Abstract
The structural features determining efficient biosynthesis, stability in the membrane and, after solubilization, in detergents are not well understood for integral membrane proteins such as G protein-coupled receptors (GPCRs). Starting from the rat neurotensin receptor 1, a class A GPCR, we generated a separate library comprising all 64 codons for each amino acid position. By combining a previously developed FACS-based selection system for functional expression [Sarkar C, et al. (2009) Proc Natl Acad Sci USA 105:14808–14813] with ultradeep 454 sequencing, we determined the amino acid preference in every position and identified several positions in the natural sequence that restrict functional expression. A strong accumulation of shifts, i.e., a residue preference different from wild type, is detected for helix 1, suggesting a key role in receptor biosynthesis. Furthermore, under selective pressure we observe a shift of the most conserved residues of the N-terminal helices. This unique data set allows us to compare the in vitro evolution of a GPCR to the natural evolution of the GPCR family and to observe how selective pressure shapes the sequence space covered by functional molecules. Under the applied selective pressure, several positions shift away from the wild-type sequence, and these improve the biophysical properties. We discuss possible structural reasons for conserved and shifted residues.
Keywords: deep sequencing, directed evolution, protein stability, stability in detergents
Very few residues are strictly conserved in the family of G protein-coupled receptors (GPCRs), the eukaryotic seven-transmembrane (TM) receptors that regulate many cellular events in response to chemically diverse ligands. GPCRs undergo conformational changes in response to agonist binding, and need to maintain a delicate balance between stability in the membrane, flexibility required for signaling, and the subsequent steps of receptor inactivation and degradation or recycling (1). These constraints limit stability and at least partly explain the paucity of structural information from this large family, despite herculean efforts.
Structural studies have been reported only recently (2–4), mostly for naturally stable receptors or including engineered domain insertions and/or trial-and-error optimization of the protein sequence (5, 6; summarized and reviewed in ref. 7). The limited number of solved receptor structures and the redundancy of the datasets do not reflect the functional diversity of GPCRs and still limit general conclusions about their activation mechanism, and thus about fundamental rules for agonist and antagonist design.
Most GPCRs are not amenable to functional and structural studies, because their biophysical properties are imposing major roadblocks to earlier steps in the characterization process, expression, purification, and detergent stability.
We wished to determine the critical information content in the GPCR sequence and structure for their biophysical properties and compare this to the conserved sequence features of the whole family and to experimentally test and expand previously proposed architectural rules about membrane proteins. Here, rat neurotensin receptor 1 (rNTR1)-D03 (termed here D03) was used as a model. D03 is a variant of rNTR1 that had been obtained previously by in vitro evolution of the wild type (8). D03 displays higher functional expression and detergent stability than wild type (5,000 vs. 500 receptors per cell), thus allowing reliable detection and interpretation of small changes in expression levels and detergent stability. However, despite increased expression level of D03 and improved detergent stability of the evolved variant D03 (8) or the engineered variant NTS1-7m (9), the critical structural features might not yet have been identified, requiring an in-depth and comprehensive mutagenic analysis.
Previous mutagenesis studies have relied on either error-prone PCR (8, 10) or spiked oligonucleotides (11) and could thus not cover the complete mutant space. More importantly, the studies using reporter genes have not quantitated stability or expression level (8, 10, 11). We have therefore undertaken a complete analysis of mutant space to explore the biosynthesis and biophysical properties of a GPCR.
Results and Discussion
Directed Evolution System for GPCRs.
