Abstract
Bacterial pathogens are becoming increasingly resistant to antibiotics. As an alternative therapeutic strategy, phage therapy reagents containing purified viral lysins have been developed against Gram-positive organisms but not against Gram-negative organisms due to the inability of these types of drugs to cross the bacterial outer membrane. We solved the crystal structures of a Yersinia pestis outer membrane transporter called FyuA and a bacterial toxin called pesticin that targets this transporter. FyuA is a β-barrel membrane protein belonging to the family of TonB dependent transporters, whereas pesticin is a soluble protein with two domains, one that binds to FyuA and another that is structurally similar to phage T4 lysozyme. The structure of pesticin allowed us to design a phage therapy reagent comprised of the FyuA binding domain of pesticin fused to the N-terminus of T4 lysozyme. This hybrid toxin kills specific Yersinia and pathogenic E. coli strains and, importantly, can evade the pesticin immunity protein (Pim) giving it a distinct advantage over pesticin. Furthermore, because FyuA is required for virulence and is more common in pathogenic bacteria, the hybrid toxin also has the advantage of targeting primarily disease-causing bacteria rather than indiscriminately eliminating natural gut flora.
Keywords: colicin, muramidase, TonB-dependent transport, plague
Bacteriophages are viruses that have a prolific ability to infect, multiply within, and subsequently eliminate large numbers of bacteria. Their potential for use in microbial therapy was recognized shortly after their discovery in the late 1910s (1, 2). Phage therapy was initially developed, but later fell out of favor, when small molecule antibiotics became cheaper and easier to manufacture in large homogeneous quantities. More recently, the emergence of antibiotic resistant bacterial strains has renewed interest in using phages or their components in therapy, agriculture, food, and water treatment (3). Reagents have been developed using phage derived proteins called lysins to clear in vivo infections of Gram-positive bacteria such as Streptococcus pneumoniae (4) and Bacillus anthracis (5). Clearance occurs because lysins hydrolyze the surface exposed peptidoglycan that forms the cell wall of Gram-positive bacteria causing cell rupture ; however, a major limitation of this approach is that it cannot be applied to Gram-negative bacteria because viral lysins cannot cross the outer membrane without help from accessory proteins or membrane-disrupting agents. The outer membrane effectively shields peptidoglycan from degradation by the externally added lysin.
We solved the crystal structure of a protein toxin from Y. pestis that showed us how to engineer a phage lysin capable of killing Gram-negative bacteria. This protein, called pesticin, belongs to a class of bacterial proteins known as bacteriocins (referred to as colicins when they target E. coli). Bacteriocins are produced by bacteria to kill related bacterial strains in times of stress. Bacteria producing the bacteriocin protect themselves by expressing an immunity protein to inhibit activity of the toxin (6). Y. pestis produces pesticin to promote virulence. Pesticin, pesticin immunity protein, and plasminogen activator (Pla) are encoded on a ∼10 kb plasmid called pPCP1 (7). Pla facilitates invasion in bubonic plague and, as such, is an important virulence factor (8, 9). When strains lose the pPCP1 plasmid, they are killed by pesticin thus ensuring maximal virulence in the bacterial population.
Bacteriocins belong to two classes. Type A bacteriocins depend on the Tol system to traverse the outer membrane, whereas type B bacteriocins require the Ton system. For both classes, the primary interaction that triggers the translocation process occurs between the bacteriocin and a specific outer membrane receptor protein. Pesticin has been classified as a type B bacteriocin because killing depends on the products of the tonB, exbB, and exbD genes and, like other type B bacteriocins, pesticin has a TonB box motif located near its N-terminus (10). In addition to tonB, exbB, and exbD, these bacteria must express a receptor gene—fyuA—in order to be killed by pesticin. FyuA is an integral outer membrane protein that acts as the receptor for pesticin (11). It is also a major virulence factor for some Yersinia strains (12, 13) and certain pathogenic E. coli (14, 15). Although Y. pestis also expresses FyuA in its outer membrane, it is protected from pesticin-induced cell death by the product of the pesticin immunity gene that is always found adjacent to the pesticin gene (10). Interaction between FyuA and pesticin is necessary to trigger the import of pesticin across the outer membrane into the periplasm. Once inside the periplasm, pesticin kills the cell by degrading peptidoglycan (16) through a muramidase (lysozyme) activity (17).
