Abstract
When nerve cells communicate, vesicles from one neuron fuse with the presynaptic membrane releasing chemicals that signal to the next. Similarly, when insulin binds its receptor on adipocytes or muscle, glucose transporter-4 vesicles fuse with the cell membrane, allowing glucose to be imported. These essential processes require the interaction of SNARE proteins on vesicle and cell membranes, as well as the enigmatic protein Munc18 that binds the SNARE protein Syntaxin. Here, we show that in solution the neuronal protein Syntaxin1a interacts with Munc18-1 whether or not the Syntaxin1a N-peptide is present. Conversely, the adipocyte protein Syntaxin4 does not bind its partner Munc18c unless the N-peptide is present. Solution-scattering data for the Munc18-1:Syntaxin1a complex in the absence of the N-peptide indicates that this complex adopts the inhibitory closed binding mode, exemplified by a crystal structure of the complex. However, when the N-peptide is present, the solution-scattering data indicate both Syntaxin1a and Syntaxin4 adopt extended conformations in complexes with their respective Munc18 partners. The low-resolution solution structure of the open Munc18:Syntaxin binding mode was modeled using data from cross-linking/mass spectrometry, small-angle X-ray scattering, and small-angle neutron scattering with contrast variation, indicating significant differences in Munc18:Syntaxin interactions compared with the closed binding mode. Overall, our results indicate that the neuronal Munc18-1:Syntaxin1a proteins can adopt two alternate and functionally distinct binding modes, closed and open, depending on the presence of the N-peptide, whereas Munc18c:Syntaxin4 adopts only the open binding mode.
Keywords: membrane fusion, protein interactions, small-angle neutron scattering, small-angle X-ray scattering
Membrane trafficking is an essential and highly regulated process whereby cargo-carrying vesicles are directed to dock and fuse with specific cell membranes. The process requires soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) proteins on vesicle and target membranes. These interact to form high-affinity SNARE complexes required for docking and membrane fusion (1, 2). Structurally, the SNARE complex is a parallel four-helix bundle comprising helices contributed from several partner SNAREs (3). This helical bundle promotes vesicle fusion by bringing the vesicle and plasma membranes into close proximity and by providing the energy required for membrane fusion (4, 5).
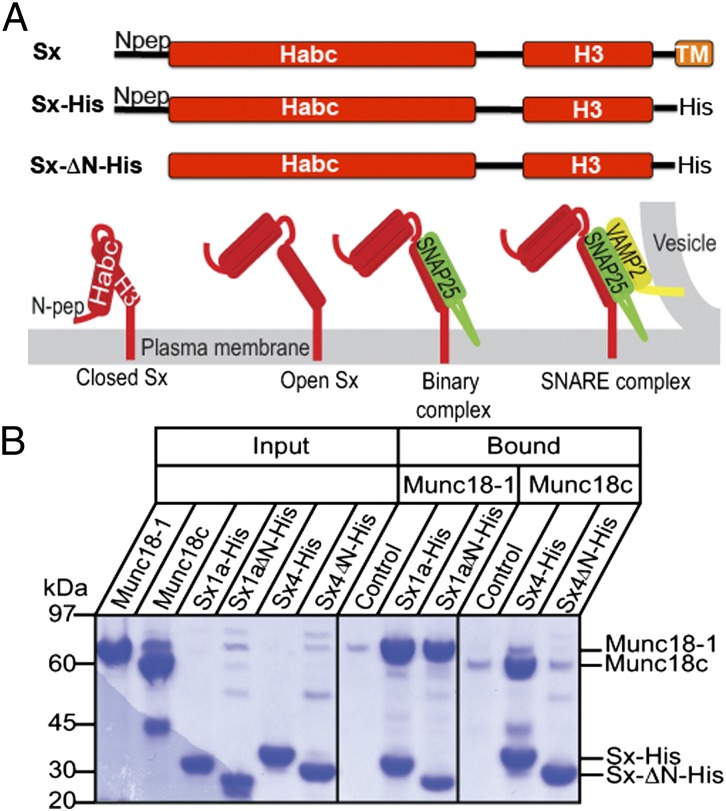
In the neuronal synapse, neurotransmitters are released from nerve cells by SNARE-mediated fusion of vesicles with the plasma membrane in response to action potentials. In this process, one of four SNARE helices is contributed by Syntaxin (Sx)1a, a protein localized at the plasma membrane by a transmembrane helix. In addition to its membrane-anchored SNARE helix (H3), Sx1a has an Habc domain that can bind to the H3 helix (6) forming a closed Sx1a conformation (Fig. 1A). This closed conformation inactivates Sx1a by preventing H3 interacting with SNARE partners, SNAP25 on the plasma membrane and vesicle associated membrane protein 2 (VAMP2, also known as synaptobrevin) on the vesicle membrane. Conversely, when the intramolecular Habc interaction is removed, Sx1a can adopt an open conformation and H3 is then free to participate in SNARE complex assembly by forming a SNARE binary complex with SNAP25 and a SNARE ternary complex with VAMP2, leading to membrane fusion (Fig. 1). Proteins that differentially promote the closed or open Sx1a conformation could thus negatively or positively regulate synaptic SNARE complex formation, membrane fusion, and neurotransmission. In the homologous fusion system in adipocytes and muscle, glucose-transporter (GLUT)4-containing vesicles are trafficked to the plasma membrane to facilitate glucose uptake in response to insulin signaling. The equivalent Sx in this system is Sx4.
Fig. 1.