A total of 376 DNA libraries were generated by separately randomizing amino acid positions 43–418 of rNTR1-D03 (8) into all 64 codons. Sequencing of individual clones from every second library was performed before selection to confirm the library design and quality. This analysis of about half of all libraries showed an even distribution of the four bases in all codon positions (Table S1), consistent with full randomization. Libraries were expressed in Escherichia coli, exposed to a fluorescence-labeled agonist of rNTR1, BODIPY-neurotensin, in a buffer facilitating ligand penetration through the outer membrane (8). After ligand binding to the native GPCR located in the inner membrane, the 1% highest-binding cells were selected by FACS. This selection for ligand binding directly selects for efficient production, insertion, and correct folding of the GPCR in the inner membrane of E. coli. The system was tested and adjusted in a proof-of-principle experiment using the 64-codon library of residue Y3477.31 that is critical for binding of the agonist neurotensin (12) (Figs. S1 and S2). We use the sequential numbering in plain text and the Ballesteros–Weinstein numbering as a superscript; here, the first number denotes the helix in sequential order, and the second number defines the position within the helix, where the most conserved position of a helix is denoted as x.50, counting downward toward the N terminus and upwards to the C terminus (13).
Standard Sanger full-length sequencing of 6–20 selected clones per randomized position ensured that no spurious additional mutations influenced the phenotype (4,298 sequences; Fig. S3). Very few mutations were found outside the randomized positions. Ultradeep 454 sequencing yielded 890,381 high-quality reads, each covering about 180–250 bp (SI Text, Figs. S4 and S5, and Table S2). After processing, 518,228 sequences deviating from the wild type by exactly one codon in a randomized position remained, covering 363 of the 376 positions of rNTR1-D03 at an average of 1,428 independent sequences per randomized position.
Because libraries randomized in different positions had to be combined for 454 sequencing, only 63 of the 64 codons could be quantified. The frequency of the wild-type codon was obscured (Fig. S5). However, we could determine the wild-type codon conservation from the frequencies of synonymous codons. Based on these results, the mutational tolerance in each position was addressed by calculation of rmsd (Fig. 1). The rmsd (see SI Text and Eq. S1 for detailed description) compares the observed amino acid frequency distribution after selection with the input amino acid frequency distribution (that of an unbiased NNN codon randomization). The rmsd reflects the selective pressure shaping the amino acid distribution in a given position. Sanger and 454 datasets agree well. On average, synonymous codons were used with equal frequency (Fig. S6), indicating a purely phenotypic selection at the protein level, e.g., membrane insertion, folding, and stability within the membrane and deleterious effects of misfolded proteins on the host organism. Thus, we did not detect any effects of rare codons.
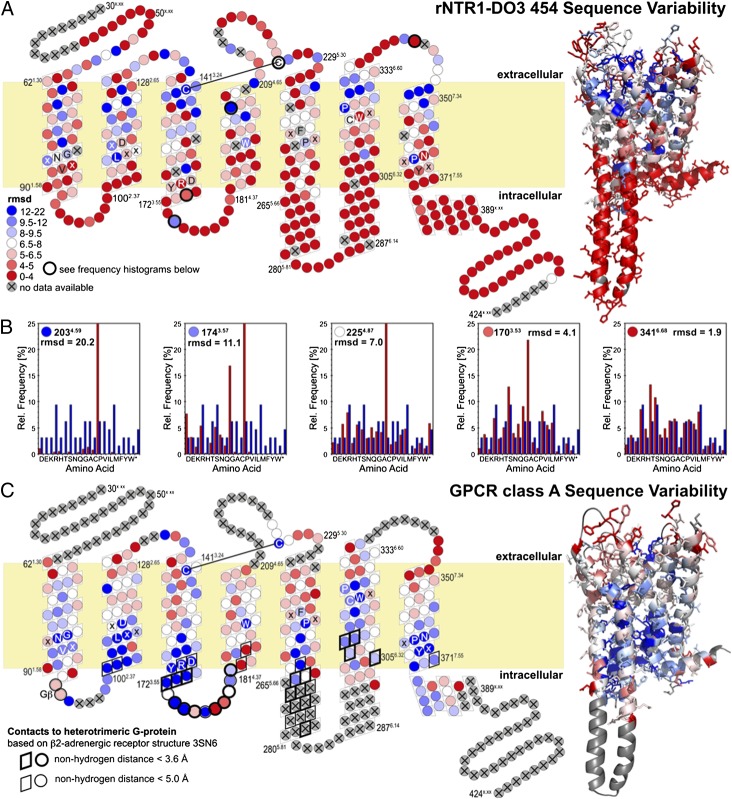
Fig. 1.