Here, we report the first production of an engineered lysin that can directly kill Gram-negative bacteria. We solved the 2.1 Å structure of pesticin and used the structure to guide engineering of a “hybrid lysin.” Pesticin consists of two domains: one that targets and binds to FyuA, and the other that degrades peptidoglycan. The engineered hybrid consists of T4 lysozyme, which is an archetypal lysin, attached to the FyuA-targeting domain derived from pesticin. To visualize the import machinery better, we solved the structure of FyuA at 3.2 Å and the hybrid toxin at 2.6 Å resolution. FyuA is a TonB-dependent iron transporter (18) consisting of a 22-strand β-barrel with a plug domain inserted in the pore. The hybrid toxin resembles pesticin and retains the same two-domain architecture. We show that the hybrid lysin crosses the outer membrane and kills cells not only in a model E. coli system but also in bacterial pathogens. Importantly, killing only affects cells that produce FyuA (11). The action of the hybrid lysin is unaffected by Pim (10), which is a protein that some Yersiniae produce to inhibit peptidoglycan degrading enzymes. The hybrid lysin can, therefore, potentially be used against any strain expressing FyuA.
Results
Structure Determination of Y. pestis FyuA and Pesticin.
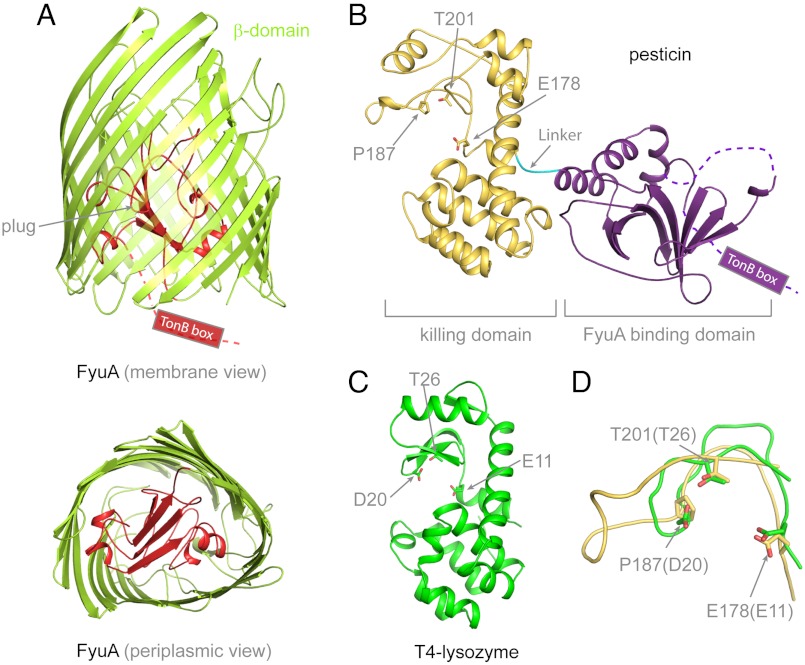
We began this project by determining the structure of FyuA, a 71 kDa outer membrane transporter that normally transports ferric yersiniabactin (Fe-Ybt) and is important for virulence in certain Yersinia and E. coli strains. FyuA was expressed in E. coli, purified and crystallized using the detergents LDAO and C8E4. Crystals diffracted to 3.2 Å resolution, and the structure was solved by single wavelength anomalous dispersion (Table S1). The structure of FyuA has features typical of the TonB-dependent transporter family (Fig. 1A) (18) including a membrane-spanning 22-strand β-barrel whose pore is blocked by a plug domain. On the periplasmic side of the membrane, the individual β-strands of the barrel are connected by short turns, whereas on the extracellular side strands are connected by long extracellular loops L1–L11. A portion of L8 is disordered (residues 487–492). The TonB box motif (STLVVTA, residues 8–14) is essential for interactions with TonB-ExbB-ExbD (13, 19) and is only partially visible in the structure (residues 1–12 are disordered). Mutations in the TonB box prevent transport of siderophores and bacteriocins (6).
Fig. 1.