Sx constructs and interactions. We used Sx1a and Sx4 with the transmembrane anchor replaced with a polyhistidine tag (Sx-His) and with the N-peptide removed (SxΔN-His). (A) Schematic showing different forms of membrane-anchored Sx: closed Sx, open Sx, binary complex with SNARE partner SNAP (green), and ternary complex with SNARE partners [SNAP25 in green; VAMP2 (or synaptobrevin) in yellow]. The four-helix bundle of the SNARE ternary complex is required for membrane fusion, and open Sx is thought to be required to form SNARE complex. (B) Sx1a, Sx1aΔN, Sx4, and Sx4ΔN were immobilized on metal-affinity resin via the C-terminal polyhistidine tags and incubated with detagged Munc18-1 or Munc18c. The gels at right and center show the proteins that were pulled down (bound) on equivalent amounts of resin after extensive washing. Resin-bound Sx1a and Sx1aΔN are both able to pull down Munc18-1, whereas Sx4 but not Sx4ΔN pulls down Munc18c. (Left) Proteins used in the experiment. Control shows the negative control interaction of Munc18 with resin. These data are representative of three replicates.
Sec1/Munc18 (SM) proteins are also highly conserved and probably regulate all eukaryotic vesicle transport processes, although a variety of interactions, binding modes, and functions have been described. Thus, SM proteins are thought to play a major inhibitory role by keeping Sxs in the closed conformation, as exemplified by a crystal structure of the neuronal Munc18-1 in complex with Sx1a (7). A closed binding mode is also observed in the crystal structure of a primordial Munc18:Sx complex suggesting that closed binding, and inhibition of Sx, is important in the pathway leading to SNARE assembly (8).
SM proteins have also been shown to interact with SNARE binary and SNARE ternary complexes (9–11), and these interactions require an open Sx binding mode as well as the presence of an N-peptide, the 10–30 residues at the very N terminus of Sx (11–14). The two binding modes (closed and N-peptide) are thought to be associated with different aspects of SM protein function. For example, in the synaptic fusion system, Munc18-1 may act as a chaperone by binding closed Sx1a during trafficking to the plasma membrane (14–17). Munc18-1 can then bind and stimulate SNARE complex assembly at the plasma membrane, to facilitate membrane fusion (17, 18). Similarly, in Caenorhabditis elegans, the closed SM:Sx binding mode is required for chaperoning the Sx protein UNC64 to the plasma membrane whereas the N-peptide binding mode is proposed to be required for membrane fusion in vivo (19, 20). Dual chaperone/activation roles for the SM protein Vps45p have also been shown in yeast (21). In vitro, fusion assays have shown that Munc18-1 can either inhibit vesicle fusion or stimulate vesicle fusion, with the contribution of the N-peptide varying depending on the experimental setup (18, 22–24). Rathore et al. showed that the Sx N-peptide recruits Munc18-1 for assembly of the SNARE:SM complex and does not need to be attached to Sx to exert this effect (25). Finally, the central cavity of the SM protein has been linked to binding of closed Sx1a and to binding of the core helical bundle of SNARE complexes (24, 26, 27).
Thus, SM proteins have been shown to interact with Sx in a closed conformation and to interact with binary (Sx1a:SNAP25 in the case of Munc18-1) and ternary SNARE complexes (Sx1a:SNAP25:VAMP2), the last two requiring an open Sx conformation. The only two crystal structures of SM proteins in complex with a full-length cytoplasmic Sx (lacking the transmembrane domain) reveal a closed binding mode (7, 8, 28) and NMR data also suggest a closed binding mode (29). Furthermore, there are no experimentally determined structures of SM proteins interacting with SNARE complexes. However, intriguing SM protein conformational changes were revealed in the crystal structures of Munc18-1 complexed with a noncognate N-peptide (30) and Munc18c (the adipocyte SM protein) with a cognate N-peptide (31). These structural changes were speculated to show how SM proteins might transit from binding closed Sx to binding open Sx (30).
Using protein pull-down experiments, chemical cross-linking, isothermal titration calorimetry, and small-angle scattering, we investigated the binding of two SM:Sx pairs: neuronal Munc18-1:Sx1a and adipocyte Munc18c:Sx4, sharing ∼40–50% identity. We found that in the absence of the N-peptide, Sx1a binds tightly to Munc18-1 in a closed conformation consistent with the published crystal structure. In the presence of the N-peptide, the interaction affinity is similar but a different, more extended Sx1a conformation is bound to Munc18-1. On the other hand, Munc18c did not bind tightly to Sx4 lacking the N-peptide but did bind to Sx4 when the N-peptide was present; the binding mode in solution is similarly extended. We modeled the low-resolution structures of the complexes using scattering and cross-linking data that together suggest an open binding mode.
Results
Differential Role of the Sx N-Peptide.
We used isothermal titration calorimetry (ITC) to evaluate the binding of Munc18 proteins to cognate Sxs with and without the N-peptide. The Munc18-1 results recapitulate those of others (14, 28, 32) showing that Sx1a and Sx1a lacking its N-peptide (Sx1aΔN) bind to Munc18-1 with similar high affinities (Kd, 1.4 and 10 nM, respectively; see Table S1 for full details).
We also investigated the interaction between the homologous proteins from adipocytes, Munc18c and Sx4, that regulate the trafficking of GLUT4 vesicles to the cell surface in response to insulin signaling. We found that the Munc18c:Sx4 interaction (Kd 95 nM; Table S1) is weaker than Munc18-1:Sx1a. Strikingly, we were unable to detect any interaction between Munc18c and Sx4ΔN by ITC, even with very high concentrations of Sx4ΔN (Fig. S1). Circular dichroism spectra of Sx4ΔN and Sx4 are equivalent, showing that they are both helical (Fig. S2A), and Sx4ΔN has a sigmoidal melting curve consistent with temperature denaturation cooperativity indicating it is likely to be correctly folded (Fig. S2B). We also used pull-down experiments to verify the result that Munc18-1 interacts tightly with Sx1a and Sx1aΔN, whereas Munc18c interacts with Sx4 but not Sx4ΔN (Fig. 1). The two homologous systems thus differ in their ability to interact with Sx lacking an N-peptide: Munc18-1 binds Sx1aΔN with an affinity almost equal to that of its interaction with Sx1a, whereas Munc18c interacts tightly with Sx4 only when its N-peptide is present.
