Abstract
Although the Drosophila Y chromosome is degenerated, heterochromatic, and contains few genes, increasing evidence suggests that it plays an important role in regulating the expression of numerous autosomal and X-linked genes. Here we use 15 Y chromosomes originating from a single founder 550 generations ago to study the role of the Y chromosome in regulating rRNA gene transcription, position-effect variegation (PEV), and the link among rDNA copy number, global gene expression, and chromatin regulation. Based on patterns of rRNA gene transcription indicated by transcription of the retrotransposon R2 that specifically inserts into the 28S rRNA gene, we show that X-linked rDNA is silenced in males. The silencing of X-linked rDNA expression by the Y chromosome is consistent across populations and independent of genetic background. These Y chromosomes also vary more than threefold in rDNA locus size and cause dramatically different levels of PEV suppression. The degree of suppression is negatively associated with the number and fraction of rDNA units without transposon insertions, but not with total rDNA locus size. Gene expression profiling revealed hundreds of differentially expressed genes among these Y chromosome introgression lines, as well as a divergent global gene expression pattern between the low-PEV and high-PEV flies. Our findings suggest that the Y chromosome is involved in diverse phenomena related to transcriptional regulation including X-linked rDNA silencing and suppression of PEV phenotype. These results further expand our understanding of the role of the Y chromosome in modulating global gene expression, and suggest a link with modifications of the chromatin state.
Keywords: evolution, microarray, mutation accumulation, Y dominance
The Y chromosome in Drosophila melanogaster is gene-poor, heterochromatic, and largely degenerate owing to the lack of recombination in males and the reduced effective population size of the Y (1–3). Beyond carrying a handful of factors essential for male fertility, the Y chromosome has been considered to have little function. Despite this relative lack of functional DNA, recent studies have shown that Y-linked genetic variation in Drosophila modulates the expression of hundreds of genes across the genome (4–6). Although it is presumed that this transacting transcriptional regulation is related to epigenetic modification of chromatin state by the Y chromosome, the functional basis for this effect has remained elusive (5).
The D. melanogaster Y chromosome contains fewer than 20 single-copy coding genes (7–9), with the bulk of the chromosome composed of repetitive DNA. Among the best-studied Y-linked loci is the rDNA locus (designated bobbed), which physically accounts for ∼10% of the entire Y chromosome and is homologous to the X-linked rDNA locus. The rDNA locus in D. melanogaster is a tandemly repeated array consisting of hundreds of units, each of which encodes 18S, 5.8S, and 28S ribosomal RNA genes (Fig. 1A). In an rDNA cistron, many of the rDNA units cannot form functional ribosomes because of insertions of the site-specific retrotransposons R1 and R2 (10). Partial deletions of the X-linked rDNA locus result in a “bobbed” phenotype in homozygous females, characterized by slow development and shorter bristles. Of note, this bobbed phenotype is not seen in males carrying a bobbed X chromosome and a normal Y chromosome (11), suggesting that rDNA transcription from the Y chromosome in males can compensate for X-linked rDNA deletions. However, the functional interactions between the X-linked and Y-linked rDNA loci are not well known, beyond the fact that they facilitate the X–Y pairing during meiosis (12).
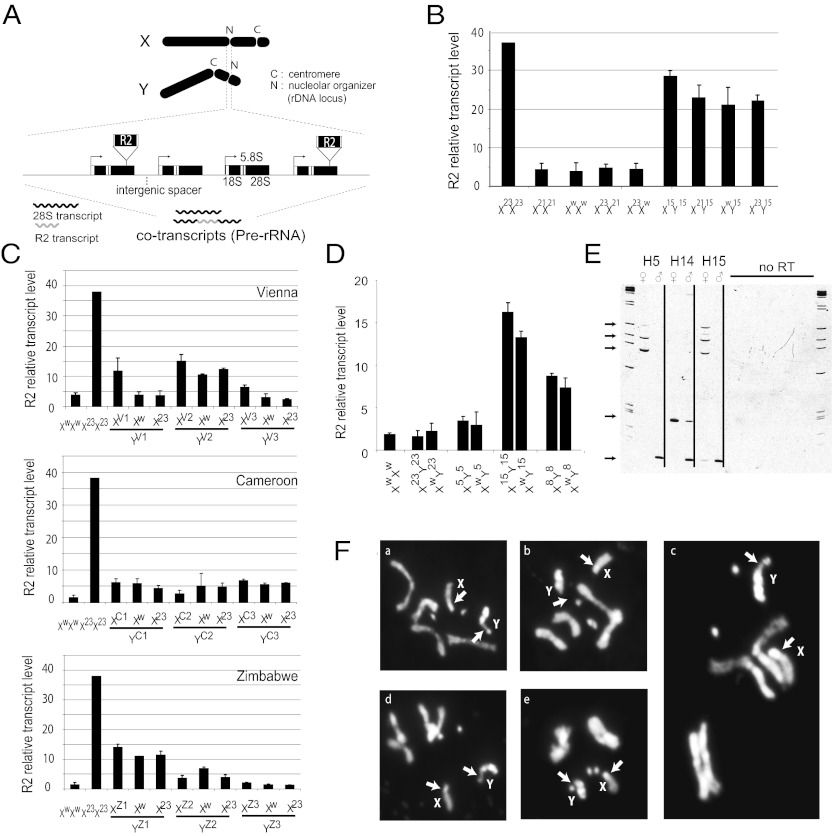
Fig. 1.
