Abstract
In the diploid cells of most organisms, including humans, each chromosome is usually distinguishable from its partner homolog by multiple single-nucleotide polymorphisms. One common type of genetic alteration observed in tumor cells is uniparental disomy (UPD), in which a pair of homologous chromosomes are derived from a single parent, resulting in loss of heterozygosity for all single-nucleotide polymorphisms while maintaining diploidy. Somatic UPD events are usually explained as reflecting two consecutive nondisjunction events. Here we report a previously undescribed mode of chromosome segregation in Saccharomyces cerevisiae in which one cell division produces daughter cells with reciprocal UPD for the same pair of chromosomes without an aneuploid intermediate. One pair of sister chromatids is segregated into one daughter cell and the other pair is segregated into the other daughter cell, mimicking a meiotic chromosome segregation pattern. We term this process “reciprocal uniparental disomy.”
Keywords: genome stability, aneuploidy
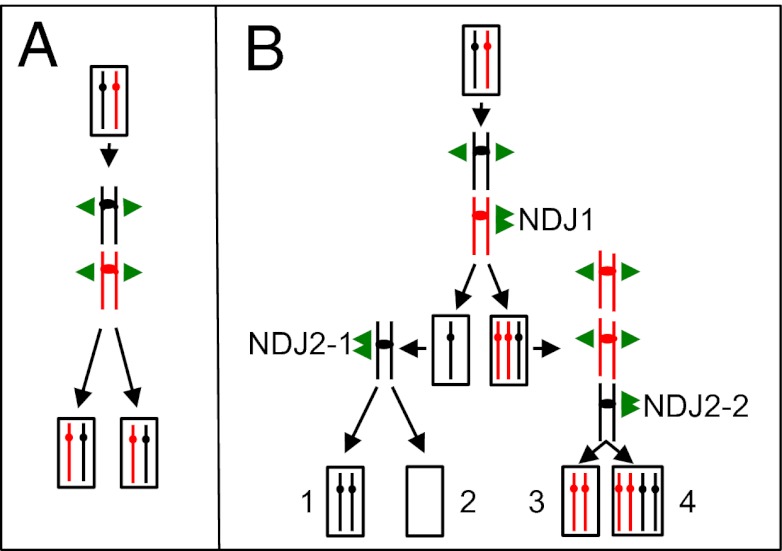
One of the basic tenets of biology is that mitosis results in two daughter cells that are identical to the original mother cell. This genetic identity reflects the duplication of each homolog to form a pair of sister chromatids, followed by a chromosome disjunction process in which the centromere is split and the two sister chromatids are disjoined into different daughter cells (Fig. 1A). In diploid cells, failure of correct chromosome disjunction produces cells that lack one homolog copy (monosomy) or have an extra copy (trisomy). In the yeast Saccharomyces cerevisiae, as in mammals, aneuploid cells grow slowly relative to euploid cells (1–3).
Fig. 1.
Normal chromosome disjunction vs. NDJ leading to UPD. The two homologs are depicted as red or black lines, with the centromeres shown as circles or ovals. The single green arrows indicate normal segregation of the two sister chromatids into different daughter cells, and the double green arrows indicate NDJ of two chromatids into a single daughter cell. (A) Normal mitotic chromosome disjunction in a diploid. The daughter cells have the same chromosomes as the original mother cell. (B) Pathways of mitotic chromosome NDJ resulting in UPD. In the first NDJ event (NDJ1), the black homolog disjoins normally and the red homolog undergoes NDJ. As shown on the left, the monosomic strain undergoes a second NDJ event (NDJ2-1), generating one UPD cell (1) and one cell lacking both copies of the homolog (2). On the right, the trisomic cell undergoes a second NDJ (NDJ2-2), producing one cell with UPD (3) and one cell with four copies of the homolog (4).
In general, the two homologs of a diploid cell are heterozygous for many SNPs. For example, in humans, each pair of homologs is heterozygous for an average of 106 SNPs (4, 5). Regions of chromosomes or entire chromosomes can undergo loss of heterozygosity (LOH), however. The term “uniparental disomy” (UPD) has been used to describe LOH events in diploid cells in which a region of a chromosome or an entire chromosome is derived from the homolog of only one parent (6). UPD events have been associated with a variety of disorders involving recessive mutations or genetic imprinting (7, 8) and are common in tumor cells (9). Somatic LOH events involving a portion of a chromosome are likely to be produced by a different mechanism from those involving the entire chromosome (mitotic recombination/deletion and nondisjunction [NDJ], respectively). In this paper, we restrict the definition of UPD to mitotic LOH events that involve the entire chromosome (isodisomy), although meiotic UPD events and segmental UPD also have been observed and implicated in human disease (8).