Crystal structures of Y. pestis FyuA and pesticin. (A) The crystal structure of FyuA consists of two domains: the β-barrel in green and the plug domain in red. The TonB box of FyuA was mostly disordered and not observed in the crystal structure. In the periplasmic view of FyuA, the location of the TonB box is not shown for clarity. (B) The crystal structure of pesticin consists of two distinct domains: The predominantly β-sheet N-terminal domain (FyuA binding domain) in purple and encompasses residues 1–164. The mainly α-helical C-terminal domain (residues 168–357) in gold (killing domain). A short linker connects the two domains and is shown in cyan that spans residues 165–167. The TonB box of pesticin was disordered and not observed in the crystal structure. The residues mutated in this study are labelled and shown in stick representation and include residues in the presumed active site of pesticin. (C) To illustrate that the C-terminal domain of pesticin is structurally similar to T4 lysozyme, the crystal structure of T4 lysozyme (green) (PDB code = 2LZM) is represented in the same orientation as the C-terminal domain of pesticin. Similarity between the two proteins is primarily structural; there is little sequence identity except for a partial conservation of active site residues that is apparent after a structural alignment. (D) Superimposition of the pesticin (gold) and T4 lysozyme (green) active sites shows that the catalytic threonine and glutamate residues are conserved in pesticin, whereas the catalytic aspartate in lysozyme is replaced by a proline in pesticin. Therefore, pesticin shares a functional resemblance to T4 lysozyme, but the active sites are not identical. For clarity, representation of secondary structure elements was omitted in D.
To learn more about how pesticin kills bacteria expressing FyuA, we solved the 2.1 Å crystal structure of pesticin (40 KDa) using multiwavelength anomalous dispersion (Table S1). The structure of pesticin consists of two domains joined by a short linker (Fig. 1B). The N-terminal domain is formed by a seven-strand antiparallel β-sheet with two α-helices inserted between strands 5 and 6 and an additional α-helix following strand 7. The C-terminal domain of pesticin is primarily α-helical, except for a small irregular antiparallel three-strand β-sheet (Fig. 1B). The N-terminal 13 residues are disordered in the structure. This region includes the TonB box motif (DTMVV, residues 3–7) (10) which is necessary for type B bacteriocins to be translocated across the outer membrane. Other regions that could not be resolved include residues 25–34 (25–32 in noncrystallographic symmetry related copy B) and the C-terminal residue.
Pesticin Shares Structural Similarity with Phage T4 Lysozyme.
Pesticin does not resemble any other known bacteriocin in sequence or structure (6); so, we analyzed the pesticin fold using the DALI server (20) to search for other structural homologs. DALI analysis revealed a striking similarity between the C-terminal domain of pesticin and phage T4 lysozyme (21) that had not been postulated prior to the structure determination because the sequence similarity is low (Fig. 1C and Fig. S1). T4 lysozyme is an archetypal lysin, and members of the T4 lysozyme family have an essential catalytic triad corresponding to E11, D20, and T26 (Fig. 1 B–D) (22). Two of these residues are conserved in pesticin: E178 and T201. We confirmed that E178 and T201 are active site residues by mutating them to alanine, which completely abolished pesticin activity using a bactericidal assay (Fig. S2).
Structure Determination of a Pesticin-T4 Lysozyme Hybrid.
The structural and functional resemblance between the two proteins suggested that we could create a hybrid lysin by fusing the N-terminal domain of pesticin to T4 lysozyme, an idea that only became clear upon analyzing the pesticin structure. The hybrid lysin thus consists of a Y. pestis “FyuA-targeting” domain and a T4 phage “killing” domain (Fig. S3). We crystallized the hybrid lysin and solved the structure by molecular replacement to 2.6 Å (Table S1 and Fig. S4A). Like pesticin, the hybrid lysin consists of two domains folded similarly to wild type pesticin (Fig. S4 D and E). The individual domains have an rmsd of 0.5 Å with their counterparts in pesticin. Two slightly different conformations were observed within the asymmetric unit of the hybrid structure with the only difference being a small rotation of the T4 lysozyme domain relative to the targeting domain; however, the position and orientation of the lysozyme killing domain of the hybrid was rotated ∼90 degrees relative to the killing domain of wild type pesticin.
Pesticin and the Hybrid Lysin Specifically Target Cells Expressing FyuA.