Multiple lines of evidence suggest that the X-linked rDNA is silenced in males. (A) Schematic of the X and Y chromosomes in D. melanogaster and the ribosomal loci containing the retrotransposable element R2. At bottom is a magnified rDNA locus containing R2 elements that insert into specific sites in the 28S rRNA genes. R2 is cotranscribed with the rRNA units; thus, R2 transcription can be used to represent rRNA gene transcription. (B) High levels of expression for the H23 X rDNA locus can be repressed by a low-expressing rDNA loci on another X; however, high levels of expression from the H15 Y rDNA locus cannot be shut off by an X rDNA locus. (C) Nine wild Y chromosomes from three different geographic populations demonstrated preferential Y-linked rDNA expression, also suggesting X-linked rDNA silencing in the males of those crosses. The rDNA locus in w1118 had low transcriptional levels, whereas H23 had high expression levels. (D) Four Y chromosomes with different levels of rDNA and R2 expression were introgressed into the w1118 background. The bar graph shows similar levels of the Y-linked rDNA and R2 expression in the introgressed background compared with their original levels. (E) RT-PCR results demonstrate the expression of specific 5′ truncated R2 elements in males and females. All arrows except the bottom one indicate silencing transcription of specific X-linked R2 copies in males. The bottom arrow shows a transcribed specific R2 copy on the Y chromosome. (F) Premetaphase chromosomes from adult brain tissue stained with DAPI. Cytological analyses found that only the Y chromosome formed a secondary constriction site, indicative of active transcription of the rDNA locus in the previous interphase in males. The Y chromosomes of H8 (a and c), H15 (d and e), and H23 (b) all demonstrated secondary constriction at the rDNA loci, whereas the X chromosome loci did not. Arrows indicate the locations of the rDNA loci on the X and Y chromosomes.
It is clear that the rDNA locus can have transacting regulatory effects on the transcription of rRNA genes; the phenomenon of nucleolar dominance occurs when one rDNA locus is inactivated by another. Observed mostly in interspecific hybrid plants, nucleolar dominance also has been reported in some hybrids of Drosophila and Xenopus (13). In D. melanogaster, silencing of one X-linked rDNA locus by that of an X chromosome of different origin has been reported in interspecific hybrid females (14, 15). Examples in intraspecies crosses of Drosophila simulans females have been reported as well (16). The mechanisms of nucleolar dominance are likely related to chromatin modifications (17), implying a potential role for the rDNA locus in modulating the chromatin state. The relationship between Y-linked and X-linked rDNA loci and the mechanism of related chromatin modifications in males remain unclear, however.
Another form of transcriptional silencing—position-effect variegation (PEV)—has been shown to be influenced by Y-linked variation in males (5, 18). In PEV, euchromatic genes juxtaposed to a heterochromatin boundary as a result of chromosomal rearrangement are transcriptionally repressed in some cells but not others, based on variations in the degree of heterochromatin spreading. In artificially constructed D. melanogaster lines carrying varying amounts of Y chromosomal DNA (18), the amount of Y chromosomal content was found to correlate with the degree of suppression of a PEV marker gene. In a later study, introgression lines carrying naturally occurring Y chromosomes also demonstrated varying suppression of PEV marker genes (5). These results suggest that the Y chromosome plays a role in modulating the chromatin state of the genome.
Here we use a set of mutation-accumulation lines (the Harwich lines) derived from a single founder more than 550 generations ago (Fig. S1 and Materials and Methods) to dissect in more detail the potential effects of this hypothesized modulation of genomic chromatin state by the Y chromosome. We first show that the X-linked rDNA locus is silenced in males, suggesting that the Y-linked rDNA locus is transcriptionally dominant in D. melanogaster. We then demonstrate that PEV varies across these lines and is correlated with features of the rDNA locus and patterns of global gene expression. Together, these results suggest that Y-linked variation is involved in modulating at least two processes of transcriptional silencing, probably related to chromatin modification. We argue that the Y chromosome plays an important role in modulating global heterochromatin patterns and thus the global transcriptional program, and ultimately organismal phenotypes.
Results and Discussion
Evidence for X-Chromosomal rDNA Silencing in Males.