Sequence variability of rNTR1-D03 (A) and GPCR class A consensus (C). Positions in a snake plot (Left) and homology model of rNTR1-D03 (Right) are color coded in a gradient from red for high mutational tolerance to blue for low tolerance (see SI Text for details). Letters indicate conserved sequence motifs in each helix. Positions that were not randomized (flexible N terminus and C terminus) or for which no 454 data were obtained (13 transmembrane positions) are coded in gray. (B) Histograms of observed amino acid frequencies (red) compared with the amino acid frequency distribution expected for an unbiased library (blue) illustrate the significance of the different rmsd levels.
According to the ultradeep sequencing analysis after selection, positions are classified as conserved, as shift positions, or as not significant, the latter describing a broad amino acid distribution. A shift position focuses the selection on an amino acid (or very few) different from the wild type. Both conserved and shift position are subclassified as robust or weak, where weak effects are observed only after correction for codon bias, and robust effects under any condition (details are given in SI Text).
Very few amino acids are globally conserved within the GPCR family, and amino acid distributions are broad for most positions in the 454 results (Fig. S7). Structurally relevant glycines and prolines, the conserved disulfide bond and very few other residues are immutable in rNTR1 and GPCRs in general (Fig. 2 and Fig. S8). The different selection pressures, toward efficient biosynthesis, membrane insertion, and folding to form a functional ligand binding site in E. coli (rNTR1-D03 libraries), or toward finely regulated signaling competence in response to different ligands (in the case of the GPCR family evolved in nature), are reflected in different patterns of sequence constraints. In general, the GPCR class A consensus shows a strong conservation of the intracellular helix ends, pointing to their importance for downstream signaling interactions. G protein contact residues, conserved in the GPCR family, are variable in our results, whereas residues involved in ligand contact are conserved in our dataset. Residues equally conserved in both systems and those shifting away from the rNTR1 sequence presumably reflect the constraints of efficient folding and stability. The statistically robust 454 data set provides a unique opportunity to assess the relevance of structural rules (14–16), deduced from early membrane protein structures and statistical analysis of primary sequences (Fig. S9). Aromatic residues are often found to contact the periphery of the membrane, whereas aliphatics contact the hydrophobic interior or the bilayer. Basic residues are found at the cytoplasmic ends of transmembrane helices. rNTR1 obeys this last rule, displaying a strong accumulation and preference of basic amino acids at the cytoplasmic helix ends, especially at TM1/IL1 and TM5. TM5 is connected to TM6 by a long intracellular loop, and the increase in basic amino acids in TM5 might help to define the helix length and position the helix within the lipid bilayer through interactions with the phospholipid head groups.
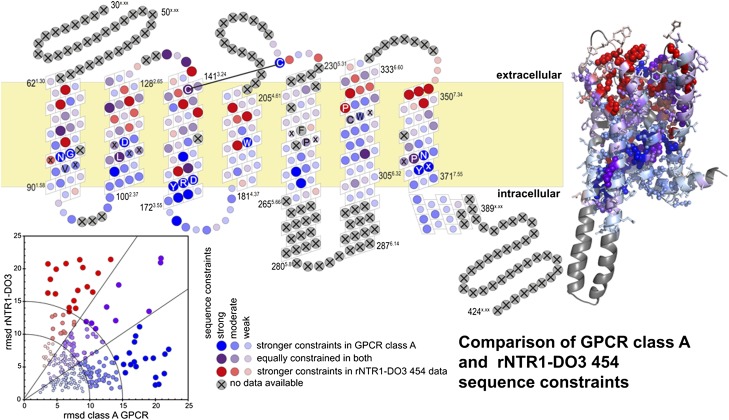
Fig. 2.
