPK reaction mixture was mixed with 40 μL of PCR mixture (final concentrations: 200 μM each of dGTP, dATP, and dTTP; 2 pg/μL DNA template, 0.5 μM DNA primers, and 0.05 U/μL Taq DNA polymerase and PCR reaction buffer). PCR controls with 1, 2, 4, 6, 12, and 15 μM dCTP (i.e., the final dCTP concentration that would result if NDPK converted all dNDP to dNTP) were prepared. All samples were subjected to PCR cycling, and the DNA formed was analyzed by fluorescence intensity measurement and gel electrophoretic analysis, as described in SI Materials and Methods.
Screen for RNR Inhibitors in a 96-Well Format.
Reactions with class I RNR from P. aeruginosa in a total reaction volume of 30 μL were prepared in separate wells of a PCR plate. The RNR reactions contained 0.25 μM NrdA, 1 μM NrdB, 5 mM ATP, 20 mM magnesium-acetate, 30 mM DTT, and 1.5 mM 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate in 30 mM Tris-buffer (pH 7.5).
Compounds from the NCI/Developmental Therapeutics Program Open Chemical Repository (diversity set II containing 1,364 compounds, http://dtp.cancer.gov/) were added (final concentration of 100 μM) to the wells of columns 2–11. In addition, eight control samples without test compound and eight with 3.5 mM hydroxyurea were set up in the wells of columns 1 and 12, respectively. All samples contained 1% DMSO.
After setup, samples were incubated for 30 min and CDP (final concentration of 100 μM) was then added to start the reactions. Reactions proceeded for 60 min at ambient temperature and were then quenched by heating. The chosen incubation time gave a substrate conversion of 40–50% in positive control samples, which is suitable for screening purposes. After heat quenching, the samples were allowed to cool off, NDPK was added (final concentration of 25 mU/μL), samples were incubated overnight, and the reaction was quenched by heating.
After cooling, 70 μL of PCR mixture (see above) lacking dCTP was added to each well and samples were subjected to PCR cycling as described above. After PCR, 10 μL from each well was transferred to a black 96-well plate, 190 μL of SYBR green in TAE was added to each well, and the fluorescence intensity recorded as described in SI Materials and Methods.
Data Analysis.
For calculation of the Z-factor 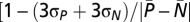 , where σP and σN are SDs of the positive and negative controls and
, where σP and σN are SDs of the positive and negative controls and  and
and  are the average of positive and negative controls], performance measures of the method of one series with negative control samples (3.5 mM hydroxyurea and 1% DMSO) and one series with positive control samples (only 1% DMSO) were prepared and incubated as described above. The Z-factor for individual 96-well plates fluctuated from 0.6 to 0.9. Software (Marvin and Instant JChem, version 5.3.8) from Chemaxon (www.chemaxon.com) was used for structure drawing and database management.
are the average of positive and negative controls], performance measures of the method of one series with negative control samples (3.5 mM hydroxyurea and 1% DMSO) and one series with positive control samples (only 1% DMSO) were prepared and incubated as described above. The Z-factor for individual 96-well plates fluctuated from 0.6 to 0.9. Software (Marvin and Instant JChem, version 5.3.8) from Chemaxon (www.chemaxon.com) was used for structure drawing and database management.
Dose–Response Analysis.
Dose–response analyses were performed for compounds that inhibited RNR activity >90% in the primary screen at a concentration of 100 μM, where inhibition by 3.5 mM hydroxyurea was defined as 100%.
In dose–response experiments, compounds were tested in the conventional assay (SI Materials and Methods) at concentrations starting at 100 μM and diluted in 11 steps (1:2.25 in each). Other reaction constituents were as described above in the screen for RNR inhibitors in a 96-well format. For compounds with satisfactory dose–response behavior, IC50 values were determined by fitting a four-parameter dose–response model (B + (B − T)/(1 + 10(log[IC50] − log[I])h), where B is the lower plateau, T is the top plateau, and h is the slope) to the data by nonlinear regression. Statistical analysis of model-to-data fit was performed with SOLVERSTAT, and reported SEs are calculated by the program (37).
Antimicrobial Activity.
Compounds with confirmed activity in the dose–response analysis were tested for bacterial growth inhibition by means of a disk diffusion test (38), MIC (39), and MBC (39) as described in SI Materials and Methods.
In Vivo Effects of RNR Inhibitors.
Compounds with antimicrobial activity were tested for their effects on expression of RNR genes by quantitative real-time PCR (SI Materials and Methods). Compounds dissolved in DMSO were added to P. aeruginosa cultures, grown in minimal media enriched with vitamin B12, at early log phase. The relative fold-changes in mRNA expression of RNR genes were subsequently monitored over time with respect to the time point of inhibitor addition using quantitative real-time PCR according to the comparative cycle threshold (ΔΔCt) method (40). Control samples, to which only DMSO were added, were also prepared.