To determine the level and status of rDNA transcription, we took advantage of the non-LTR retrotransposable element R2, which inserts specifically into the 28S rRNA gene and consequently blocks the function of that particular unit. Because R2 cotranscribes with the 28S rRNA gene (Fig. 1A; ref. 19), it can be used as a sensitive marker for judging whether the rDNA transcription from either the X-linked or the Y-linked locus is dominant or additive. Using this approach, we found that ribosomal RNA genes are transcribed differently (as large as 40-fold) both between males and females of the Harwich population and across individual lines (Fig. S2). Among the 15 Harwich lines, some lines (e.g., H23) have high R2 transcript levels in females relative to males, whereas other lines (e.g., H15) have high R2 transcript levels in males relative to females.
We carried out crosses among the Harwich lines and also a laboratory strain, w1118, which has extremely low levels of R2 transcription in both males and females. Females of a high-R2 transcription line, H23, were crossed to males of a low-R2 transcription line (either H21 or w1118) (Fig. 1B, Left). In F1 females, which had one X chromosome with a low transcription level and another X chromosome with a high transcription level, the rRNA genes derived from the high-transcription X were silenced. Similar X–X interactions were reported previously as an example of intraspecific nucleolar dominance (16).
In contrast to the pattern that we found in females, with silencing of the X chromosome with higher R2 transcription, in males the X-linked rRNA genes were consistently silenced regardless of transcription level. Males of the high-R2 transcription line H15 were crossed to three types of females, from H23 (high R2 transcription in females), w1118 (low R2 transcription), and H21 (low R2 transcription). In all cases, the F1 males exhibited R2 expression levels indistinguishable from those of the H15 males (Fig. 1B and Fig. S2), indicating X-linked rDNA silencing. To confirm this result, we expanded our analysis to include males from three different geographic populations (Vienna, Cameroon, and Zimbabwe). These males were crossed with either low-R2 transcription w1118 or high-R2 transcription H23 females. In virtually all cases, the male progeny exhibited expression levels similar to the male parent (Fig. 1C). These results again strongly suggest that X-linked rDNA is silenced in males.
By backcrossing Harwich Y chromosomes into the w1118 background, we could also show that the X-linked rDNA silencing is independent of genetic background and stable over generations. After nine generations of backcrossing, the R2 transcription levels of males remained consistent with the transcription levels in the original Harwich lines (Fig. 1D). One exception is that this X-linked rDNA silencing was not consistently observed when the Y chromosomes were introgressed into a female background with compound X chromosomes (Fig. S3). However, given the complexity of the regulatory situation when three rDNA loci are present in the flies, the relevance of this observation in a normal XY context is not clear.
Finally, we examined two sources of molecular evidence for X-linked rDNA silencing in males. We first monitored the transcription of specific 5′ truncated R2 elements, which allowed us to directly distinguish transcription from the rDNA of the X and Y chromosomes. Comparing males and females from three Harwich lines (H5, H14, and H15), we found that X-linked R2 elements either were not transcribed or had dramatically suppressed transcription in males (Fig. 1E). We then examined somatic cells for the presence or absence of a secondary constriction on the X or Y chromosome. The rDNA locus generates the nucleolus seen in all interphase cells. As cells approach metaphase, RNA polymerase I is diminished, and the nucleolus disappears. A signature of the nucleolus temporarily remains in premetaphase, however, because the previous transcription activity of the rDNA locus causes a secondary constriction at the site. This constriction site is in addition to the primary constriction at the centromere. Our cytological analysis demonstrated only a single secondary constriction site associated with nucleolar rDNA transcription in males, on the Y chromosome (Fig. 1F), indicating X-linked rDNA transcriptional silencing. Although the molecular basis for X-linked rDNA silencing is not clear, in other cases it involves changes in chromatin, such as histone modifications (17).
Y-Linked rDNA Variation Associated with PEV.
The rDNA locus on the X chromosome in Drosophila species evolves rapidly in size, mainly as a result of unequal crossing over between sister chromatids (20, 21), suggesting that the Y-linked rDNA also may evolve sufficiently rapidly to accumulate detectable variation over the 550 generations of evolution since the establishment of the Harwich lines. To further isolate the role of the Y chromosome-linked features, we introgressed the 15 Harwich Y chromosomes into an otherwise identical isogenic background (Bloomington Stock Center line 4361, with genotype y[1];bw[1];e[4];ci[1]ey[R]; Fig. S4). We then determined variations in the copy number of rDNA units (using quantitative PCR; Materials and Methods) in the 15 Harwich Y chromosomes by crossing the males of the 15 Harwich Y introgression lines with females containing a compound and bobbed X chromosome. The size of the rDNA locus on the Y varied more than threefold, from 160 to 500 units, with an average size of 330 units (Fig. 2A). The Y-linked rDNA locus in these lines appeared to be larger than the X-linked rDNA locus, which ranges in size from 150 to 300 units (20). The average size of an rDNA unit is ∼13 kb, including the rRNA genes and intergenic spacers (22, 23), and the total size of the Y-linked rDNA locus ranges from 2 to 6 Mb among the lines, representing approximately one-tenth of the Y chromosome. This amount of variation at the rDNA locus could have a significant epigenetic influence genome-wide (24).