Correlation between sequence constraints in natural GPCRs and in the rNTR1-D03 454 sequencing experiment. Colors indicate whether the amino acid distribution in a given position is equally constrained in both systems (purple), more constrained in natural class A GPCR sequences (blue), or in the deep sequencing from rNTR1-D03 (red). The size of the circles indicates the constraints imposed by the respective system; large circles indicate positions with high constraints.
Of particular interest are the shift positions (Fig. 3). Strong shift positions are exclusively located within the TMs. Both lipid-exposed side chains and those pointing into the helical interspace are observed to shift. Shifted residues form clusters, defining specific locations important for protein folding and stability, with the shift mutations releasing a previous constraint present in the wild-type sequence. Clustered mutations represent different solutions to the same problem, and may thus not be additive.
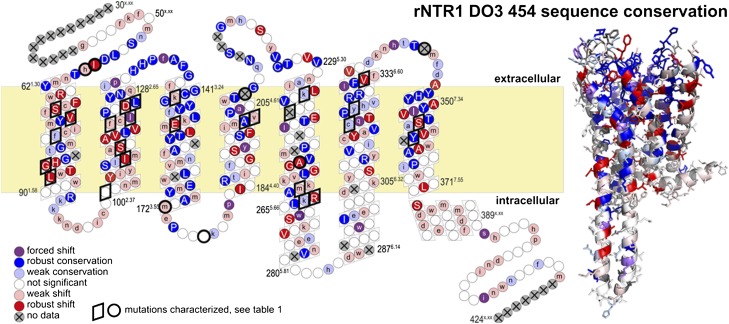
Fig. 3.
Sequence shifts. Positions are colored according to conservation (blue) or shift (red) away from the rNTR1-D03 sequence. Uppercase letters refer to robust effects and lowercase letters to effects obscured by codon bias in the raw amino acid consensus. The strength of the conservation or shift is given by the rmsd of the amino acid distribution from the input distribution as shown in Fig. 1A and listed in Fig. S7. For the highlighted positions, the influence of mutations on the performance of the receptor has been further characterized (Table 1).
We find an accumulation of shifts in TM1 and TM2, whereas TM3, known to be involved in structural rearrangements upon activation (1, 17), remains highly conserved. TM1 shows three independent gains of aromatic amino acids pointing toward the lipid interface at the N terminus, at V65W1.33, L66F1.34, or A69F1.37. This gain of aromatic side chains might enhance protein integration into the lipid bilayer during positioning of the first TM helix in the lipid bilayer. Considering that membrane protein biosynthesis is crucially dependent on correct helix folding, targeting, and insertion into the lipid bilayer, we hypothesize that the accumulation of shifts in the N terminus and the most N-terminal helices is an adaptation to that process, and this effect might be highly relevant for increased GPCR expression levels (Fig. 3). This particular importance of the N-terminal helices was not apparent from previous analyses of residue preferences in membrane proteins.
In TM7, we find a loss of aromatic residues at positions F346A7.26, F350A7.34, and F358V7.42, approximately within two helical turns from each other. According to our model of rNTR1, F3587.42 is located close in space to the proline-induced kink in TM6, and the shift F358V7.42 might relieve a steric constraint and lead to optimized helix packing. Because TM7 contributes specifically to ligand binding, a comparison with the GPCR consensus is more difficult. Y3477.31, for example, is crucial for ligand binding in rNTR1 (12). The GPCR consensus shows an additional proline at position 3627.46 that will substantially influence the structural architecture of TM7, but is absent in rNTR1.