Sx1a and Sx1aΔN Have Different Binding Modes for Munc18-1.
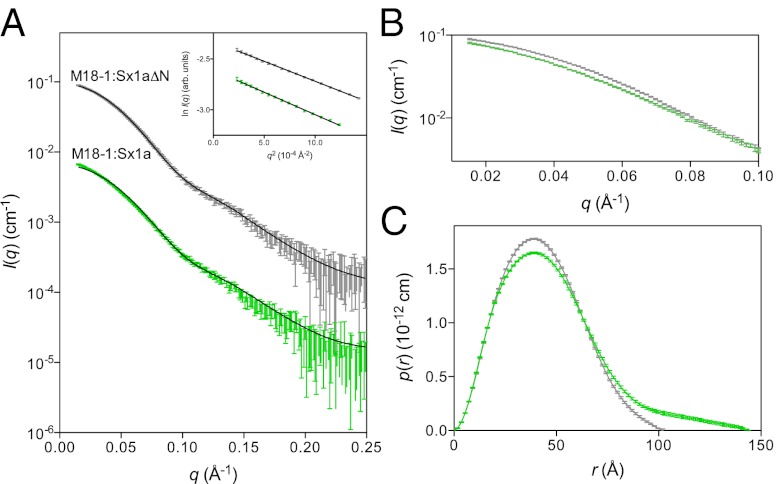
We next used small-angle X-ray scattering (SAXS) to investigate whether the low-resolution solution structures of Munc18-1:Sx1a and Munc18-1:Sx1aΔN complexes were similar. However, comparison of the scattering data and pair-distance distribution functions, p(r), indicated significant differences (Fig. 2). Specifically, the p(r) profiles (Fig. 2C) have a large difference in the maximum linear dimension (Dmax) of the two complexes: the maximum dimension of the Munc18-1:Sx1aΔN complex is ∼100 Å, whereas the Munc18-1:Sx1a complex is 50% larger, ∼150 Å. This difference indicates that the presence of the Sx1a N-peptide modulates the way Sx1a binds to Munc18-1.
Fig. 2.
Munc18-1:Sx1a and Munc18-1:Sx1aΔN complexes differ. (A) SAXS data for Munc18-1:Sx1a (green) and Munc18-1:Sx1aΔN (gray). Inset shows the Guinier regions are linear. The calculated scattering profile from the closed Munc18-1:Sx1a crystal structure (solid line) is overlaid on the Munc18-1:Sx1aΔN SAXS data, showing an excellent correspondence (χ2 = 0.6). By comparison, the Munc18-1:Sx1a scattering data fit less well to the crystal structure profile (χ2 = 3.5). Data are shown on an absolute scale, where the Munc18-1:Sx1a scattering data have been offset by a factor of 10−1 for clarity. Error bars represent propagated counting statistics. (B) Comparison of the low-angle portion of the scattering data for Munc18-1:Sx1aΔN and Munc18-1:Sx1a indicates a significant deviation, indicating differences between their structures. Data were normalized by protein concentration for this comparison. (C) Pair-distance distribution function, p(r), for Munc18-1:Sx1aΔN and Munc18-1:Sx1a derived from the scattering data using GNOM (46) indicates that Dmax, the maximum dimension of the complex, is significantly larger for Munc18-1:Sx1a than for Munc18-1:Sx1aΔN.
Sx1aΔN Binds to Munc18-1 in a Closed Conformation.
The Munc18-1:Sx1a crystal structure defines a closed Sx1a conformation bound to Munc18-1 (7). We compared the calculated scattering profiles from the Munc18-1:Sx1a closed crystal structure with solution scattering data from Munc18-1:Sx1aΔN and Munc18-1:Sx1a. As shown in Fig. 2A, the predicted scattering profile of the Munc18-1:Sx1a crystal structure matches well to the SAXS data for the complex lacking the N-peptide interaction (Munc18-1:Sx1aΔN), whereas the match to the SAXS data where the N-peptide is present is much poorer (Munc18-1:Sx1a data). Furthermore, the radius of gyration (Rg) and Dmax values calculated from the crystal structure match those from Munc18-1:Sx1aΔN scattering data but not those from Munc18-1:Sx1a data (Table 1). Dynamic light scattering confirmed the different Rg values for the two complexes (Fig. S2C). These data suggest that when the Sx1a N-peptide interaction is absent the closed binding mode of the Munc18-1:Sx1a crystal structure can be adopted in solution. The presence of a native N-peptide interaction may thus change the way Sx1a recognizes Munc18-1 in solution.
Table 1.
Structural parameters for Munc18:Sx complexes
| Protein or complex | Rg (Å) | Dmax (Å) |
| Munc18:Sx complex | ||
| Munc18-1:Sx1a (X-ray scattering) | 37.8 ± 0.3 | 145 |
| Munc18-1:Sx1aΔN (X-ray scattering) | 33.0 ± 0.3 | 100 |
| Munc18-1:Sx1a (predicted X-ray scattering, crystal structure)* | 32.9 | 110 |
| Munc18c:Sx4 (X-ray scattering data) | 38.4 ± 0.3 | 145 |
| Munc18 within the complex | ||
| Munc18-1 (100% D2O data of Munc18-1:Sx1a) | 23.7 ± 0.3 | 70 |
| Munc18c (100% D2O data of Munc18c:Sx4) | 23.4 ± 0.3 | 70 |
| Munc18-1 (crystal structure, 100% D2O)* | 22.5 | 80 |
| Sx within the complex | ||
| Sx1a (40% D2O data of Munc18-1:Sx1a) | 39.1 ± 0.6 | 145 |
| Sx4 (40% D2O data of Munc18c:Sx4) | 39.5 ± 0.5 | 145 |
| Sx1a (crystal structure, 40% D2O)* | 32.0 | 110 |
Rg is the radius of gyration of the complex or molecule, Dmax is the maximum dimension of the complex or molecule. The error in Dmax is approximately ± 10 Å. Values in bold show that Munc18-1:Sx1aΔN in solution and Munc18-1:Sx1a(closed) crystal structure are more compact than Munc18-1:Sx1a and Munc18c:Sx4 in solution and that this is attributable to a change in conformation of Sx.