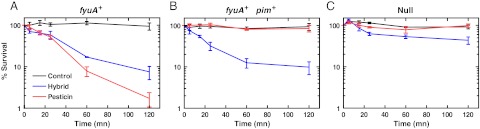
To evaluate the engineered hybrid lysin, we needed a method to assess wild type pesticin activity. We established a bactericidal assay using E. coli cells expressing Y. pestis FyuA from a pET20b vector. As expected, we found that pesticin was able to kill E. coli cells expressing FyuA (Fig. 2A), whereas control cells containing unmodified pET20b were completely resistant (Fig. 2C). We then used this assay to determine whether our pesticin-lysozyme hybrid would kill E. coli cells expressing FyuA. The hybrid lysin was able to kill fyuA+ cells in the bactericidal assay (though to a lesser extent than pesticin). For the fyuA- cells (Null), the sample treated with pesticin survived similarly to the untreated control, whereas the sample treated with hybrid lysin showed low levels of cell death. The level of killing, however, was small compared to the level seen when the hybrid lysin was added to cells expressing FyuA. These data indicate that killing by the hybrid largely depends on FyuA expression (Fig. 2 A and C). We also confirmed that fyuA+ cells are not killed by the addition of purified T4 lysozyme (Fig. S2). Because cell death arises from peptidoglycan degradation in the periplasm, our results indicate that T4 lysozyme was transported across the outer membrane when attached to the N-terminal domain of pesticin.
Fig. 2.
Bactericidal assays demonstrate that pesticin and the hybrid lysin kill E. coli cells expressing FyuA. Pesticin or hybrid lysin at a concentration of 100 μg/mL was added to E. coli T7 express cells in PBS at 37 °C while shaking. Control cultures had no proteins added. Samples were removed at several time points, plated, and surviving bacterial colonies were counted. These experiments were repeated at least three times. The standard error of the mean is shown. (A) Cells expressing fyuA are killed by pesticin and the hybrid lysin. (B) Cells expressing fyuA and pim are killed by the hybrid lysin but not by pesticin due to protection conferred by the pesticin immunity protein. Note that the immunity protein cannot protect against killing by the hybrid lysin. (C) E. coli T7 express cells transformed with pET20b (Null) are resistant to killing by pesticin or the hybrid lysin because they do not express fyuA on the cell surface.
We tested pesticin’s ability (at high and low concentrations) to inhibit the growth of fyuA+ cells and found that each concentration inhibited growth similarly (Fig. S5). In contrast, colicin Ia is self-inhibitory at high concentrations because colicin Ia requires two copies of its receptor Cir, one for initial cell binding and the other for translocation across the outer membrane (23). As a result, at high concentration colicin Ia binds all the available Cir on the cell surface and then cannot find free Cir for translocation. Because high concentrations of pesticin inhibited growth similarly compared to low concentrations, this suggests that pesticin only requires one copy of FyuA to bind and enter the cell or that another unknown receptor could be involved.
The Hybrid Lysin Is Not Inactived by Pesticin Immunity Protein.
We next asked whether Pim could inhibit the toxicity of our hybrid lysin. Y. pestis strains producing pesticin are protected from cell death by Pim. To test whether the hybrid lysin could overcome the natural immunity conferred by Pim, we constructed a vector that expresses the fyuA and pim genes concomitantly. Fig. 2B shows that the expression of Pim protects against killing by pesticin: E. coli cells transformed with this vector were not killed by pesticin at a concentration of 100 μg/mL. In contrast, adding 100 μg/mL of the hybrid lysin to cells expressing Pim and FyuA resulted in cell death (Fig. 2B). Therefore, the pesticin-lysozyme hybrid can overcome the natural immunity of Pim-producing strains giving it an advantage over pesticin by extending its potential therapeutic application to a wide variety of Yersiniae.
Yersinia and UPEC Strains Are Sensitive To Pesticin and the Hybrid Lysin.