GPCRs share high conservation of key signature motifs in every helix (highlighted in Fig. 2; Table S3) that are common to the GPCR family and reflect structural features (e.g., proline-determined helical architecture) or the common signaling process [E/DRY motif in TM3 (E1663.49, R1673.50, and Y1683.51 in rNTR1) for receptor activation] (18). As explained above, the Ballesteros–Weinstein nomenclature (added in superscript) refers to 50 as the most conserved position of a helix. Unexpectedly, we find that the key residues of the first three helices are shift positions, i.e., N82H1.50, D113S2.50, and R167L3.50. N82H1.50 and D113S2.50 are close in space according to our homology model, and these mutations might be alternative solutions to the same problem of an unsatisfied salt bridge. D1132.50 was shown to be responsible for the sodium-sensitivity of the rNTR1 receptor (19). R167L3.50 is part of the E/DRY motif for which a crucial role, the “ionic lock,” in receptor activation has been proposed (17). The fact that we find GPCR signature residues shift is rather surprising, and suggests that functionally required conformational flexibility of the receptor may limit its biophysical stability. In vivo, these signature residues might be required to maintain the delicate balance of receptor-signaling networks, and even facilitate GPCR degradation after endocytosis. When studied in an isolated system, however, these residues are identified as roadblocks for recombinant overexpression and detergent stability. The phenotype of rNTR1-D03 is determined by nine mutations, originating from random mutagenesis on the wild type (8). Among these mutations, we find R167L3.50 to improve functional expression but to decrease signaling competence (8). Interestingly, we find the D03 mutations to be mostly confirmed after full randomization (Figs. S3 and S7). For R1673.50, we find only mild preference of aliphatic residues over R, allowing us to reintroduce R1673.50 and thus to restore signaling capability without compromising functional expression levels, as shown for D03 (8).
Several mutations, based on the D03 background, were selected for further characterization (Table 1). For all mutants tested, expression levels were as high as or higher than rNTR1-D03, indicating that no false-positive selection result was observed. A86L1.54, I253A5.54, and F358V7.42 significantly increase the stability of the receptor in n-dodecyl-β-d-maltopyranoside (DDM), a mild detergent used for solubilization and functional studies of many GPCRs, increasing the apparent Tm by 7.5, 3.5, and 4°, respectively, in the absence of ligand (Table 1). F358A7.42, a nonenriched negative control, had no effect on receptor stability, clearly showing the specific effect of the valine substitution. These results clearly demonstrate the power of the full randomization and selection applied here, in contrast to an alanine scan (9); the latter would only have investigated I253A5.54, but discarded F358A7.42 as unfavorable, and thus would not have discovered F358V7.42.
Table 1.
Detergent stability in DDM and expression levels of single-shift mutants
| Mutation | Conservation | Rmsd 454 | Tm, °C (n = 2)* | Tm, ΔD03, °C | Receptors per cell relative to D03, % (n = 3) |
| T68S | Robust shift | 16.7 | 28.6 | −2.5 | 99 ± 43 |
| E124D | Robust shift | 15.1 | 28.0 | −3.2 | |
| V57I | Robust shift | 11.9 | 31.2 | 0.1 | |
| S83G | Robust shift | 10.2 | 32.0 | 0.9 | 144 ± 9 |
| T354S | Robust shift | 8.6 | 28.7 | −2.4 | 129 ± 1 |
| C332V | Robust shift | 7.2 | 28.7 | −2.5 | 174 ± 7 |
| N262R | Robust shift | 6.5 | 28.8 | −2.4 | |
| D113A | Robust shift | 6.4 | 28.1 | −3.0 | 116 ± 8 |
| D113S | Robust shift | 6.4 | 29.6 | −1.5 | 145 ± 19 |
| D150E | Robust shift | 6.4 | 30.5 | −0.6 | |
| I70V | Robust shift | 6.3 | 28.7 | −2.4 | 114 ± 26 |
| A110L | Robust shift | 5.7 | 28.4 | −2.7 | 82 ± 17 |