Compounds that were found to increase the expression of RNR genes were tested for their effect on cellular dNTP levels. P. aeruginosa cultures were grown in minimal media until the OD600 reached 0.35, whereupon inhibitors dissolved in DMSO were added or only DMSO was added to control samples. Cultivation was continued for another 30 min, the bacterial cells then harvested, and dNTPs were extracted and quantified (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank C. Frumerie, D. Lundin, and J. Morrison for helpful discussions. We are grateful to MariAnn Westman for experimental help with protein expression and purification. This work was supported by the Swedish Research Council (Grants VR-M 2009-585 and VR-MH K2011-56X-20677), the Royal Swedish Academy of Sciences (the Hierta–Retzius Foundation), and the Magnus Bergvall Foundation.
Footnotes
Conflict of interest statement: The authors have submitted a patent application for the method presented in the paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113051109/-/DCSupplemental.
References
- 1.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Torrents E, Sahlin M, Sjöberg B-M. The ribonucleotide reductase family—Genetics and genomics. In: Andersson KK, editor. Molecular Anatomy and Physiology of Proteins: Ribonucleotide Reductase. Nova Science Publishers, New York; 2008. pp. 17–77. [Google Scholar]
- 3.Lundin D, Torrents E, Poole AM, Sjöberg B-M. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics. 2009;10:589. doi: 10.1186/1471-2164-10-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rofougaran R, Crona M, Vodnala M, Sjöberg B-M, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J Biol Chem. 2008;283:35310–35318. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- 5.Cerqueira NM, Pereira S, Fernandes PA, Ramos MJ. Overview of ribonucleotide reductase inhibitors: An appealing target in anti-tumour therapy. Curr Med Chem. 2005;12:1283–1294. doi: 10.2174/0929867054020981. [DOI] [PubMed] [Google Scholar]
- 6.Cooperman BS. Oligopeptide inhibition of class I ribonucleotide reductases. Biopolymers. 2003;71:117–131. doi: 10.1002/bip.10397. [DOI] [PubMed] [Google Scholar]
- 7.Sen G, Mukhopadhyay S, Ray M, Biswas T. Quercetin interferes with iron metabolism in Leishmania donovani and targets ribonucleotide reductase to exert leishmanicidal activity. J Antimicrob Chemother. 2008;61:1066–1075. doi: 10.1093/jac/dkn053. [DOI] [PubMed] [Google Scholar]
- 8.Nurbo J, et al. Design, synthesis and evaluation of peptide inhibitors of Mycobacterium tuberculosis ribonucleotide reductase. J Pept Sci. 2007;13:822–832. doi: 10.1002/psc.906. [DOI] [PubMed] [Google Scholar]
- 9.Dutia BM, Frame MC, Subak-Sharpe JH, Clark WN, Marsden HS. Specific inhibition of herpesvirus ribonucleotide reductase by synthetic peptides. Nature. 1986;321:439–441. doi: 10.1038/321439a0. [DOI] [PubMed] [Google Scholar]
- 10.Cohen EA, Gaudreau P, Brazeau P, Langelier Y. Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature. 1986;321:441–443. doi: 10.1038/321441a0. [DOI] [PubMed] [Google Scholar]
- 11.Wnuk SF, Robins MJ. Ribonucleotide reductase inhibitors as anti-herpes agents. Antiviral Res. 2006;71:122–126. doi: 10.1016/j.antiviral.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Thelander L, Sjöberg BR, Eriksson S. Ribonucleoside diphosphate reductase (Escherichia coli) Methods Enzymol. 1978;51:227–237. doi: 10.1016/s0076-6879(78)51032-x. [DOI] [PubMed] [Google Scholar]
- 13.Yeh YC. A simple and sensitive assay procedure for ribonucleotide reductase system. Anal Biochem. 1978;86:175–183. doi: 10.1016/0003-2697(78)90332-9. [DOI] [PubMed] [Google Scholar]
- 14.Shewach DS. Quantitation of deoxyribonucleoside 5′-triphosphates by a sequential boronate and anion-exchange high-pressure liquid chromatographic procedure. Anal Biochem. 1992;206:178–182. doi: 10.1016/s0003-2697(05)80030-2. [DOI] [PubMed] [Google Scholar]
- 15.Hofer A, Ekanem JT, Thelander L. Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J Biol Chem. 1998;273:34098–34104. doi: 10.1074/jbc.273.51.34098. [DOI] [PubMed] [Google Scholar]
- 16.Tholander F, Sjöberg B-M. Method for determining the amount of dNTP. 2011. International Application No.: PCT/SE2011/050315.