To confirm that our observations in the E. coli model system were consistent with the response of bacteria that naturally express FyuA, we tested Y. pestis and Y. pseudotuberculosis for their sensitivity to pesticin and the hybrid lysin (Fig. S6). Pesticin killed Y. pseudotuberculosis and Y. pestis KIM10+ but not Y. pestis KIM6+. The presence of the pPCP1 plasmid (24) that encodes pesticin and Pim renders KIM6+ insensitive to pesticin action. On the other hand, Y. pestis KIM6+ was sensitive to killing by the hybrid lysin, though to a lesser degree compared to Y. pestis KIM10+. These data indicate that the hybrid can circumvent the action of Pim in Y. pestis. A possible explanation for why the hybrid lysin does not kill Y. pestis KIM6+ as efficiently as Y. pestis KIM10+ could be the presence of the outer membrane protease Pla in the KIM6+ strain but not in the KIM10+ strain. It is possible that Pla can degrade the hybrid lysin on the cell surface before it can be transported to the periplasm. Because the hybrid lysin kills E. coli in the presence or absence of Pim at similar levels (Fig. 2), we think it is unlikely that Pim partially deactivates the hybrid lysin in Y. pestis but not in E. coli.
The bactericidal activities of pesticin and hybrid lysin were also tested against E. coli clinical isolates. We tested both proteins against three uropathogenic E. coli (UPEC) laboratory strains and eighteen clinical isolates whose fyuA genotype had been previously determined (25). Of these, all fyuA+clinical isolates and laboratory strains were susceptible to pesticin and hybrid causing 98% and 90% reduction in bacterial viability, respectively (Fig. S7). Importantly, the bactericidal effect of pesticin and hybrid lysin was observed against recurrent and asymptomatic E. coli clinical isolates indicating that the origin of the strains does not affect bactericidal activity of these proteins; however, neither pesticin nor hybrid lysin is bactericidal against fyuA- strains indicating that these toxin proteins retain specificity for pathogenic subtypes expressing fyuA.
Investigation of Pesticin and Hybrid Action by Cryo-Electron Microscopy.
Next, we used cryo-electron microscopy to visualize changes in cell morphology upon addition of T4 lysozyme, hybrid lysin, and pesticin proteins. Intact cells are too thick to yield useful information except around their periphery where the trilaminar envelope is clearly visualized. In an untreated E. coli fyuA+ control sample, almost all observed cells (∼90%) were intact and had normal envelope morphology; however, a small but significant fraction of cells (∼8%) was visibly perturbed and represent the background level of cell disruption that occurs in an untreated culture (Fig. 3A and Fig. S8A). Addition of hybrid lysin had a dramatic effect; ∼45% of observed cells exhibited gross disruption characterized by vesiculation of the inner and outer cell membranes and loss of cellular integrity (Fig. 3 B and C and Fig. S8 A and E). This kind of vesiculation was not seen when cells were treated with either T4 lysozyme or pesticin (Fig. S8 A, C, and D). In contrast to hybrid lysin treatment, the distribution of cell morphologies observed after T4 lysozyme treatment was not altered to a statistically significant degree though the percentage of disrupted cells (∼16%) was slightly higher than the untreated control. Treatment with pesticin resulted in significant but less dramatic disruption of ∼21% of cells (Fig. S8A). Pesticin disruption appeared distinct from hybrid protein disruption and is characterized by the appearance of holes in the outer membrane and eventual cell rupture (Fig. S8 C and D). The distinct morphological characteristics of cells disrupted by the hybrid lysin vs. pesticin may indicate that the two proteins are bacteriolytic through differing mechanisms of action; but, further experiments are required to confirm this.
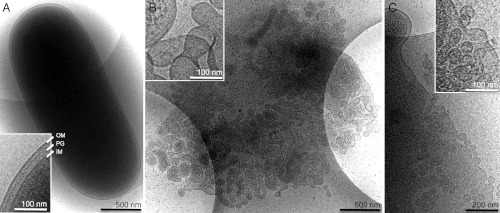
Fig. 3.
Cryo-EM of fyuA+ E. coli cells treated with the hybrid lysin. Electron micrographs of fyuA+ E. coli cells plunge-frozen on holey-carbon mesh coated sample grids. Cells were incubated with (A) no treatment (control) or (B and C) 100 μg/mL hybrid lysin for 10 min before being plunge-frozen. Extensive membrane breakdown of the treated cell in (B) is apparent. As a result, the cell is weakened and, consequently, spreads and flattens in the thin film of buffer while largely preserving its 3D surface area. As a result, it projects a larger 2D area in the micrograph. Also, as it is so much thinner, the membrane vesiculation is evident all over the cell, not just around the edges. The insets show typical regions of the corresponding cell envelopes at higher magnification. The intact inner membrane (IM), peptidoglycan layer (PG), and outer membrane (OM) are indicated and labeled in (A). Characteristic vesiculation of inner and outer cell membranes is shown in (B) and (C).