| I253A | Robust shift | 5.2 | 34.4 | 3.3 | 175 ± 19 |
| A86L | Robust shift | 4.9 | 38.4 | 7.3 | 149 ± 25 |
| R143K | Weak shift | 12.9 | 30.8 | −0.4 | |
| L119F | Weak shift | 10.1 | 31.9 | 0.8 | 134 ± 21 |
| I260A | Weak shift | 7.1 | 28.8 | −2.4 | |
| I202L | Weak shift | 5.3 | 28.0 | −3.1 | 121 ± 10 |
| I202S | Weak shift | 5.3 | 30.8 | −0.3 | 75 ± 27 |
| N58R | Weak shift | 4.2 | 30.9 | −0.3 | |
| C172R | Weak shift | 2.9 | 28.6 | −2.5 | |
| T101R | Not significant | 3.4 | 29.4 | −1.8 | 131 ± 5 |
| A177H | Not significant | 3.1 | 29.2 | −1.9 | 123 ± 13 |
| K263R | Weak conservation | 4.7 | 30.8 | −0.4 | 152 ± 17 |
| Y324L | Weak conservation | 6.3 | 24.6 | −6.6 | 164 ± 21 |
| C320L | Weak conservation | 7.0 | 26.8 | −4.4 | 152 ± 10 |
| K235R | Weak conservation | 8.7 | 30.8 | −0.4 | 148 ± 21 |
| F75L | Weak conservation | 10.8 | 27.5 | −3.6 | 108 ± 7 |
| F358V | Robust conservation | 4.8 | 34.8 | 3.7 | 179 ± 23 |
| F358A | Robust conservation | 4.8 | 29.7 | −1.5 | |
| L125V | Robust conservation | 7.2 | 28.1 | −3.1 | 143 ± 14 |
| A201S | Robust conservation | 14.0 | 27.2 | −4.0 | 180 ± 26 |
| M121L | Forced shift | 11.7 | 29.5 | −1.7 | 158 ± 10 |
| M208V | No data | 29.6 | −1.5 | 139 ± 7 | |
| V240L | No data | 28.4 | −2.7 | ||
| F342A | No data | 30.1 | −1.0 | ||
| D03 | 31† | 100 ± 13 |
*Average error, 2 °C.
†n = 8.
For these three shifts, we find a correlation between increased functional expression levels, which report on the biophysical properties in the bilayer, and stability in detergents. This overlap is intriguing regarding the fact that selection for functional expression reflects the sum of correct biosynthesis; efficient and functional membrane integration; stability in the lipid bilayer; resistance to degradation; and the potential influence of misfolded species on survival and growth of the host organism, whereas stability within a detergent micelle is governed by the influence of the small detergent molecules on the structure, potentially penetrating into the core, or allowing some access to solvent molecules. The chemical nature of the detergent, protein packing, and helical stability will all influence the observed stability, and some of the selected mutations have improved stability in detergents and functional expression.
I253A5.54 is located in the core of TM5, facing TM3 and TM6. Previous functional and structural studies on GPCRs have shown an important role for TM5 in receptor activation (1), because TM5 contains crucial residues that, upon binding of agonist, induce local structural rearrangements that presumably lock the receptor in the activated state (1, 20). I253A5.54, located one helical turn away from the structurally conserved proline in TM5, might reduce conformational flexibility of the receptor, thereby increasing stability. In contrast to that, we assume that A86L1.54 and F358V7.42 improve detergent stability by addressing helix packing. Based on our homology model, A86L1.54 and F358V7.42 are solvent accessible, and a compact helix fold might reduce the susceptibility toward detergent penetration into the helical core.
Both conformational stabilization and optimized helix interactions reduce the susceptibility of the receptor toward denaturation by detergents, and these effects are certainly not rNTR1 specific, but of general importance for membrane protein stabilization. However, not all of the mutations that increase functional expression also affect stability, or the reverse (Table 1). We thus hypothesize that a large number of shift residues improve interactions within the receptor that are critical for efficient and correct biosynthesis, thus increasing functional expression in the lipid bilayer, whereas only a subset of shift residues affect interactions within the folded receptor, thus leading to higher detergent resistance.