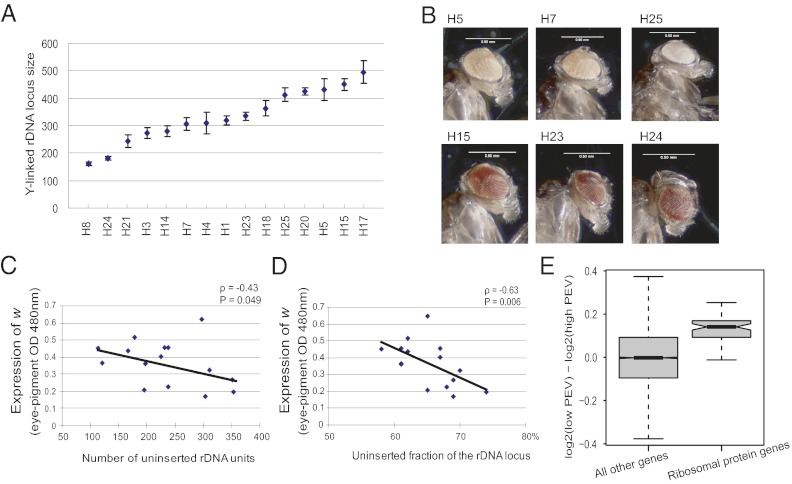
Fig. 2.
Y-linked rDNA variation correlates with PEV phenotype. (A) The Y-linked rDNA varies more than threefold in size. (B) Representative images showing the variation in levels of eye pigmentation among the Harwich Y introgression lines crossed to the w[m4h] PEV background. (C) The number of transposon uninserted rDNA units is negatively correlated with the PEV phenotype (expression of the w gene measured by eye pigment absorbance at 480 nm). (D) The uninserted fraction of the rDNA locus is negatively correlated with the PEV. (E) Boxplot showing distribution the of log2(low PEV expression) − log2(high PEV expression) for genes encoding ribosomal proteins (based on the Ribosomal Protein Gene database, accessed at http://ribosome.med.miyazaki-u.ac.jp/), compared with the distribution of the same fold change for genes not encoding ribosomal proteins. Expression of ribosomal genes is significantly higher in the low-PEV lines (P < 2.2e-16, Mann-Whitney U test).
To further explore the possible epigenetic effects of variation across these Y chromosomes, we examined the phenomenon of PEV, which has been shown to be affected by variation in the Y chromosome (5, 18). As a marker for PEV, we used the classic w[m4h] system, in which the white gene is relocated from its normal euchromatic region to the pericentromeric heterochromatin near the base of the X chromosome. The spread of heterochromatin causes variegated expression of the white gene, resulting in eye-color mosaicism, a mottled pattern of red and white spots or sectors (25, 26). We observed dramatic variation in eye pigmentation across the Harwich Y chromosomes introgressed into a w[m4h] genetic background (Fig. 2B and Fig. S5). Differences in eye pigmentation were weakly (and nonsignificantly) correlated with rDNA locus size (ρ = –0.22, P = 0.21, Spearman rank correlation test) (Fig. S6), an observation consistent with a previous report on Y chromosomes carrying partial deletions of Y-linked rDNA (24).
On the other hand, we found that both the number and the fraction of the rDNA locus that is free of R1 and R2 retrotransposable element insertions, termed uninserted rDNA units (20, 21, 27), are significantly and strongly correlated with the PEV phenotype [number of uninserted rDNA units: ρ= –0.43, P = 0.049, Spearman rank correlation test, (Fig. 2C); uninserted fraction of the rDNA locus: ρ= –0.63, P = 0.006, Spearman rank correlation test (Fig. 2D)]. On insertion, R1 and R2 elements render the corresponding rDNA units nonfunctional, and only the uninserted units form normal ribosomal RNA that functions in protein synthesis. It has been suggested that only a fraction of the entire rDNA locus is transcribed, depending on the levels of transposon insertion (16). Previous studies found that heterchromatin formation is disrupted in mutants with small rDNA arrays (28), and that partial deletions of the rDNA array result in reduced heterochromatin-induced gene silencing (29). The association of uninserted rDNA units with the suppressed expression of the white gene that we observed is consistent with those studies. Our results also suggest that the uninserted fraction of the rDNA locus may be more important for heterochromatin formation than the overall size of the rDNA array.
A recent study suggested that an increase in rRNA transcription indicates reduced heterochromatin in the genome (30). Although R2 serves as a good marker for tracking if R2-inserted rDNA units are transcribed, using R2 transcript levels to directly measure the total rRNA gene transcript levels is insufficient, because R2 inserts into only a fraction of the rDNA units, and the transcribed fraction of the total units is not known. Therefore, we used gene expression levels of ribosomal protein genes as a proxy for rRNA gene expression. The expression data for ribosomal protein genes were derived from microarray analyses comparing low-PEV lines (predominantly red eyes, lines H15 and H23) with high-PEV lines (predominantly white eyes, lines H5 and H7). As a class, ribosomal protein genes (n = 74) were expressed at significantly higher levels (median increase in expression, 10%) in the low-PEV group than in the high-PEV group (P < 2.2e-16, Mann-Whitney U test) (Fig. 2E). Given that the PEV phenotype is caused by the spread of heterochromatin into the adjacent w[m4h] gene, a reduction in heterochromatin would be expected to allow greater expression of w[m4h]. The relationships between PEV and both the uninserted fraction of rDNA units and the expression levels of ribosomal protein genes might indicate a reduction in heterochromatin in lines with low PEV. The mechanism of such a chromatin modulation remains unclear, however.