It has been suggested that UPD in somatic cells reflects two independent NDJ events (8, 9). In one pathway to UPD formation, NDJ1 yields a monosomic cell, and a subsequent NDJ event (NDJ2-1) produces UPD; in another pathway, NDJ1 yields a trisomic cell that undergoes a second NDJ (NDJ2-2) to produce UPD (Fig. 1B). Although these pathways are plausible models for the production of UPD, there is no direct experimental evidence that UPD is generated by these mechanisms. Here we provide evidence that UPD in S. cerevisiae occurs through a different pathway than those shown in Fig. 1.
Two difficulties impede the analysis of somatic LOH events in yeast and other eukaryotes. First, these events are rare; for example, the frequency of LOH resulting from mitotic recombination in yeast is approximately 10−5 for an average chromosome arm (10), and the frequency of chromosome loss is approximately 10−6 (11, 12). Second, the methods developed to select LOH events usually detect a subset of the potential classes of events. We previously developed a genetic method for selecting reciprocal mitotic crossovers on chromosome V in yeast (10), as described in further detail below. In strains with a crossover, we can map the position of the recombination event using microarrays that can detect LOH of heterozygous SNPs (13). In the analysis described here, we find that approximately 5% of the LOH events observed using this system result not from crossovers, but rather from a unique and unexpected pathway that produces reciprocal UPD.
Results
Experimental System.
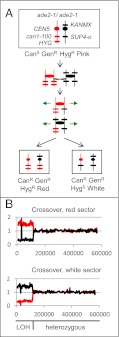
The diploid strain (SLA11.7) that we used to select LOH events (mostly representing mitotic crossovers) is shown in Fig. 2A. One chromosome V homolog contains a recessive ochre mutation in the CAN1 gene (can1-100); cells with the WT CAN1 gene are sensitive to the drug canavanine (CanS). At the allelic position on the other homolog is the SUP4-o gene, which encodes an ochre suppressor. In addition, the strain is homozygous for the ade2-1 ochre mutation on chromosome XV. In the absence of the ochre suppressor SUP4-o, ade2-1 strains form red colonies instead of the white colonies observed for WT strains. In diploid strains with one copy of the SUP4-o gene, pink colonies are formed. Thus, the SLA11.7 strain is CanS and forms pink colonies. It is also heterozygous for the dominant drug resistant markers HYG, resulting in resistance to hygromycin (HygR), and KANMX, resulting in resistance to geneticin (GenR). A reciprocal crossover between the can1-100/SUP4-o markers and the centromere results in a canavanine-resistant (CanR) red/white-sectored colony in which the red sector is both GenR and HygR and the white sector is GenR but sensitive to hygromycin (HygS) (Fig. 2A).
Fig. 2.
Detection of reciprocal crossovers leading to LOH events. (A) Pattern of marker segregation resulting from a reciprocal crossover. At the top of the panel, we show the arrangements of markers on chromosome V in the diploid strain SLA11.7. The chromosome is shown as a vertical line, the location of the markers by short horizontal lines, and the centromere by circles; red and black signify that the chromosomes were derived from the W303a-related haploid and the YJM789-related haploid, respectively. As discussed in the text, the SUP4-o–encoded tRNA partially suppresses the red colony phenotype associated with the ade2-1 mutation and suppresses the canavanine-resistance phenotype associated with can1-100. A reciprocal crossover event can produce two canavanine-resistant daughter cells. One daughter cell is canavanine-resistant and forms a red sector because it lacks the SUP4-o gene, and the other is canavanine-resistant because it lacks any CAN1 gene and forms a white sector because it has two copies of SUP4-o. Both sectors retain one copy of the KANMX gene and thus are geneticin-resistant. (B) Analysis of genomic DNA isolated from red and white sectors resulting from a reciprocal crossover (sectored colony PG311-16B; ref. 13). Genomic DNA was purified from each sector of a canavanine-resistant red/white -sectored colony and examined by microarrays capable of distinguishing LOH at a resolution of 1 kb throughout the genome. We measured the normalized hybridization of genomic DNA from the experimental strains and the control diploid to oligonucleotides with SNPs specific to W303a or YJM789 (the haploid strains used to construct the diploid). The y-axis shows the ratio of hybridization (experimental/control) for SNP-specific oligonucleotides, with red and black lines indicating hybridization to W303a- and YJM789-specific oligonucleotides, respectively. The x-axis shows Saccharomyces Genome Database (SGD) coordinates. The red sector has an LOH event in which W303a-derived SNPs become homozygous, and the white sector has the reciprocal LOH event. This pattern is consistent with a reciprocal crossover occurring near SGD coordinate 120,000. The “spike” of hybridization near SGD coordinate 30,000 (Fig. 3B) is an artifact reflecting the substitution of the can1 gene with SUP4-o (13).