Transport Across the Outer Membrane.
Our bactericidal experiments establish that pesticin and the hybrid lysin require FyuA for transport across the outer membrane, and the electron microscopy images allow visualization of cell destruction once the toxin has entered the periplasm; however, the import pathway remains unclear. If pesticin and the hybrid lysin were transported through the FyuA β-barrel, then unfolding of the bacteriocin would likely be required because the dimensions of the pore do not appear to accommodate fully folded pesticin or hybrid (Fig. 4). To probe the import pathway further, we created a thermostable hybrid lysin by fusing a fully active T4 lysozyme mutant containing two disulfide bonds (26) to the FyuA-binding domain of pesticin. The disulfide bonds would presumably prevent unfolding of the T4L domain under conditions of the bactericidal assay. To confirm proper fold and disulfide bond formation, the thermostable bacteriocin was crystallized and the structure solved by molecular replacement to 1.8 Å resolution (Table S1 and Fig. S4 B and C). Although the killing domain of this protein is unlikely to unfold, the thermostable hybrid killed fyuA+ E. coli almost as effectively as the “wild type” hybrid with 0.8% survival for the hybrid lysin compared to 10% survival for the thermostable hybrid lysin. This result suggests that the import pathway does not require complete unfolding of the bacteriocin.
Fig. 4.
The pore of the FyuA β-domain is too small to transport directly folded pesticin or the thermostable hybrid across the outer membrane. The β-domain of FyuA is shown in light green and the plug domain in red, yersiniabactin (ybt) in spheres, pesticin (pst) in gold, and thermostable pst-hybrid (TS-hybrid) in dark green with the location of the engineered disulfide shown as yellow spheres (Fig. S4). For comparison, dimensions of the FyuA pore, Ybt, Pst, and TS-hybrid are shown with units in nanometers. The top row represents a periplasmic view up through the pore of the FyuA beta domain, whereas the middle row represents a membrane (side) view. The bottom row shows an overlay of FyuA with Ybt and Pst illustrating the size restrictions preventing direct transport of Pst (assuming the plug domain is fully ejected).
Discussion
T4 lysozyme has long been known to lyse bacteria by disrupting the peptidoglycan layer found between the outer and inner membranes of E. coli; however, this process works from “inside-out” because T4 lysozyme is normally generated within the cytoplasm of a phage-infected E. coli cell where it subsequently facilitates cell lysis and the release of assembled phages. By attaching an FyuA “targeting” domain to the N-terminus of T4 lysozyme, we have been able to make the process work in the opposite direction. This engineered version of T4 lysozyme can be simply added to the external medium and will kill Gram-negative bacteria expressing FyuA without the need for added accessory proteins. Our data show that the engineered viral lysin can traverse the outer membrane in order to reach the peptidoglycan layer. This approach overcomes one of the major limitations to the general use of purified lysins in therapy, i.e., their ability to lyse only Gram-positive bacteria where the peptidoglycan is exposed to the exterior and their lack of effect on Gram-negative bacteria (27).
By evading the pesticin immunity protein, our hybrid toxin can kill a broader range of pathogenic bacteria giving it a distinct advantage compared to wild type pesticin. Furthermore, Y. pestis uses pesticin to maintain maximal virulence by producing pesticin, Pim, and Pla from the same plasmid (pPCP1). Isolates of Yersinia pestis that do not express Pla due to loss of this plasmid become much less virulent (28, 29) and, because they no longer produce Pim, can be eliminated by pesticin-producing virulent strains. Thus, by creating the hybrid lysin, we have effectively subverted a weapon that Y. pestis uses to maintain maximal virulence and turned it, at least in principle, on itself. In addition, Pim is a periplasmic protein suggesting that pesticin first crosses the outer membrane and is then inactivated by Pim in the periplasm (10). Because replacing the C-terminal domain of pesticin with T4 lysozyme restores killing of E. coli that express pim, inactivation likely involves an interaction between Pim and the C-terminal domain of pesticin.