Global Gene Expression in High-PEV and Low-PEV Lines.
Our observation that PEV varies considerably among the Harwich Y chromosome introgression lines suggests that Y-chromosomal modifiers of heterochromatic state have evolved among the Harwich lines within the past 550 generations. These introgression lines also afford the opportunity to ask whether Y-linked regulatory variation (4) has evolved among the Harwich Y chromosomes as well. We selected two representative lines from the predominantly red-eyed (low-PEV) and predominantly white-eyed (high-PEV) Harwich Y chromosome introgression lines: H15 and H23 for the low-PEV lines and H5 and H7 for the high-PEV lines. We measured global gene expression using two-color microarrays across these four lines (Fig. S7).
Of note, we found strong evidence that global patterns of gene expression differ significantly between the low-PEV and high-PEV flies. First, there was a bias toward up-regulation in genes differentially expressed in red-eyed and white-eyed flies, with twice as many genes up-regulated in red-eyed flies (93 vs. 46, with a 10% false discovery rate) (Fig. 3A). This difference is in contrast to comparisons within PEV classes, where equal numbers of genes were up-regulated and down-regulated (13 and 13 for H5 vs. H7 and 60 and 52 for H15 vs. H23). Second, clear functional coherence was evident, as indicated by Gene Ontology (GO) enrichment, among genes differentially expressed in PEV groups, with several functional categories enriched in both the set of genes up-regulated and the set of genes down-regulated in the low-PEV group (Table S1). The finding of enriched pheromone-response and odorant-binding genes is of interest in view of a previous report (31) and their location near euchromatin–heterochromatin boundaries. Third, hierarchical clustering of our four samples based on whole-genome gene expression levels strongly clustered the two low-PEV lines and the two high-PEV lines into separate groups (Fig. 3B). Finally, a correlation matrix of all pairwise fold-change ratios among the four lines (H5, H7, H15, and H23; Fig. S8) clearly showed that the interclass comparisons (i.e, H15 vs. H5, H15 vs. H7, H23 vs. H5, and H23 vs. H7) are more similar than the intraclass comparisons (i.e, H5 vs. H7 and H15 vs. H23). Taken together, these results suggest that the high-PEV and low-PEV classes represent significantly diverged Y chromosomes that affect global gene expression in different ways. This global gene expression divergence emerged very quickly owing to the accumulation of Y-linked regulatory variation within only 550 generations.
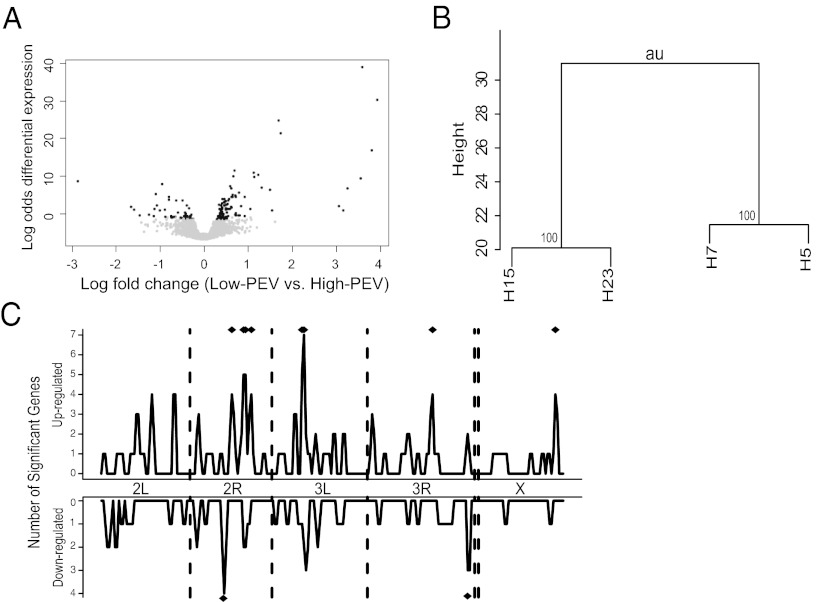
Fig. 3.