SLA11.7, constructed by mating two sequence-diverged haploid strains (W303a and YJM789), is heterozygous for approximately 55,000 SNPs (14). Mitotic crossovers result in LOH of markers that are centromere-distal to the exchange (Fig. 2A), and we used oligonucleotide-containing microarrays (15) to detect LOH. The details of the microarray construction and analysis of LOH using these microarrays have been described by St. Charles et al. (13). In brief, we prepared microarrays in which 13,000 of the 55,000 heterozygous SNPs were assayed (average genome density of one SNP/kb). We used four 25-base oligonucleotides for each SNP, two for the Watson and Crick strands of the W303a form of the SNP and two for the YJM789 form; the SNP was located in the middle of the oligonucleotide. We competitively hybridized genomic DNA isolated from red or white sectors with differentially labeled genomic DNA from the heterozygous control strain. In strains heterozygous for a SNP at a particular genomic location, we observed a normalized hybridization signal of ∼1. In strains with an LOH event involving a heterozygous SNP, we observed increased hybridization to the oligonucleotides specific to one haploid strain and decreased hybridization to the oligonucleotides specific to the other haploid strain (13). Fig. 2B shows the patterns of hybridization of genomic DNA isolated from the red and white sectors of a CanR colony, with the red line representing hybridization to the W303a-specific oligonucleotides and the black line representing hybridization to the YJM789-specific oligonucleotides. The pattern of hybridization observed in this sectored colony is that expected from a reciprocal crossover located approximately 120 kb from the left telomere.
Evidence for Reciprocal UPD.
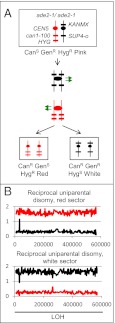
The rate of CanR red/white-sectored colonies was ∼1.6 × 10−6/division. If these colonies reflect a reciprocal mitotic crossover, then the expected phenotype of the red sector is CanR GenR HygR, and the expected phenotype of the white sector is CanR GenR HygS (Fig. 2A). Of 106 CanR red/white-sectored colonies examined, 101 had these phenotypes. In five colonies, however, the red sector was CanR GenS HygR, and the white sector was CanR GenR HygS. These phenotypes indicate a “reciprocal UPD” (RUD) event (Fig. 3A).
Fig. 3.
Diagnosis of RUD. (A) Pattern of marker segregation resulting from a RUD event. The chromosomes are depicted in the same way as in Fig. 2A. In the RUD event, unlike the reciprocal crossover shown in Fig. 2A, the cells in the red sector are geneticin-sensitive. (B) Analysis of genomic DNA isolated from red and white sectors from a RUD sectored colony [SLA11.7(43)]. Genomic DNA was purified from each sector of a canavanine-resistant red/white-sectored colony and examined as described for Fig. 2B. The red sector exhibits the hybridization patterns consistent with UPD for the W303a-derived chromosome, and the white sector has the patterns expected for UPD for the YJM789-derived chromosome.
To confirm this conclusion, we examined genomic DNA isolated from the red and white sectors by SNP microarrays. The red sector had two copies of the W303a-derived homolog, whereas the white sector had two copies of the YJM789-derived homolog (Fig. 3B). This observed hybridization pattern is not consistent with monosomy for cells of the red or white sectors, given that monosomic strains have a hybridization ratio of ∼1 for one homolog and a low hybridization ratio for the other homolog. It also should be emphasized that there is no evidence for mitotic recombination between the two homologs. Such events would be detectable by a local region of heterozygosity. However, we cannot rule out the possible existence of a recombination event occurring in chromosome regions in which there are no SNPs, such as the telomeres. In total, we observed 11 RUD events, including 10 events derived from SLA11.7 and one event derived from the closely related strain PG311. The frequency of RUD for chromosome V was ∼0.8 × 10−7/cell division. These results demonstrate that cells with UPD can be formed by a mechanism that does not proceed through an aneuploid intermediate.