This engineered lysin is specifically targeted to the TonB dependent transporter FyuA that is a major virulence factor of several pathogenic bacterial species. Importantly, FyuA is also primarily associated with pathogenic bacteria, and not normal gut flora, allowing our hybrid lysin to target disease-causing bacteria specifically. The physiological role of FyuA is to transport chelated iron against a concentration gradient into the bacterial cell. Iron uptake by this route is, in some cases, essential. FyuA was shown to be required for virulence in a mouse model of plague infection (12, 13). Together with other genes that make up a genetic feature known as the “Yersinia high pathogenicity island”, fyuA is also prevalent in E. coli that are pathogenic to humans including the E. coli strain O104:H4 that caused a recent outbreak of hemolytic uremic syndrome in Germany (14, 15). High frequencies of fyuA have been demonstrated in genomes of E. coli isolated from blood or urine in disease states like sepsis (30), urosepsis (31), urinary tract infections e.g., pyelonephritis (32), and diarrhea (15). In another study, the presence of the fyuA trait was most closely associated with the ability of E. coli isolates to kill mice in a lethality assay (33). Additionally, FyuA appears to be critical for efficient biofilm formation by urinary tract infectious E. coli in human urine (34). Given its clear clinical relevance, FyuA has been proposed as a particularly useful target for therapeutic intervention (35), and our hybrid lysin thus provides an FyuA-specific tool to potentially combat bubonic and pneumonic plague as well as various E. coli infections. It should be noted that the bactericidal activity of the hybrid toxin will likely need to be improved for it to be medically relevant. Future studies could focus on increasing the efficacy of our hybrid lysin against pathogenic bacteria that carry the fyuA gene perhaps through subtle modifications of the existing construct or by substituting alternate killing domains. From a more general medical perspective, the hybrid lysin approach offers a possibility to target pathogenic Gram-negative bacteria that express specific TonB dependent transporters and to avoid selective pressure on native bacterial flora. Indiscriminate killing of the entire bacterial flora—pathogenic or not—by wide spectrum antibiotics may be contributing to the continuing rise of antibiotic resistance.
Materials and Methods
The complete methods are presented in the SI Material and Methods. The pesticin, pim, and fyuA genes were amplified from Y. pestis (strain 195/P) genomic DNA and cloned into pET vectors (Novagen) for expression. Seleno-methionine derivatized FyuA or pesticin was produced using SelenoMet medium (Molecular Dimensions Limited). FyuA, pesticin, and hybrid lysin were purified using immobilized metal affinity, anion exchange, and size exclusion chromatography steps. FyuA was concentrated to 12 mg/mL and crystallized from 2.4 M ammonium formate and 0.1 M Hepes at pH 6.8. Pesticin, hybrid lysin, and thermostable hybrid lysin were concentrated to 12 mg/mL and crystallized from 20% wt/vol PEG3350 and 0.25 M CaCl2. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) and General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) beamlines at the Advanced Photon Source (Argonne, IL). Cryo-electron microscopy was performed on E. coli cells transformed with pET20b-fyuA+ by incubating cells in culture with or without the hybrid lysin for 10 min, applying samples to holey carbon grids, and plunge-freezing in liquid ethane.
Supplementary Material
ACKNOWLEDGMENTS.
We thank S. Adhya for Yersinia strains; D. Nelson for Gram-positive strains; W.G. Coleman for help with bactericidal assays; R. Sougrat for help with early electron microscopy experiments; R. Bonocora, D. Hinton, I. Hook-Barnard, C. Khursigara and K. Swartz for discussions; and S. Adhya, H.D. Bernstein, and D. Hinton for critically reading the manuscript. This work is supported by the Intramural Research Program of the NIH, NIDDK, NIAID, and NIAMS. S.K.B. and B.J.H. acknowledge support from a trans-NIH Biodefense grant from the NIAID. J.P.H acknowledges support from the Career Award for Medical Scientists from the Burroughs Wellcome Fund and NIH grants K12 HD001459-09 and P50 DK064540. We thank the respective staffs at the SER-CAT and GM/CA-CAT beamlines at the Advanced Photon Source, Argonne National Laboratory for their assistance during data collection. Use of the Advanced Photon Source was supported by the US DOE, Basic Energy Sciences, Office of Science under Contract No. W-31-109-Eng-38 (SER-CAT) and the US DOE, Basic Energy Sciences, Office of Science under contract No. DE-AC02-06CH11357 (GM/CA-CAT).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203472109/-/DCSupplemental.