*Values calculated from the crystal structure (see Materials and Methods).
Sx1a and Sx4 Are Open When Bound to Cognate Munc18s.
To investigate conformational changes suggested by SAXS, we used small-angle neutron scattering (SANS) with contrast variation (33). Sx proteins were labeled with deuterium (DSx) for measurement of Munc18:DSx complexes at two contrast points: ∼40% D2O, where scattering is dominated by DSx; and ∼100% D2O, where scattering is dominated by Munc18. This analysis allows evaluation of the bound conformations of the individual components, Munc18 and Sx, within the complex. We investigated the two homologous systems, Munc18-1 complexed with Sx1a and Munc18c complexed with Sx4. In both cases, the Sx N-peptide was present.
SAXS and SANS data for the Munc18-1:Sx1a and Munc18c:Sx4 complexes indicate they share many common features (Table 1). First, the Rg and Dmax values are very similar for both and different to values calculated from the closed Munc18-1:Sx1a crystal structure (Table 1; also see Tables S2 and S3). Second, the 100% D2O SANS data, where unlabeled Munc18 dominates scattering (see Ref. 34) for more information), indicate that Munc18-1 and Munc18c adopt similar overall shapes (Dmax ∼70 Å; Rg 23–24 Å). Finally, and most importantly, the 40% D2O SANS data, where Sx dominates scattering, suggest that the increased size of the complexes compared with the closed Munc18-1:Sx1a crystal structure is primarily due to a conformational change in Sx. Thus, the maximum dimension of Sx in both complexes is 145 Å, compared with 110 Å in the closed crystal structure. In addition, the p(r) distribution of the 40% D2O contrast point (representing the bound Sx conformation) has two peaks, indicative of two domains with centers of mass separated by 40Å, whereas the p(r) distribution for closed Sx1a models have a single peak consistent with one domain (Fig. S3A). These data suggest that in both Munc18-1 and Munc18c complexes, Sx can adopt an extended conformation with a much larger dimension than that of closed Sx, when the N-peptide interaction is present.
To help identify the spatial arrangement of specific residues in the complex, we cross-linked Munc18c:Sx4 and Munc18-1:Sx1a and analyzed digested peptides by mass spectrometry. The observed cross-link distances were inconsistent with the closed crystal structure binding mode (Table S4 and Fig. S3 B and C), supporting the conclusion that Sx binding to Munc18 differs from the closed binding mode observed in the Munc18-1:Sx1a crystal structure.
Modeling Indicates an Open Munc18:Sx Binding Mode.
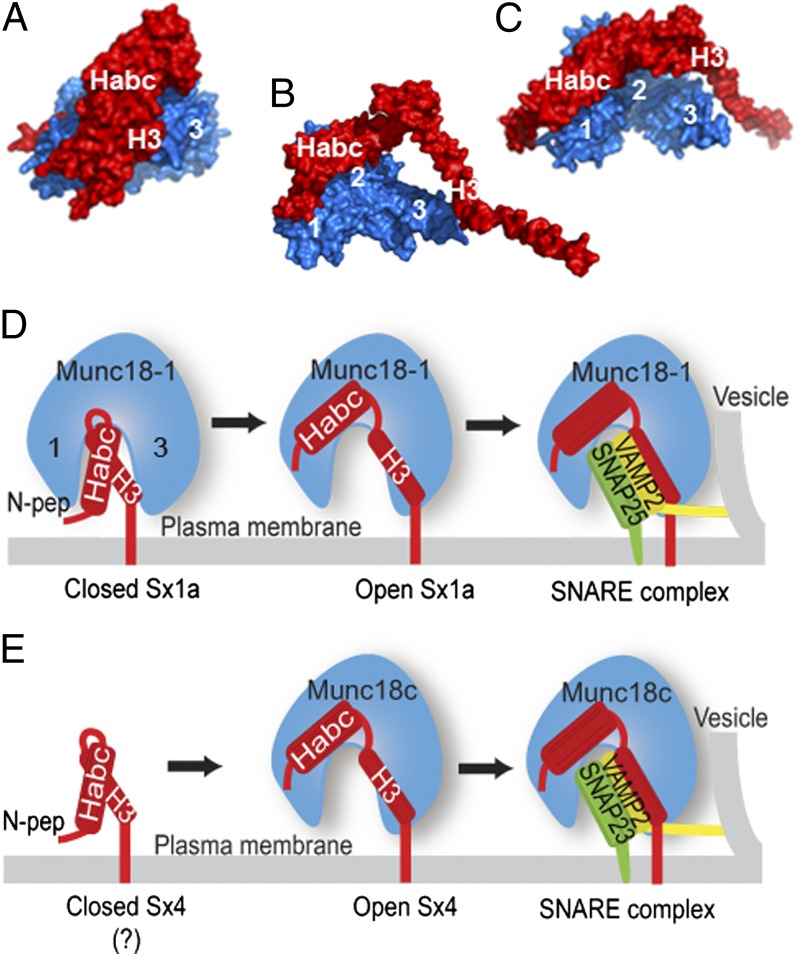
Rigid-body modeling of the low resolution solution structures of the Munc18:Sx complexes was performed by simultaneous refinement against SAXS and SANS data (35, 36) and cross-linking restraints. The refined models represent excellent fits to these data (Fig. 3 and Fig. S4), and indicate an open Sx conformation (Fig. 4). In both Munc18-1:Sx1a and Munc18c-Sx4 models, the Habc domain of the Sxs is located on a concave surface between domains 1 and 2 of Munc18 that is created by rotation of domain 1. This is in contrast to the binding mode of Habc in the crystal structure, where it is located in the Munc18 arch-shaped cavity. The SNARE H3 helix extends away from Habc across the central Munc18 cavity. However, the modeled H3 position varies in the two complexes and is not definitive in either model because H3 appears to sample a range of conformations (Fig. S5). This conformational heterogeneity presents a challenge for modeling. Thus, we were unable to obtain a model that satisfied the scattering data and all of the cross-links between the Sx H3 helix and Munc18c, whereas refinement that excluded the H3:Munc18c cross-links yielded a model in excellent agreement with the scattering data. This result implies that the H3 helix samples several open conformations, with the presented model in Fig. 4 a likely representative of the dominant conformation. Thus, many models can be generated that fit the data, and there may be other ways to fit the data that we have not considered, but none of the models generated during refinement adopted the closed Sx binding mode. Thus, the neutron scattering and cross-linking data clearly indicate that the majority of Sx molecules are bound in an open conformation to Munc18 and the position of the Habc domain appears to be well defined by the scattering and cross-linking data.