One potential artifactual explanation of our results is that the red/white sectors are generated by spore–spore matings in a small fraction of cells that undergo meiosis. This possibility is highly unlikely, for two reasons. First, the diploid strains used in our study were deleted for MATα (one of the two mating type loci in S. cerevisiae) and thus are incapable of undergoing meiosis (16). Second, meiosis in yeast is associated with high levels of meiotic recombination (more than one crossover per chromosome per meiosis), and no chromosome V recombination events were observed in any of the 11 RUD events. We also examined LOH on all yeast chromosomes in the 11 strains with RUD events (Table 1). None of these strains had more than two LOH events per genome, a much lower frequency than expected for meiotic products.
Table 1.
Unselected genomic alterations in sectored colonies with RUD events
| Strain name | Sector color | Event type (chromosome) |
| SLA11.7(2) | Red | Partial trisomy* (extra copy of W303a-derived chromosome XII) |
| SLA11.7(2) | White | None |
| SLA11.7(5) | Red | Gene conversion (chromosome XII)† |
| SLA11.7(5) | White | None |
| PG311(8A) | Red | Gene conversion (chromosome II) |
| Trisomy (extra copy of YJM789-derived chromosome XII) | ||
| Trisomy (extra copy of W303a-derived chromosome XIII) | ||
| PG311(8A) | White | None |
| SLA11.7(21) | Red | None |
| SLA11.7(21) | White | None |
| SLA11.7(21B) | Red | Monosomy (loss of YJM789-derived chromosome I) |
| SLA11.7(21B) | White | Trisomy (extra copy of YJM789-derived chromosome I) |
| SLA11.7(43) | Red | None |
| SLA11.7(43) | White | None |
| SLA11.7(50) | Red | None |
| SLA11.7(50) | White | None |
| SLA11.7(76) | Red | None |
| SLA11.7(76) | White | None |
| SLA11.7(209) | Red | Gene conversion (chromosome VII) |
| Trisomy (extra copy of W303a-derived chromosome IX) | ||
| SLA11.7(209) | White | Gene conversion (chromosome I) |
| Gene conversion (chromosome VII, identical to red sector event) | ||
| Trisomy (extra copy of W303a-derived chromosome IX) | ||
| SLA11.7(251) | Red | None |
| SLA11.7(251) | White | None |
| SLA11.7(264) | Red | None |
| SLA11.7(264) | White | None |
Sectored colonies with RUD events were examined for loss or duplication of sequences throughout the genome using SNP arrays. For most of the sectored colonies, we pooled multiple colonies derived from each side of the sectors; however, for sectored colonies SLA11.7 (2), SLA11.7 (5), and PG311 (8A), we analyzed only a single colony derived from the red and white sectors.
*In this strain, the microarrays indicated that there were two or three copies of chromosome XII. This pattern could be produced as a consequence of an NDJ event that occurred in the red sector in an early cell division with selection for cells with the extra copy of XII and selection against cells missing a copy of XII.
†Interstitial regions of LOH were classified as gene conversion events.
Frequency of Chromosome NDJ in SLA11.7.
One explanation for RUD events is that they represent two independent NDJ events in which one pair of sister chromatids disjoins into one daughter cell and the second pair of disjoins into the other daughter cell. As described in Methods, we measured the rate of loss of the SUP4-o–containing copy of chromosome V (2.8 × 10−7/division) and, in separate experiments, the rate of loss of the can1-100–containing copy of chromosome V (6.3 × 10−7/division). If the rate of chromosome loss is equivalent to the rate of NDJ, then the expected rate of RUD resulting from two independent NDJ events in one cell cycle is one-half of the product of the NDJ rates for each chromosome, or 8.8 × 10−14. Because this rate is six orders of magnitude lower than the observed RUD rate of 0.8 × 10−7/division, we conclude that RUD events are not the consequence of two independent NDJ events. However, this conclusion is valid only if the cells that undergo RUD have the same frequency of NDJ as other cells in the population, as discussed in more detail below. Importantly, the rate of RUD is only approximately 10-fold less than the rate of single-chromosome NDJ events.