Global gene expression divergence in low-PEV and high-PEV lines. (A) Volcano plot showing more genes up-regulated in red-eyed, low-PEV flies (H15 and H23) compared with white-eyed, high-PEV flies (H5 and H7) than genes down-regulated in that comparison. Black dots indicate differentially expressed genes, whereas gray dots indicate genes that are not differentially expressed. The dots with X-axis values >0 indicate genes that are up-regulated in low-PEV flies; those with values <0 indicate down-regulated genes in low-PEV flies. The GEL50 value as a proxy of power, measuring the gene expression level at which there is a 50% chance of detection of statistical significance, is 1.85-fold in this study (37). (B) The expression data for the two low-PEV and two high-PEV lines were hierarchically clustered using the R package pvclust. A dendrogram shows that H15 and H23 are clustered and H5 and H7 are clustered for global gene expression. (C) To estimate spatial clustering along the genome, the number of significantly up-regulated or down-regulated genes was calculated separately in each 1-MB sliding window across the genome, with a step size of 500 kb. To estimate significance, a number of genes equal to the actual number of significant genes were randomly sampled 10,000 times to get a null expectation for each window. Windows marked with a diamond are significantly elevated at a nominal P < 0.01.
One attractive hypothesis is that epigenetic regulation of X-linked and autosomal gene expression by the Y chromosome might be mediated at the level of chromatin by Y-linked rDNA, microsatellites, and other noncoding repetitive sequences. Although a direct test of this hypothesis is beyond the scope of the present work, the chromatin-level hypothesis predicts that genes affected by Y-linked regulatory variation should be physically clustered along chromosomes (32). To test this hypothesis, we calculated the number of genes significantly up-regulated (or down-regulated) in high-PEV vs. low-PEV lines for sliding windows of 1 Mb with a step size of 500 kb (Fig. 3C). Several regions of the genome showed a significant excess of genes with expression affected by the Y chromosome (nominal P value of < 0.01). Taken together, these results imply a link between the chromatin modification mechanisms that drive PEV and the global patterns of gene expression regulation implicated in Y-linked regulatory variation.
Conclusions
In summary, we have presented biological evidence for X-linked rDNA silencing in males of D. melanogaster. We also have shown that the uninserted rDNA units are correlated with the suppression of PEV phenotype. In addition, hundreds of genes are differentially expressed among the Y introgression lines, and global gene expression patterns clearly diverge between the low-PEV and high-PEV flies.
The Y chromosomes that we analyzed in this study here have accumulated mutations for 550 generations. Although our findings indicate a considerable divergence at the Y-linked rDNA locus, they almost certainly have diverged in other ways as well. The Y chromosome contains abundant microsatellite repeats, degenerated transposon sequences, and other repetitive elements that also likely have evolved over this time scale and have affected global patterns of gene expression. For example, abundant microsatellite sequences residing on the Y chromosome may compete for binding with transcription factors, or degenerated transposon sequences accumulated in the heterochromatic regions may provide a source of siRNAs that modify chromatin structure. Such Y-linked noncoding sequence variation may jointly impact such phenotypes as PEV, X-linked rDNA silencing, and global patterns of gene expression. Future work is needed to disentangle the effects of individual repetitive elements on the Y chromosome.
Materials and Methods
Fly Sources.
The Harwich mutation-accumulation lines were originally subdivided from a single founder line and then maintained for 100 generations with 10 pairs of unselected flies per generation (33). After the initial 100 generations, the lines were further maintained by mass transfer every generation in regular laboratory conditions (20). The effective population size (Ne) is approximately 20. The w1118 line was from a stock maintained in the laboratory. The nine geographic lines (three Vienna lines, three Cameroon lines, and three Zimbabwe lines) were gifts from James Fry, University of Rochester. The compound X chromosome (attached X^XY) stocks C(1)DX, y and C(1)RM, the Y introgression background line 4361, and the PEV background line B6175 were all obtained from the Bloomington Drosophila Stock Center.
RNA Blots.
Total RNA was extracted from 25 adult females or 50 adult males. For this, 10 μg of RNA was separated on 1.2% (wt/vol) agarose, 2.2 M formaldehyde gels. Then the RNA was transferred to GeneScreen Plus and hybridized with an anti-sense RNA probe from the 5′ end of the R2 element as described previously (21). Relative levels of the 3,600-nt full-length R2 transcript were scanned using a PhorsphorImager screen and quantified using ImageQuant (GE Healthcare). As a control for RNA loading and quality, all R2 hybridization signals were standardized by monitoring the level of the ribosomal protein gene rp49 hybridization on the same blots. PCR products containing promoter sequences for T7 polymerase were generated for synthesis of rp49 antisense RNA probe. P-32–labeled antisense RNA was generated using T7 polymerase under the conditions suggested by the supplier (Invitrogen). The primers for the rp49 transcript were 5′-CAGCATACAGGCCCAAGATC-3′ and 5′-GTAATACGACTCACTATAGGGCAGTAAACGCGGTTCTGCATG-3′.
DNA Blots.