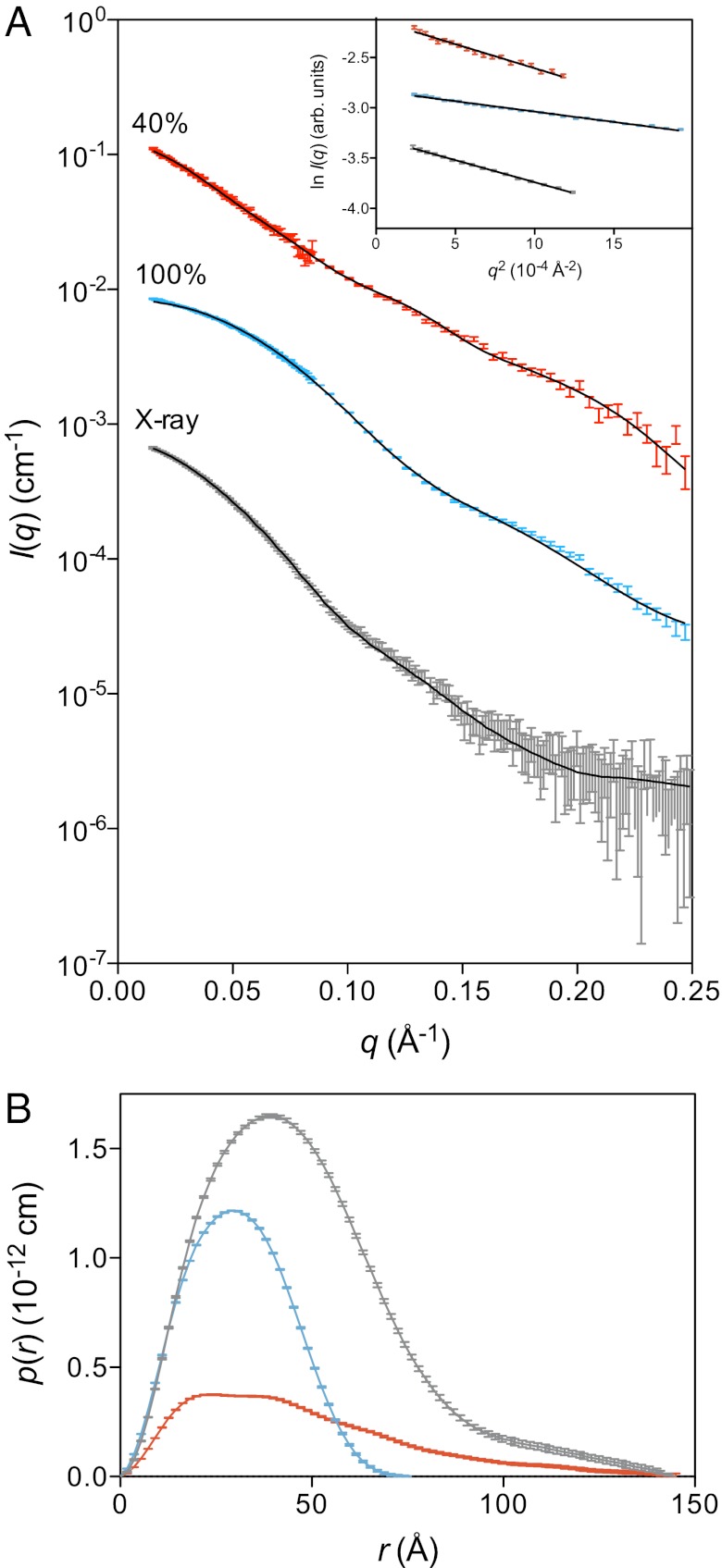
Fig. 3.