Explaining our RUD events by the models shown in Fig. 1B would require that aneuploid cells revert to diploidy very rapidly. Consequently, we examined the frequency with which a monosomic derivative of SLA11.7 returned to diploidy. The monosomic strain retaining the SUP4-o–containing homolog was isolated as a pink colony that was CanR GenR HygS; we confirmed that this strain had a single copy of chromosome V by microarrays. Given that strains with one copy of SUP4-o form pink colonies and derivatives with two copies of SUP4-o result in white colonies, we measured the frequency of pink/white colonies to estimate the rate at which the monosomic strain duplicated chromosome V. Although the observed rate of chromosome duplication was high (3.8 × 10−3/division) relative to the frequency of chromosome loss, it was much too low for the mechanisms shown in Fig. 1B to be a plausible explanation of the RUD events.
Genome Instability in Strains That Have Undergone RUD.
Although the frequency of RUD events is much too high to represent two independent NDJ events in SLA11.7, it is possible that the strains that undergo RUD have a transient or permanent alteration that increases genome instability. This suggestion of increased genomic instability in cells with RUD events is supported by our genome-wide analysis of LOH (Table 1). Unselected genomic alterations, including aneuploidy and interstitial LOH events (presumably gene conversions), were observed in 5 of the 11 sectored colonies resulting from RUD (Table 1). Five independent trisomic chromosomes and one monosomic chromosome were found in addition to four independent gene conversion events. In sectored colony SLA11.7(21B), the red sector was monosomic for chromosome I and the white sector was trisomic for chromosome I, as would be expected for a classic NDJ event. In previous work, we examined 13 sectored colonies derived from PG311 (closely related to SLA11.7; ref. 14) that had reciprocal crossovers rather than RUD and observed no aneuploidy in these strains, with only one colony exhibiting a gene conversion event (13). This difference in the number of aneuploidy events in strains with RUD and isogenic strains with crossovers is significant (P = 0.03, Fisher’s exact test), although the difference in the number of gene conversion events is not (P = 0.14).
The increased frequency of aneuploidy in cells with RUD events could reflect either a transient chromosome segregation defect occurring in some subset of cells in a WT strain or a genetic change (e.g., a mutation that increases NDJ). We tested whether strains that experienced a RUD event had subsequent high levels of genome instability in two experiments. We first subcultured cells derived from the red and white sectors of two RUD colonies [SLA11.7(21B) and SLA11.7(251)] from a single cell to a colony four times (representing approximately 100 cell divisions), and then tested whether the subcultured strains were aneuploid using the SNP microarrays. We found no alterations in ploidy; one subcultured strain had a gene conversion event on chromosome VII. Obtaining a frequency of 0.8 × 10−7 for RUD events representing two independent NDJ events would require an NDJ frequency of ∼4 × 10−4/homolog, or ∼1.3 × 10−2 NDJ events/diploid genome/division (calculated by multiplying 4 × 10−4 by 32, the number of homologs per diploid cell). The probability of not detecting NDJ in a strain subcultured 100 times [(0.987)100] is approximately 0.27, and the probability of no NDJ events in four such subcultured strains is (0.27)4, less than 1%.
We also directly measured the frequency of chromosome loss events in one strain derived from the red sector of an RUD event [SLA11.7(251)] and in two strains derived from red sectors of reciprocal crossover events [PG311(1.4) and PG311(1.7)]. Because these strains were homozygous for the can1-100 allele, we could not use the frequency of CanR cells as an assay for chromosome loss. Instead, as described in detail in Methods, we measured the rate of loss of a heterozygous URA3 gene located on chromosome V using medium containing 5-fluooroate (5-FOA) that selects against Ura+ cells (17). To calculate the rate of chromosome loss, we then determined the fraction of 5-FOA-resistant (5-FOAR) cells that had lost heterozygosity for the KANMX marker on the opposite arm of chromosome V. The rate of chromosome loss in the RUD strain (1.5 × 10−5/division) was approximately the same as that seen in the strains with reciprocal crossovers (average of 1.3 × 10−5/division). We also measured chromosome loss rates using 5-FOA in the parental diploid SL11.7; the rate was 3.1 × 10−6/division. Although the loss rates observed in the red sectors were slightly elevated (threefold to fivefold) compared with the rate in the parental strain (possibly due to the adenine auxotrophy), the persistent level of genomic instability was not significantly elevated in strains that underwent a RUD event. In addition, although these rates of chromosome loss as measured by 5-FOA resistance were high relative to the rates measured using the canavanine assay, they are similar to those reported in other experiments using the 5-FOA technique (12, 18). These results indicate that strains that have undergone RUD do not have a persistently high rate of chromosome NDJ, but likely experienced a transient genome-instability phenotype, as discussed below.