References
- 1.d’Herelle F. Sur un microbe invisible antagoniste des bacilles dysentériques. C R Acad Sci. 1917;165:373–375. [Google Scholar]
- 2.Twort FW. An investigation on the nature of ultra-microscopic viruses. Lancet. 1915;186:1241–1243. doi: 10.1017/s0022172400043606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischetti VA, Nelson D, Schuch R. Reinventing phage therapy: Are the parts greater than the sum? Nat Biotechnol. 2006;24:1508–1511. doi: 10.1038/nbt1206-1508. [DOI] [PubMed] [Google Scholar]
- 4.Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 5.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 6.Cascales E, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuenemann VJ, et al. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc Natl Acad Sci USA. 2011;108:E746–E752. doi: 10.1073/pnas.1105107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brubaker RR, Beesley ED, Surgalla MJ. Pasteurella pestis: Role of pesticin I and iron in experimental plague. Science. 1965;149:422–424. doi: 10.1126/science.149.3682.422. [DOI] [PubMed] [Google Scholar]
- 9.Eren E, Murphy M, Goguen J, van den Berg B. An active site water network in the plasminogen activator pla from Yersinia pestis. Structure. 2010;18:809–818. doi: 10.1016/j.str.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Pilsl H, Killmann H, Hantke K, Braun V. Periplasmic location of the pesticin immunity protein suggests inactivation of pesticin in the periplasm. J Bacteriol. 1996;178:2431–2435. doi: 10.1128/jb.178.8.2431-2435.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heesemann J, et al. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 12.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Bielaszewska M, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infect Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 15.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferber DM, Brubaker RR. Mode of action of pesticin: N-acetylglucosaminidase activity. J Bacteriol. 1979;139:495–501. doi: 10.1128/jb.139.2.495-501.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vollmer W, Pilsl H, Hantke K, Holtje JV, Braun V. Pesticin displays muramidase activity. J Bacteriol. 1997;179:1580–1583. doi: 10.1128/jb.179.5.1580-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fetherston JD, Lillard JW, Jr., Perry RD. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 21.Matthews BW, Remington SJ. The three dimensional structure of the lysozyme from bacteriophage T4. Proc Natl Acad Sci USA. 1974;71:4178–4182. doi: 10.1073/pnas.71.10.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroki R, Weaver LH, Matthews BW. A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme. Science. 1993;262:2030–2033. doi: 10.1126/science.8266098. [DOI] [PubMed] [Google Scholar]
- 23.Jakes KS, Finkelstein A. The colicin Ia receptor, Cir, is also the translocator for colicin Ia. Mol Microbiol. 2010;75:567–578. doi: 10.1111/j.1365-2958.2009.06966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry RD, Pendrak ML, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson JP, et al. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5:e1000305. doi: 10.1371/journal.ppat.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura M, Matthews BW. Control of enzyme activity by an engineered disulfide bond. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 27.Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005;13:491–496. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Lathem WW, Price PA, Miller VL, Goldman WE. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. 2007;315:509–513. doi: 10.1126/science.1137195. [DOI] [PubMed] [Google Scholar]
- 29.Sodeinde OA, et al. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 30.Jaureguy F, et al. Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis. Clin Microbiol Infect. 2007;13:854–862. doi: 10.1111/j.1469-0691.2007.01775.x. [DOI] [PubMed] [Google Scholar]
- 31.Bingen-Bidois M, et al. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect Immun. 2002;70:3216–3226. doi: 10.1128/IAI.70.6.3216-3226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houdouin V, et al. Phylogenetic groups and virulence factors of Escherichia coli strains causing pyelonephritis in children with and without urinary tract abnormalities. Clin Microbiol Infect. 2007;13:740–742. doi: 10.1111/j.1469-0691.2007.01748.x. [DOI] [PubMed] [Google Scholar]
- 33.Johnson JR, et al. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J Infect Dis. 2006;194:1141–1150. doi: 10.1086/507305. [DOI] [PubMed] [Google Scholar]
- 34.Hancock V, Ferrieres L, Klemm P. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology. 2008;154:167–175. doi: 10.1099/mic.0.2007/011981-0. [DOI] [PubMed] [Google Scholar]
- 35.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.