The combination of saturation mutagenesis, FACS selection, and analysis by 454 sequencing has produced unprecedented insight into the contribution of every receptor position on receptor functionality, stability in the natural bilayer, and in detergents. Membrane proteins maintain a delicate balance between expression, function, and stability. In vivo evolution of GPCRs and membrane proteins in general shows strong selective and functional pressure (21). The high tolerance toward randomization, and especially the high frequency of shifts observed in this study, including signature residues in helices, is intriguing and of utmost importance for our understanding of membrane proteins. Though our in vitro evolutionary approach covers the full mutant space in one library, in vivo evolution would require neutral intermediates, some of which might not be tolerated in case of integral membrane proteins. Our results show that rNTR1, and presumably other GPCRs, offer ample opportunity for engineering, without compromising protein activity.
Several trends uncovered here might be of general applicability: clusters that evolve toward the class A consensus indicate the validity of the rules of preferential location of aromatic amino acids (16) and positive charges (14). These trends might be easily transferable to other GPCRs. Furthermore, the importance of the N-terminal helices had not previously been highlighted: the location of some of the most prominent shifts within TM1 indicates that functional expression may be at least partially limited by cotranslational membrane insertion via the SecYEG translocon in E. coli.
Some receptor residues, among them key GPCR signature residues, might be conserved mainly to maintain a fine-tunable receptor-signaling network, because the receptor needs to be able to exist in two conformational states: signaling active and inactive. Though the mutations investigated, even when combined, maintain signaling at high agonist concentration, they show higher levels of activity at low agonist concentrations than wild type (22). Thus, selection may have favored mutants that shift the balance toward the activated state, because selection was carried out in the presence of agonist. This selection for biophysical behavior maintained receptor-activated nucleotide exchange at the G proteins (22), but not the ability of the GPCR to fully turn off in the absence of agonist.
It is interesting to note that mutations selected in E. coli also lead to improved expression in eukaryotic hosts (8, 22). Some shift mutations significantly increase the stability of rNTR1 in short-chain detergents, presumably by controlling access of detergent molecules (22).
We believe that this systematic and evolutionary approach to the biophysical properties of the GPCR family will greatly help equally systematic structure determination efforts across this important family, and also delineate the biophysical constraints on the natural evolution of this family. These results may be thus of interest for other membrane proteins, and could be the basis for rational design of membrane proteins for higher stability in detergent solution for more efficient drug screening and structural studies.
Methods
Library Design and Synthesis.
A total of 376 different libraries were generated for receptor positions 43–418 by a PCR approach using oligos with NNN-diversified codons, generating a position-specific 64-codon library with at least 10- to 20-fold oversampling. Each library was subcloned into the expression vector pRGD03 (8, 23). The absence of any bias in the randomized library was confirmed by sequencing individual variants before selection.
Library Expression and Selection.
Each library was separately expressed in E. coli DH5α for 20 h at 20 °C. After expression, cells were labeled using BODIPY-neurotensin, a fluorescence-labeled ligand of rNTR1. The outer membrane was permeabilized by a Tris·HCl salt buffer to allow for ligand binding (1–2 h at 4 °C). The 1% highest fluorescent cells were selected using FACS and recovered for sequence analysis.
Sequencing Analysis and Characterization.
Per library, 6–20 single selected clones were analyzed by Sanger sequencing. The plasmid DNA of the selected variant pool was isolated and analyzed by ultradeep 454 sequencing. Expression levels were determined by radioligand binding assays, and detergent stability was assessed with protein variants isolated from the E. coli membrane according to Sarkar et al. (8).