Discussion
We have shown that the yeast S. cerevisiae can produce two cells that experience RUD in a single mitotic division. Although the RUD events are rare (∼10−7/division), they are similar in frequency to other classes of mitotic genetic rearrangements (i.e., reciprocal crossovers and chromosome loss) in WT yeast strains. Because most aneuploid cells have a substantial growth disadvantage compared with euploid cells (19), an advantage of RUD as a pathway for producing UPD compared with the pathways diagrammed in Fig. 1B is the lack of aneuploid intermediate formation. It is possible that the UPD events observed in cells derived from many human tumors may reflect RUD instead of the consecutive NDJ events invoked previously. Further study of RUD in yeast has the potential to provide important insights into the mechanisms underlying UPD events associated with human disease.
We evaluated two plausible mechanisms for producing RUD: (i) a rare meiosis I-like segregation event for chromosome V and (ii) an infrequent transient elevation of chromosome NDJ. Meiosis I segregation is distinguished from mitotic and meiosis II segregation by two properties: establishment of a connection (usually a crossover) between the two homologs, and monopolar attachment of the kinetochores to the spindle. In our experiments, a crossover would result in LOH. Such events were not observed on chromosome V in RUD strains, although we cannot exclude the possibility of a crossover in regions very close (within 5 kb) to the telomeres. Alternatively, it is possible that a connection between the homologs is dictated by a different mechanism, such as the achiasmate meiosis I-like segregation (distributive pairing) described by Dawson et al. (20). This type of pairing requires the meiosis-specific Zip1p (21). Finally, we note that the chromosomes in spo13 diploid strains induced to undergo meiosis exhibit a mixture of reductional and equational segregation (22).
In addition to a connection between the two homologs, meiosis I segregation requires monopolar attachment of the kinetochores to the spindle. In meiosis I, the sister kinetochores are attached to microtubules emanating from the same spindle pole (monopolar attachment), whereas in meiosis II and mitosis, sister kinetochores are attached to microtubules emanating from different spindle poles (bipolar attachment). Although the molecular mechanisms that distinguish these two modes of attachment are incompletely understood, monopolar attachment is associated with cohesin binding at the core centromere (23), and the need for the monopolin complex to direct this binding has been suggested (24). Monje-Casas et al. (25) reported that the mitotic expression of the normally meiosis-specific monopolin complex was sufficient to direct meiosis I-like segregation of mitotic chromosomes. Thus, one possible scenario is that a small subset of mitotic cells expresses the meiotic proteins that allow monopolar kinetochore attachment. However, no RUD events for the other chromosomes occurred in strains with RUD for chromosome V (Table 1). This result might be a consequence of our selection regimen or might indicate that chromosome V is particularly susceptible to monopolar orientation in cells expressing low levels of the monopolins.
Thus, a meiosis I segregation pattern likely would require the expression of meiotic proteins such as Zip1p and proteins of the monopolin complex in mitotic cells. An alternative plausible mechanism is that RUD events are a consequence of transiently high levels of NDJ in a subset of WT cells. We found that the frequency of RUD events was too high to represent two independent chromosome NDJ events if these events occurred at the rates observed in a WT strain. Because the strains that have undergone RUD have significantly more unselected aneuploid events compared with strains that have not undergone RUD, the former strains must have either a transient or a permanent alteration that elevates chromosome NDJ. Our measurements of NDJ frequency in strains that have undergone RUD indicate that the change is transient rather than permanent. Even without these measurements, however, it is very unlikely that the RUD events could be a consequence of a genome-destabilizing mutation, for several reasons. First, the frequency of RUD events was very high (0.8 × 10−7) compared with the expected frequency if RUD required a new mutation and subsequent NDJ events, given the mutation rate per gene in WT cells of approximately 10−7 per division (26). Second, because all cells with RUD events were isolated from independent cultures, a mutation or mutations causing the same phenotype would have to occur repeatedly. Finally, because our experiments were performed in diploid strains, the mutations would have to be dominant, unlike most mutations.