- 17.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 18.Sheridan C. First cystic fibrosis drug advances towards approval. Nat Biotechnol. 2011;29:465–466. doi: 10.1038/nbt0611-465. [DOI] [PubMed] [Google Scholar]
- 19.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pötsch S, Sahlin M, Langelier Y, Gräslund A, Lassmann G. Reduction of the tyrosyl radical and the iron center in protein R2 of ribonucleotide reductase from mouse, herpes simplex virus and E. coli by p-alkoxyphenols. FEBS Lett. 1995;374:95–99. doi: 10.1016/0014-5793(95)01082-p. [DOI] [PubMed] [Google Scholar]
- 21.Fontecave M. Ribonucleotide reductases and radical reactions. Cell Mol Life Sci. 1998;54:684–695. doi: 10.1007/s000180050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corsino P, et al. A novel class of cyclin-dependent kinase inhibitors identified by molecular docking act through a unique mechanism. J Biol Chem. 2009;284:29945–29955. doi: 10.1074/jbc.M109.055251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahlin M, Gräslund A, Petersson L, Ehrenberg A, Sjöberg B-M. Reduced forms of the iron-containing small subunit of ribonucleotide reductase from Escherichia coli. Biochemistry. 1989;28:2618–2625. doi: 10.1021/bi00432a039. [DOI] [PubMed] [Google Scholar]
- 24.Twitchett MB, Dobbing AM, Sykes AG. New mechanistic insights into the reactivity of the R2 protein of E. coli ribonucleotide reductase (RNR) J Inorg Biochem. 2000;79:59–65. doi: 10.1016/s0162-0134(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 25.Audi SH, et al. Toluidine blue O and methylene blue as endothelial redox probes in the intact lung. Am J Physiol Heart Circ Physiol. 2000;278:H137–H150. doi: 10.1152/ajpheart.2000.278.1.H137. [DOI] [PubMed] [Google Scholar]
- 26.Moura JC, Cordeiro N. 3,7-bis(dialkylamino)phenothiazin-5-ium derivatives: Biomedical applications and biological activity. Curr Drug Targets. 2003;4:133–141. doi: 10.2174/1389450033346902. [DOI] [PubMed] [Google Scholar]
- 27.Wondrak GT. NQO1-activated phenothiazinium redox cyclers for the targeted bioreductive induction of cancer cell apoptosis. Free Radic Biol Med. 2007;43:178–190. doi: 10.1016/j.freeradbiomed.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inouye Y, et al. Biological properties of streptonigrin derivatives. I. Antimicrobial and cytocidal activities. J Antibiot (Tokyo) 1985;38:1429–1432. doi: 10.7164/antibiotics.38.1429. [DOI] [PubMed] [Google Scholar]
- 29.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: Evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolzán AD, Bianchi MS. Genotoxicity of streptonigrin: a review. Mutat Res. 2001;488:25–37. doi: 10.1016/s1383-5742(00)00062-4. [DOI] [PubMed] [Google Scholar]
- 31.Ming LJ. Structure and function of “metalloantibiotics.”. Med Res Rev. 2003;23:697–762. doi: 10.1002/med.10052. [DOI] [PubMed] [Google Scholar]
- 32.Davies BW, et al. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol Cell. 2009;36:845–860. doi: 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torrents E, Poplawski A, Sjöberg B-M. Two proteins mediate class II ribonucleotide reductase activity in Pseudomonas aeruginosa: Expression and transcriptional analysis of the aerobic enzymes. J Biol Chem. 2005;280:16571–16578. doi: 10.1074/jbc.M501322200. [DOI] [PubMed] [Google Scholar]
- 34.Grinberg I, et al. Functional analysis of the Streptomyces coelicolor NrdR ATP-cone domain: Role in nucleotide binding, oligomerization, and DNA interactions. J Bacteriol. 2009;191:1169–1179. doi: 10.1128/JB.01145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrents E, et al. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J Bacteriol. 2007;189:5012–5021. doi: 10.1128/JB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torrents E, Westman M, Sahlin M, Sjöberg B-M. Ribonucleotide reductase modularity: Atypical duplication of the ATP-cone domain in Pseudomonas aeruginosa. J Biol Chem. 2006;281:25287–25296. doi: 10.1074/jbc.M601794200. [DOI] [PubMed] [Google Scholar]
- 37.Comuzzi C, Polese P, Melchior A, Portanova R, Tolazzi M. SOLVERSTAT: A new utility for multipurpose analysis. An application to the investigation of dioxygenated Co(II) complex formation in dimethylsulfoxide solution. Talanta. 2003;59:67–80. doi: 10.1016/s0039-9140(02)00457-5. [DOI] [PubMed] [Google Scholar]
- 38.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Tech Bull Regist Med Technol. 1966;36:49–52. [PubMed] [Google Scholar]
- 39.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 40.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol. 2006;73:15.8.1–15.8.28. doi: 10.1002/0471142727.mb1508s73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.