Total DNA was extracted from 50 adult males. Southern blot analysis was performed exactly as described by Averbeck and Eickbush (20). In brief, genomic DNA was digested by restriction enzymes BamHI, ClaI, and PstI that specifically cut the rDNA and inserted elements. Genomic DNA fragments containing R1 or R2 insertions and uninserted rDNA by R1/R2 were then separated on 1% (wt/vol) agarose gel and further hybridized with a probe from the 28S rRNA gene. The fractions of uninserted and inserted rDNA units were determined using the PhosphorImager screen on a Typhoon scanner (GE Healthcare) and quantified with ImageQuant (GE Healthcare).
RT-PCR.
RT-PCR was conducted with the M-MLV reverse transcriptase under the conditions suggested by the supplier (Invitrogen). In brief, 1 μg of total RNA was incubated at 65 °C for 5 min with R2 primer (1μm) and dNTPs (0.5 mM). The primer sequence was 5′- GTATGGAAATCTATCGAAAGATACT-3′, which lies at the R2 3′ end. After rapid chilling on ice, first-strand buffer (1×), DTT (0.01 M), and RNase inhibitor (RNAseOUT; Invitrogen) were added. The reaction mixtures were then incubated at 37 °C for 2 min. Then the M-MLV reverse transcriptase (200 U) was added, and the incubation was continued for another 50 min. Standard PCR used 2 μL of the reverse-transcriptase reaction mixture. A negative control including the same reactions without the M-MLV reverse transcriptase was conducted concurrently. The primers used for standard PCR were 5′-TGCCCAGTGCTCTGAATGTC-3′ (forward), which anneals to 28S gene sequences 80 bp upstream of the R2 insertion site and 5′-GCATGCACGATTCATTGCTC-3′ (reverse), which is located 60 bp upstream from the R2 3′ end. Therefore, a nested PCR approach was used with a primer upstream of the primer used for the reverse-transcriptase reactions.
Cytology.
Brain tissue was dissected from female third-instar larvae, fixed, and squashed as described previously (34). Rehydrated tissue was stained with 0.4 μg/mL of DAPI for 2 min, washed briefly in water, and mounted in SlowFade reagent. All observations and photography were performed with a Nikon DEC512 fluorescence microscope.
Quantitative PCR.
To assess the size of the Y-linked rDNA locus, males from the Harwich Y introgression lines were crossed to females of a stock with attached X chromosomes in which the rDNA units were largely deleted on both X chromosomes. At least five biological replicates (F1 female progeny) consisting of three experimental replicates for each line were sampled. The quantitative protocol was as described by Paredes and Maggert (35), performed with an ABI Step-One Real-Time PCR analyzer (Applied Biosystems). The tRNA gene tRNAK-CTT (15 copies per haploid genome) was used as a reference for calculating the copy number of rRNA genes. The primer sequences were identical to those described by Paredes and Maggert (35).
Eye Pigment Assay.
Males from the 15 Harwich Y introgression lines were crossed to females from a stock carrying w[m4h]. Male progeny from the crosses were collected and aged in 25 °C for 3 d, then flash-frozen in liquid nitrogen. Heads were removed and homogenized with 20 μL of acidified ethanol. Eye pigment expression was measured using Nanodrop (Thermo Scientific) at a wavelength of 480 nm. Each measurement contained five heads of each line, and between 6 and 10 biological replicates were obtained per line. Pictures of eye pigmentation phenotype (PEV phenotype) were obtained with a Syncroscopy automontage system.
Microarray Processing and Gene Expression Analyses.
Four Y introgression lines, with H5 and H7 representing the high-PEV group and H15 and H23 representing the low-PEV group, were selected to perform genomic gene expression profiling analyses. The microarrays were 18,000-feature spotted cDNA arrays. RNA samples were extracted from 50 adult males that were collected and aged at 25 °C for 3 d and then flash-frozen in liquid nitrogen. Total RNA was extracted from whole flies using TriZol (Life Technologies). Microarray processing, including cDNA synthesis, labeling with fluorescent dyes (Cy3 and Cy5), and hybridization, was carried out using the Genisphere 3DNA Kit and scanned in an Axon 4000B scanner. After hybridization and scanning, raw data were background-corrected and normalized in the R package Limma, using the normexp method for background correction and the loess method for normalization. Linear models were fit in Limma. Gene Ontology analyses were conducted with GeneMerge software (36). Differences in expression between low-PEV and high-PEV lines were estimated using the contrast [(H15+H23)/2 − (H5+H7)/2], where H5 functions as the reference and is therefore defined to be 0. All results are presented after multiple test correction using the false discovery rate method implemented in Limma (38). We consider genes significant if they survive multiple test correction at an FDR of 10%. Microarray gene expression data are available from the Gene Expression Omnibus database (accession no. GSE37068).
Gene Clustering Analyses.
Samples (H5, H7, H15, and H23) were hierarchically clustered using the R package pvclust. Because H5 is treated as the reference in the design matrix and fold change values are on a log2 scale, H5 expression was defined as 0 for all genes. A distance matrix was calculated using Euclidean distance, and a hierarchical clustering was generated using Ward's minimum variance method of clustering. To estimate significance, the pvclust function was used to perform a multiscale bootstrap resampling with 10,000 replicates. Approximately unbiased P values calculated by pvclust represent the P value for the test that the hypothesis of no cluster is true; thus, high P values indicate strong support for the cluster.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM042790 (to T.H.E.) and GM084236 (to D.L.H.).