Supplementary Material
Acknowledgments
We thank Anette Schütz and Malgorzata Kisielow [Flow Cytometry Laboratory, Swiss Federal Institute of Technology Zurich (ETHZ)/University of Zurich (UZH)] for valuable technical support during FACS selections, and Jan Furrer and Michael Hohl for help during the selections for high functional expression. We also thank the following former Codon Devices, Inc., employees: Patti Aha, Subhayu Basu, Katie Black, Brad Chapman, Eric Devroe, Chris Emig, David Rappolli, and Shervin Riahi for method development and for assembling the position-specific libraries; Kathy Galle for project management; and Brian Baynes for support of this collaboration. Further, we thank the Codon Devices DNA assembly, automation, and sequencing teams for technical support of library construction and validation, which were funded by the Codon Devices, Inc., research and development budget. ET was supported by a postdoctoral fellowship by the Roche Research Foundation. This work was supported by grants from the National Center of Competence in Research Structural Biology funded by the Swiss National Science Foundation (A.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202107109/-/DCSupplemental.
References
- 1.Deupi X, Kobilka B. Activation of G protein-coupled receptors. Adv Protein Chem. 2007;74:137–166. doi: 10.1016/S0065-3233(07)74004-4. [DOI] [PubMed] [Google Scholar]
- 2.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaakola VP, et al. The 2.6 Ångstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 6.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar CA, et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci USA. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata Y, et al. Thermostabilization of the neurotensin receptor NTS1. J Mol Biol. 2009;390:262–277. doi: 10.1016/j.jmb.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, et al. Rapid identification of functionally critical amino acids in a G protein-coupled receptor. Nat Methods. 2007;4:169–174. doi: 10.1038/nmeth990. [DOI] [PubMed] [Google Scholar]
- 11.Sommers CM, Dumont ME. Genetic interactions among the transmembrane segments of the G protein coupled receptor encoded by the yeast STE2 gene. J Mol Biol. 1997;266:559–575. doi: 10.1006/jmbi.1996.0816. [DOI] [PubMed] [Google Scholar]
- 12.Barroso S, et al. Identification of residues involved in neurotensin binding and modeling of the agonist binding site in neurotensin receptor 1. J Biol Chem. 2000;275:328–336. doi: 10.1074/jbc.275.1.328. [DOI] [PubMed] [Google Scholar]
- 13.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure function relations in G protein-coupled receptors. Meth Neurosci. 1995;25:366–428. [Google Scholar]
- 14.von Heijne G. Membrane proteins: From sequence to structure. Annu Rev Biophys Biomol Struct. 1994;23:167–192. doi: 10.1146/annurev.bb.23.060194.001123. [DOI] [PubMed] [Google Scholar]
- 15.Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelkar DA, Chattopadhyay A. Membrane interfacial localization of aromatic amino acids and membrane protein function. J Biosci. 2006;31:297–302. doi: 10.1007/BF02704101. [DOI] [PubMed] [Google Scholar]
- 17.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: Beyond the ground state. Mol Pharmacol. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 19.Martin S, Botto JM, Vincent JP, Mazella J. Pivotal role of an aspartate residue in sodium sensitivity and coupling to G proteins of neurotensin receptors. Mol Pharmacol. 1999;55:210–215. doi: 10.1124/mol.55.2.210. [DOI] [PubMed] [Google Scholar]
- 20.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Schöneberg T, Hofreiter M, Schulz A, Römpler H. Learning from the past: Evolution of GPCR functions. Trends Pharmacol Sci. 2007;28:117–121. doi: 10.1016/j.tips.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Schlinkmann KM, et al. Maximizing detergent stability and functional expression of a GPCR by exhaustive recombination and evolution. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.05.039. in press. [DOI] [PubMed] [Google Scholar]
- 23.Tucker J, Grisshammer R. Purification of a rat neurotensin receptor expressed in Escherichia coli. Biochem J. 1996;317:891–899. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.