We suggest that RUD events occur in a subset of WT cells that have transient defects in one of the mechanisms affecting chromosome segregation, such as the spindle assembly or decatenation checkpoints, sister chromatid cohesion, or attachment of the microtubules to the kinetochore (27, 28). For example, in some cells, a stochastic reduction of one of the proteins involved in the spindle assembly checkpoint might lead to an elevation in the frequency with which both kinetochores of a pair of sister chromatids become attached to the same spindle pole. It is also possible that the aneuploidy is a consequence of problems in DNA replication rather than of a defect in chromosome segregation itself. Reduction in DNA polymerase alpha levels leads to greatly elevated rates of chromosome loss and mitotic recombination (29). Although most of these stochastic events would be expected to affect all chromosomes in the cell to the same extent, some mechanisms might be chromosome-specific; for example, a reduced level of cohesins might increase NDJ more significantly for some chromosomes than others.
Although we cannot rule out the possibility that some yeast chromosomes undergo a meiosis I segregation pattern, we favor the explanation that some cells have a short-lived problem in chromosome segregation, given that this latter explanation does not require the expression of meiosis-specific proteins in mitosis. In addition, our observation of unselected chromosome NDJ events that were not RUD events is more consistent with the latter explanation. Whatever the mechanistic details, it is clear that yeast has an unexpected pathway for producing UPD that is likely conserved in other eukaryotes. The tools available in yeast should allow investigation of the genetic and epigenetic mechanisms underlying UPD.
Methods
Strains.
The relevant features of the PG311 diploid genotype are shown in Fig. 1, and the construction of PG311 has been described previously (14). SLA11.7 is an isogenic derivative of PG311 in which the V261553::LEU2 insertion is replaced by V261553::KANMX by standard procedures. The locations of markers in SLA11.7 (distances from left telomere in kb) are HYG (9), can1-100/SUP4-o (32), CEN5 (152), and KANMX (261). To measure chromosome loss using 5-FOA resistance, we transformed three ura3 mutant strains [the red sector of the RUD isolate SLA11.7(251) and the red sectors of colonies resulting from reciprocal crossovers, PG311(1.4) and PG311(1.7)] with a PCR fragment containing a WT URA3 gene (generated by amplifying genomic DNA of a yeast strain with the WT gene with the primers 5′-AGAACGAAGGAAGGAGCACA and 5′-GGAGTTCAATGCGTCCATCT); the resulting diploids were heterozygous for the URA3 marker. From these diploids, we generated derivatives heterozygous for the selectable drug resistance marker KANMX on the right arm of chromosome V; this transformation was performed using a PCR fragment generated by amplifying genomic DNA of SLA11.7 with the primers 5′-TAACCTCTGCCGGAAGTGAA and 5′-AGGGGGTTGCTATGACACGAC. In the Ura+ GenR derivative of SLA11.7(251), the KANMX marker was located on the same homolog as URA3.
Identifying LOH Events by Selecting CanR Red/White Sectored Colonies.
We identified CanR red/white-sectored colonies using the medium and experimental conditions described previously (14). Sectors were screened for RUD events on standard rich growth medium plates containing 200 μg/mL of hygromycin or 300 μg/mL of geneticin. For most of our analyses, we purified multiple red and white colonies from each sector and then combined several colonies of the same color for microarray analysis.
Microarray Analysis.
DNA was isolated from each sectored colony, labeled with Cy5-dUTP, and hybridized to microarrays in competition with control DNA samples labeled with Cy3-dUTP (13). We used microarrays prepared by Agilent Technologies (Custom HD-CGH Microarrays, 2 × 105k format; G4425A) and hybridization protocols specified by the manufacturer (http://www.chem.agilent.com/en-US/Search/Library/_layouts/Agilent/PublicationSummary.aspx?whid=52010) to examine approximately 13,000 SNPs (13). Arrays were scanned with an Axon GenePix 4000B scanner (Molecular Devices).
Chromosome Loss Rates.