Footnotes
The authors declare no conflict of interest.
Database deposition: The microarray gene expression data reported in this paper have been deposited into the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE37068).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207367109/-/DCSupplemental.
References
- 1.Bull JJ. Evolution of Sex-Determining Mechanisms. London: Benjamin Cummings; 1983. [Google Scholar]
- 2.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zurovcova M, Eanes WF. Lack of nucleotide polymorphism in the Y-linked sperm flagellar dynein gene Dhc-Yh3 of Drosophila melanogaster and D. simulans. Genetics. 1999;153:1709–1715. doi: 10.1093/genetics/153.4.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 5.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci USA. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sackton TB, Montenegro H, Hartl DL, Lemos B. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci USA. 2011;108:17046–17051. doi: 10.1073/pnas.1114690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci USA. 2000;97:13239–13244. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho AB. Origin and evolution of the Drosophila Y chromosome. Curr Opin Genet Dev. 2002;12:664–668. doi: 10.1016/s0959-437x(02)00356-8. [DOI] [PubMed] [Google Scholar]
- 9.Krsticevic FJ, Santos HL, Januário S, Schrago CG, Carvalho AB. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics. 2010;184:295–307. doi: 10.1534/genetics.109.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubczak JL, Burke WD, Eickbush TH. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci USA. 1991;88:3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivertzev-Dobzhansky NP, Dobzhansky T. Deficiency and duplications for the gene Bobbed in Drosophila melanogaster. Genetics. 1933;18:173–192. doi: 10.1093/genetics/18.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai JH, McKee BD. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci. 2011;124:1955–1963. doi: 10.1242/jcs.006387. [DOI] [PubMed] [Google Scholar]
- 13.Pikaard CS. Nucleolar dominance: Uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol Biol. 2000;43:163–177. doi: 10.1023/a:1006471009225. [DOI] [PubMed] [Google Scholar]
- 14.Durica DS, Krider HM. Studies on the ribosomal RNA cistrons in interspecific Drosophila hybrids, I: Nucleolar dominance. Dev Biol. 1977;59:62–74. doi: 10.1016/0012-1606(77)90240-8. [DOI] [PubMed] [Google Scholar]
- 15.Durica DS, Krider HM. Studies on the ribosomal RNA cistrons in interspecific Drosophila hybrids, II: Heterochromatic regions mediating nucleolar dominance. Genetics. 1978;89:37–64. doi: 10.1093/genetics/89.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eickbush DG, Ye J, Zhang X, Burke WD, Eickbush TH. Epigenetic regulation of retrotransposons within the nucleolus of Drosophila. Mol Cell Biol. 2008;28:6452–6461. doi: 10.1128/MCB.01015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker S, Vitins A, Pikaard CS. Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol. 2010;22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitri P, Pisano C. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics. 1989;122:793–800. doi: 10.1093/genetics/122.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 2010;30:3142–3150. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Averbeck KT, Eickbush TH. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics. 2005;171:1837–1846. doi: 10.1534/genetics.105.047670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Eickbush TH. The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genet. 2009;5:e1000386. doi: 10.1371/journal.pgen.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellauer PK, Dawid IB. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977;10:193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini M, Manning J, Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977;10:213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- 24.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talbert PB, Henikoff S. Spreading of silent chromatin: Inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 26.Schulze SR, Wallrath LL. Gene regulation by chromatin structure: Paradigms established in Drosophila melanogaster. Annu Rev Entomol. 2007;52:171–192. doi: 10.1146/annurev.ento.51.110104.151007. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Eickbush TH. Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics. 2005;170:195–205. doi: 10.1534/genetics.104.038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA. 2009;106:17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson K, et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P-P, Hartl DL, Lemos B. Y not a dead end: Epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010;186:109–118. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackay TFC, Lyman RF, Jackson MS, Terzian C, Hill WG. Polygenic mutation in Drosophila Melanogaster: Estimates from divergence among inbred strains. Evolution. 1992;46:300–316. doi: 10.1111/j.1558-5646.1992.tb02039.x. [DOI] [PubMed] [Google Scholar]
- 34.Krider HM, Plaut W. Studies on nucleolar RNA synthesis in Drosophila melanogaster, I: The relationship between number of nucleolar organizers and rate of synthesis. J Cell Sci. 1972;11:675–687. doi: 10.1242/jcs.11.3.675. [DOI] [PubMed] [Google Scholar]
- 35.Paredes S, Maggert KA. Expression of I-CreI endonuclease generates deletions within the rDNA of Drosophila. Genetics. 2009;181:1661–1671. doi: 10.1534/genetics.108.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castillo-Davis CI, Hartl DL. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 37.Townsend JP. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinformatics. 2004;5:54. doi: 10.1186/1471-2105-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.