Sx1a is extended when bound to Munc18-1. (A) SAXS data (gray) and neutron contrast variation data at 40% D2O (red) and 100% D2O (blue) for Munc18-1:Sx1a. Data are of high quality with linear Guinier regions (Inset) and yielding estimated molecular masses consistent with a 1:1 complex (Table S3). The calculated scattering profiles for the optimized model of the complex (solid lines) are overlaid on the data, with excellent visual correspondence. The χ2 values are: 40%, 1.6; 100%, 4.5; X-ray, 0.4. The high value for the 100% D2O is primarily because of small deviations of the fit to the data at low-q and are probably attributable to the misrepresentation of unstructured regions of Munc18, which dominates the signal in the 100% data. Data are shown on an absolute scale, where the 100% D2O and X-ray scattering data have been off-set by factors of 10−1 and 10−2 for clarity. Error bars represent propagated counting statistics. (B) Pair-distance distribution function, p(r), derived from the scattering data using GNOM (46). This indicates that the increased maximum dimension of the Munc18-1:Sx1a complex is attributable to Sx1a (red curve), indicating that in the Munc18-1:Sx1a complex, Sx1a adopts an extended conformation.
Fig. 4.
An open binding mode of Munc18:Sx complexes. (A) Crystal structure of closed Sx1a (red) bound to Munc18-1. (B) Model of Munc18-1 (blue) in complex with Sx1a (red) complex, refined against solution scattering data and distance restraints, indicating that Sx1a adopts an open conformation when bound to Munc18-1. (C) Model of Munc18c (blue) in complex with Sx4 (red), refined against solution scattering data and distance restraints, indicating that Sx4 adopts an open conformation when bound to Munc18c. For both B and C, the position of the H3 helix is not definitive. (D) Schematic showing potential Munc18-1:Sx1a interactions. Munc18-1 interacts with closed Sx1a when the N-peptide is not engaged. Open Sx1a binding to Munc18-1 requires the Sx1a N-peptide. Munc18-1 binding to open Sx1a may precede SNARE complex formation with partner SNAREs SNAP25 (green) and VAMP2 (yellow). (E) It is not clear whether Sx4 exists in a closed conformation. Munc18c binds to open Sx4, and this requires an N-peptide interaction. Munc18c binding to open Sx4 may precede SNARE complex formation with SNAP23 (green) and VAMP2 (yellow).
Overall, our data suggest that the closed binding mode exemplified by the crystal structure of Munc18-1 and Sx1a is adopted in solution in the absence of the N-peptide interaction. We see no evidence for the binding of Sx4ΔN to Munc18c or for the formation of a closed binding mode for the Munc18c:Sx4 complex, indicating either that these interactions do not occur in Munc18c:Sx4 or that they are very weak. In contrast, when the Sx N-peptide is present, Sx appears to adopt an extended conformation in Munc18-1:Sx1a and Munc18c:Sx4 complexes. These low-resolution solution structures of the complexes indicate a possible binding mode of open Sx to Munc18 and suggests this is driven by the presence of an N-peptide interaction.
Discussion
The data presented here suggest that in solution under the conditions we used, Munc18 proteins can recognize an open conformation of Sx, in the absence of SNARE partners SNAP and VAMP. The Sx N-peptide influences the formation and conformation of this open Munc18:Sx complex. For the neuronal Munc18a:Sx1 system, the binding preference for Sx1a with or without N-peptide is minimal (Table S1), so that subtle changes in conditions may favor closed binding even in the presence of a native N-peptide.
The solution structures derived from refinement against SAXS, SANS (40%), SANS (100%), and cross-linking data for Munc18-1:Sx1a are in striking contradiction to the closed conformation observed in crystal structures and NMR data of this complex (29). If, as our data suggest, open and closed Sx1a can bind with almost equal affinity to Munc18-1, it is possible that the closed form of the complex is preferentially selected by crystallization. This is because the flexible SNARE helix of the open form is not conducive to crystal formation. Crystallization can select more compact shapes when extended conformations of the same protein or complex are present in solution (see, for example, Refs. 37 and 38). However, it is difficult to explain why a closed rather than an open conformation was observed in NMR studies of the Munc18-1:Sx1a complex (29). Perhaps the conditions or constructs we used favor the open conformation of Munc18-1:Sx1a and vice versa for the NMR data. Clearly, more work is required to resolve these differences.
The relatively minor energetic difference between Sx1a and Sx1aΔN suggests that a small energetic input could convert Munc18-1:Sx1a from a closed fusion-incompetent to an open fusion-competent binding mode. Thus, in neurons, membrane fusion may be a delicate balance between these two binding modes, tipped one way or the other by accessory factors. This could also impact on the observed role of Munc18-1 (inhibitory or stimulatory) in in vitro assays, using different experimental setups. For example, Söllner and colleagues (22) found that Munc18-1 preincubated with Sx1a:SNAP25 liposomes inhibits fusion with subsequently added VAMP2 liposomes. However, when SNAP25 and VAMP2 liposomes were added simultaneously to preformed Munc18-1 complex with Sx1a liposomes, this resulted in a marked stimulation of fusion that was N-peptide dependent (22). Previous work (28, 39) has shown that Munc18-1 slows SNARE complex formation from mixtures of soluble forms of SNARE proteins, when analyzed kinetically by fluorescence polarization, FRET and NMR. Our present data point to the possibility that two Sx1a binding modes of almost equal affinity might contribute to different functional outcomes under different experimental situations. Again, more work is needed to resolve these differences.
For Sx4, the role of the N-peptide is striking: Sx4 lacking the N-peptide has no measurable affinity for Munc18c under the conditions we used. The N-peptide thus appears to be essential for binding of Sx4 to Munc18c, although other parts of Sx4 must interact with Munc18c because the binding affinity of the N-peptide alone is lower than that of the full-length protein (30).
The finding that Sx1a N-peptide binding to Munc18-1 may favor a closed-to-open conformational change suggests that the open interaction mode might precede SNARE complex formation (Fig. 4). The best-fit structural models of open Sx1a and Sx4 place Habc outside the arch-shaped Munc18 cavity, so that the cavity could then accommodate the SNARE four-helix bundle (Fig. 4). This same cavity has been implicated in the binding of SNARE complexes (24, 26, 27). The Munc18 binding site of the Sx H3 helix is not definitive in our model; indeed, it is likely to be very flexible. In the context of the native membrane-anchored protein, where H3 is tethered at its C terminus by membrane insertion, we expect this flexibility to be considerably reduced.