Chromosome V loss rates leading to canavanine resistance were calculated by measuring the rates of unsectored CanR red and white/pink (indistinguishable under conditions used) colonies derived from SLA11.7 using fluctuation analysis (method of the median; ref. 30), and by determining the fraction of these CanR derivatives that had lost heterozygosity for an SNP located on the opposite chromosome arm from the can1 locus. Two methods were used to assess for LOH on the right arm of chromosome V. For the URA3 V261553::KANMX derivative of SLA11.7(251) and the parental SLA11.7 strain, 5-FOAR derivatives that were also geneticin-sensitive represented chromosome loss. For the other strains, LOH was assessed using an SNP located near the right end of chromosome V. The YJM789-derived and W303a-derived haploids differ at Saccharomyces Genome Database (SGD) coordinates 560715 and 560716, resulting in an EcoRI site at this position in YJM789 that is absent in W303a. We can detect LOH by PCR amplification of the region flanking the site, followed by treatment with EcoRI and gel electrophoretic analysis of the resulting fragments (14). The following primers were used for this analysis: 5′-TTCTCAGCCGTACAATCATGC and 5′-AAACTCCTTCCAAAGGGTCTGG.
Acknowledgments
We thank all members of the T.D.P. laboratory and R. Farber, S. Jinks-Robertson, L. Argueso, K. Bloom, D. Koshland, P. Hieter, and J. Sekelsky for discussions and advice. This work was supported by National of Institutes of Health Grants GM24110, GM52319, and 5RC1ES18091 (to T.D.P.) and National Cancer Institute Cancer Biology Training Grant 5T32CA059365-17 (to S.L.A.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Gerstein AC, Chun H-J, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 3.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelak K, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel E. A new genetic concept: Uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- 7.Spence JE, et al. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–226. [PMC free article] [PubMed] [Google Scholar]
- 8.Yamazawa K, Ogata T, Ferguson-Smith AC. Uniparental disomy and human disease: An overview. Am J Med Genet C Semin Med Genet. 2010;154C:329–334. doi: 10.1002/ajmg.c.30270. [DOI] [PubMed] [Google Scholar]
- 9.Tuna M, Knuutila S, Mills GB. Uniparental disomy in cancer. Trends Mol Med. 2009;15:120–128. doi: 10.1016/j.molmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Barbera MA, Petes TD. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiraoka M, Watanabe K, Umezu K, Maki H. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics. 2000;156:1531–1548. doi: 10.1093/genetics/156.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein HL. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics. 2001;159:1501–1509. doi: 10.1093/genetics/159.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St. Charles J, et al. High-resolution genome-wide analysis of irradiated (UV and gamma rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics. 2012;190:1267–1284. doi: 10.1534/genetics.111.137927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee PS, et al. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 2009;5:e1000410. doi: 10.1371/journal.pgen.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gresham D, et al. Optimized detection of sequence variation in heterozygous genomes using DNA microarrays with isothermal-melting probes. Proc Natl Acad Sci USA. 2010;107:1482–1487. doi: 10.1073/pnas.0913883107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopper AK, Hall BD. Mating type and sporulation in yeast, I: Mutations which alter mating-type control over sporulation. Genetics. 1975;80:41–59. doi: 10.1093/genetics/80.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 18.Craven RJ, Greenwell PW, Dominska M, Petes TD. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002;161:493–507. doi: 10.1093/genetics/161.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheltzer JM, Amon A. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson DS, Murray AW, Szostak JW. An alternative pathway for meiotic chromosome segregation in yeast. Science. 1986;234:713–717. doi: 10.1126/science.3535068. [DOI] [PubMed] [Google Scholar]
- 21.Newnham L, Jordan P, Rockmill B, Roeder GS, Hoffmann E. The synaptonemal complex protein, Zip1, promotes the segregation of nonexchange chromosomes at meiosis I. Proc Natl Acad Sci USA. 2010;107:781–785. doi: 10.1073/pnas.0913435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugerat Y, Simchen G. Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics. 1993;135:297–308. doi: 10.1093/genetics/135.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang GI, Murray AW. Estimating the per-base pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen MP, Nelson ZW, Hetrick ED, Gottschling DE. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics. 2008;179:1179–1195. doi: 10.1534/genetics.108.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stirling PC, et al. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011;7:e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase: A model for chromosome-fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 30.Kokoska RJ, Stefanovic L, DeMai J, Petes TD. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol Cell Biol. 2000;20:7490–7504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]