Our findings suggest that SM protein regulatory roles may vary considerably, even among closely related proteins. Munc18-1 and Munc18c share ∼50% identity yet Munc18-1:Sx1a appears to form two functionally distinct high affinity interactions, whereas Munc18c:Sx4 has just one binding mode. This could help explain the apparently conflicting in vivo data on the role of Munc18 and SM proteins more generally (40): thus, Munc18-1 may have negative and positive regulatory roles, whereas others have positive regulatory roles only. Disturbing the interaction between Munc18-1 and the Sx1a N-peptide could trigger switching between open and closed bound conformations. In contrast, blocking Sx4 N-peptide binding to Munc18c would prevent the formation of the Munc18c:Sx4 complex altogether. Finally, these data highlight the importance of dynamic protein interactions in complex formation and the power of complementary methods for probing these dynamic states.
Materials and Methods
SAXS.
Munc18:Sx complexes in 25 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM Tris (2-carboxyethyl) phosphine (TCEP) were concentrated to between 2–6 mg/mL. A serial dilution of the concentrated protein sample was made with the flow-through to yield samples with protein concentrations of 1.4, 2.8, and 5.6 mg/mL (Munc18-1:Sx1aΔN); 1.1, 2.2, and 3.3 mg/mL (Munc18-1:Sx1aΔN); and 0.7, 1.4, and 2.8 mg/mL (Munc18c:Sx4). The flow-through also served as the solvent blank. All samples and buffers were centrifuged at 10,000 × g for 10 min to remove large particles. The concentration of protein complexes was determined by A280 measurements using a Nanodrop (Thermo Scientific), and extinction coefficient was determined using ProtParam (41).
Data were collected on the SAXS/WAXS beamline at the Australian Synchrotron by flowing the sample (50 μL) past the beam in 1.5 mm quartz capillaries (Hampton Research) at room temperature measuring 10–15 frames with a 1-s exposure time. The sample to detector distance was 1630 mm and wavelength was λ = 1.000 Å, giving a q range spanning 0.012–0.620 Å−1. Data reduction was carried out using SAXS15ID software (42), averaging 10 frames and correcting for solvent scattering, sample transmission, detector sensitivity, and background radiation. Data were placed on an absolute scale, by normalization against a water standard.
Data quality was assessed by the concentration dependence of the scattering data, linearity of the Guinier region of the data, and estimated molecular mass of the protein complex. Data were measured for all protein complexes between 1–6 mg/mL. At all concentrations, the Guinier plot (lnI(q) vs. q2) was linear and yielded I(0) values normalized by concentration that showed no systematic trend. A systematic trend of normalized I(0) is indicative of either repulsive interparticle interactions (decrease with increasing concentration) or attractive interparticle interactions (increase with increasing concentration). Estimated molecular masses were determined for each protein complex (43), where the program MULCh (44) was used to calculate the partial specific volume (v) and contrast (Δρ) at each contrast point. The estimated molecular masses of each complex are well within 5% of the expected mass of the complex. Taken together, the protein solutions are homogeneous and free of significant interparticle interactions, and the data are of high quality.
Neutron Scattering.
Purified Munc18c:DSx4 and Munc18-1:DSx1a protein complexes were concentrated to 3–6 mg/mL and exchanged into 25 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM TCEP containing either 40% or 100% (vol/vol) D2O. A sample of the dialysate served as the solvent blank in each case. All samples and buffers were centrifuged at 10,000 × g for 10 min to remove large particles from the solution. The concentration of all protein complexes was determined by A280 measurements using a Nanodrop (Thermo Scientific) or a Cary IE UV/visible spectrophotometer (Varian).
Neutron contrast variation data were collected on D22 at the Institut Laue Langevin. A neutron wavelength of 6 Å was used at sample to detector distances (2.0 m and 8.0 m for Munc18-1:Sx1a and 2.0 m and 5.6 m for Munc18c:Sx4) covering a q range of ∼0.01–0.40 Å−1. All samples were kept at 6 °C throughout data collection. Data for the 100% (vol/vol) D2O samples and buffers were collected for 45 min at both detector distances, and data for the 40% (vol/vol) D2O sample and buffer were collected for 60 min at both detector distances. Samples and buffers were contained in rectangular quartz cuvettes with a 1-mm path length (Hellma; 100-1-40-QS). The dimension of the aperture at the cell was 10 × 7 mm.
Data reduction was performed using GRASansP (45) with corrections made for solvent scattering, sample transmission, detector sensitivity, and background radiation. Data were placed on an absolute scale, by normalization against the direct beam, and data from two different detector positions were merged, yielding scattering profiles at each contrast point. Incoherent scattering was adjusted by subtracting a constant from the merged data, so that the p(r) calculated from the scattering data using GNOM (46) yielded p(0) = 0 [where p(0) was unrestrained]. All manipulations of the scattering data were performed using Primus (47). For all data, the Guinier region was linear, and the estimated molecular masses of each complex were well within 5% of the expected mass of the complex, indicating the data are of high quality.
Other.
Methods for isothermal titration calorimetry, deuteration, pull-downs, CD spectroscopy, cross-linking, modeling, and scattering profile calculation from crystal structures are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jill Trewhella, Kevin Jack, and Nathan Cowieson for help with SAXS and Alun Jones for mass spectrometric measurements. This work was supported by National Health and Medical Research Council (NHMRC) Program 535921. A.E.W. is an NHMRC Peter Doherty Fellow (569864); B.M.C. is an Australian Research Council (ARC) Future Fellow (FT100100027), D.E.J. is an NHMRC Senior Principal Research Fellow (1019680), and J.L.M. is an ARC Australian Laureate Fellow (FL0992138) and honorary NHMRC Fellow (455829). Protein deuteration at the National Deuteration Facility was partly funded by the National Collaborative Research Infrastructure Strategy of the Australian Government. Recombinant expression of Munc18c was performed at the University of Queensland Protein Expression Facility. We also acknowledge financial support from the Access to Major Research Facilities Program of the Australian Government (08/09-N-47) and access to the SAXS/WAXS beam-line at the Australian Synchrotron.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116975109/-/DCSupplemental.
References
- 1.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Li F, et al. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 2007;14:890–896. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]
- 5.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 6.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misura KMS, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 8.Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci USA. 2011;108:15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan R, Dai H, Rizo J. Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- 10.Rodkey TL, Liu S, Barry M, McNew JA. Munc18a scaffolds SNARE assembly to promote membrane fusion. Mol Biol Cell. 2008;19:5422–5434. doi: 10.1091/mbc.E08-05-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khvotchev M, et al. Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J Neurosci. 2007;27:12147–12155. doi: 10.1523/JNEUROSCI.3655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malintan NT, et al. Abrogating Munc18-1-SNARE complex interaction has limited impact on exocytosis in PC12 cells. J Biol Chem. 2009;284:21637–21646. doi: 10.1074/jbc.M109.013508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han L, et al. Rescue of Munc18-1 and -2 double knockdown reveals the essential functions of interaction between Munc18 and closed syntaxin in PC12 cells. Mol Biol Cell. 2009;20:4962–4975. doi: 10.1091/mbc.E09-08-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arunachalam L, et al. Munc18-1 is critical for plasma membrane localization of syntaxin1 but not of SNAP-25 in PC12 cells. Mol Biol Cell. 2008;19:722–734. doi: 10.1091/mbc.E07-07-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rickman C, Medine CN, Bergmann A, Duncan RR. Functionally and spatially distinct modes of munc18-syntaxin 1 interaction. J Biol Chem. 2007;282:12097–12103. doi: 10.1074/jbc.M700227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen JS, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 19.McEwen JM, Kaplan JM. UNC-18 promotes both the anterograde trafficking and synaptic function of syntaxin. Mol Biol Cell. 2008;19:3836–3846. doi: 10.1091/mbc.E08-02-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JR, et al. Binding of UNC-18 to the N-terminus of syntaxin is essential for neurotransmission in Caenorhabditis elegans. Biochem J. 2009;418:73–80. doi: 10.1042/BJ20081956. [DOI] [PubMed] [Google Scholar]
- 21.Bryant NJ, James DE. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J. 2001;20:3380–3388. doi: 10.1093/emboj/20.13.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schollmeier Y, Krause JM, Kreye S, Malsam J, Söllner TH. Resolving the function of distinct Munc18-1/SNARE protein interaction modes in a reconstituted membrane fusion assay. J Biol Chem. 2011;286:30582–30590. doi: 10.1074/jbc.M111.269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diao J, et al. Single-vesicle fusion assay reveals Munc18-1 binding to the SNARE core Is sufficient for stimulating membrane fusion. ACS Chem Neurosci. 2010;1:168–174. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107:22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L, Kümmel D, Coleman J, Melia TJ, Giraudo CG. Dual roles of Munc18-1 rely on distinct binding modes of the central cavity with Stx1A and SNARE complex. Mol Biol Cell. 2011;22:4150–4160. doi: 10.1091/mbc.E11-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han GA, et al. Munc18-1 domain-1 controls vesicle docking and secretion by interacting with syntaxin-1 and chaperoning it to the plasma membrane. Mol Biol Cell. 2011;22:4134–4149. doi: 10.1091/mbc.E11-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen XC, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41:43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu SH, et al. Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci USA. 2011;108:1040–1045. doi: 10.1073/pnas.0914906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu SH, Latham CF, Gee CL, James DE, Martin JL. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2007;104:8773–8778. doi: 10.1073/pnas.0701124104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deák F, et al. Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming. J Cell Biol. 2009;184:751–764. doi: 10.1083/jcb.200812026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitten AE, Trewhella J. Small-angle scattering and neutron contrast variation for studying bio-molecular complexes. Methods Mol Biol. 2009;544:307–323. doi: 10.1007/978-1-59745-483-4_20. [DOI] [PubMed] [Google Scholar]
- 34.Timmins PA, Zaccai G. Low resolution structures of biological complexes studied by neutron scattering. Eur Biophys J. 1988;15:257–268. doi: 10.1007/BF00256476. [DOI] [PubMed] [Google Scholar]
- 35.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petoukhov MV, Svergun DI. Joint use of small-angle X-ray and neutron scattering to study biological macromolecules in solution. Eur Biophys J. 2006;35:567–576. doi: 10.1007/s00249-006-0063-9. [DOI] [PubMed] [Google Scholar]
- 37.Vestergaard B, et al. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Rochel N, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 39.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toonen RF, Verhage M. Vesicle trafficking: Pleasure and pain from SM genes. Trends Cell Biol. 2003;13:177–186. doi: 10.1016/s0962-8924(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press, New York, NY; 2005. pp. 571–607. [Google Scholar]
- 42.Cookson DJ. 2007. SAXS15ID - Software for Acquiring, Processing and Viewing SAXS/WAXS Image Data at ChemMatCARS (Argonne, IL)
- 43.Orthaber D, Bergmann A, Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J Appl Cryst. 2000;33:218–225. [Google Scholar]
- 44.Whitten AE, Cai SZ, Trewhella J. MULCh: Modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J Appl Cryst. 2008;41:222–226. [Google Scholar]
- 45.Dewhurst C. 2003. GRASansP Version 2, User’s Manual (Institut Laue-Langevin, Grenoble, France)
- 46.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 47.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.