Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is highly prevalent in China and other Asian countries, as a major cause of cancer-related mortality. ESCC displays complex chromosomal abnormalities, including multiple structural and numerical aberrations. Chromosomal abnormalities, such as recurrent amplifications and homozygous deletions, directly contribute to tumorigenesis through altering the expression of key oncogenes and tumor suppressor genes.
Methodology/Principle Findings
To understand the role of genetic alterations in ESCC pathogenesis and identify critical amplification/deletion targets, we performed genome-wide 1-Mb array comparative genomic hybridization (aCGH) analysis for 10 commonly used ESCC cell lines. Recurrent chromosomal gains were frequently detected on 3q26-27, 5p15-14, 8p12, 8p22-24, 11q13, 13q21-31, 18p11 and 20q11-13, with frequent losses also found on 8p23-22, 11q22, 14q32 and 18q11-23. Gain of 11q13.3-13.4 was the most frequent alteration in ESCC. Within this region, CCND1 oncogene was identified with high level of amplification and overexpression in ESCC, while FGF19 and SHANK2 was also remarkably over-expressed. Moreover, a high concordance (91.5%) of gene amplification and protein overexpression of CCND1 was observed in primary ESCC tumors. CCND1 amplification/overexpression was also significantly correlated with the lymph node metastasis of ESCC.
Conclusion
These findings suggest that genomic gain of 11q13 is the major mechanism contributing to the amplification. Novel oncogenes identified within the 11q13 amplicon including FGF19 and SHANK2 may play important roles in ESCC tumorigenesis.
Introduction
Esophageal cancer is one of the most aggressive malignancies originated in the gastrointestinal tract, and ranks as the sixth leading cause of cancer-related deaths in the world [1]. Its incidence varies greatly among different regions worldwide, with China as a high-risk area. In some districts of north and central China, its incidence exceeds 100 cases/per 100,000 per year [2]. Histologically, esophageal cancer is classified as esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC). Most of the cases reported in US are esophageal adenocarcinomas, however in China and other Asian countries, ESCC is the predominant type that accounts for about 90% of all cases. Despite advances in multimodal therapies, ESCC remains a serious cancer-care problem in many countries with very low 5-year survival rates (<30%) [3]. Thus, it is of great clinical value to look for sensitive and specific biomarkers for the early detection and prognosis of this malignancy, as well as novel therapeutic targets.
Genomic amplifications and deletions contribute to human tumorigenesis by altering the expression levels of critical oncogenes and tumor suppressor genes (TSGs). In spite of its high prevalence, ESCC has not been studied as intensively as its adenocarcinoma counterpart. Efforts have been put to identify gross copy number alterations of both ESCC cell lines and tumors, including karyotyping, fluorescence in situ hybridization (FISH), conventional comparative genome hybridization (CGH) and loss of heterozygosity (LOH) analyses. According to available data published by now, the most commonly cited chromosomal amplifications in ESCC are 3q, 4q, 5p, 8p, 7q, 9q, 10q21, 11q13-q22, 18p11.3, 20q and 22qtel [4]–[12]. Amplifications harboring oncogenes, e.g. 11q13 (CCND1, EMS1), 3q26 (EIF5A2), 21q22 (ETS2), 8q24 (MYC), have been consistently observed in more than one studies [4]–[12]. Chromosomal losses recurrently involve 3p, 5q, 9p, 13q, 18q and 21q, in which target genes such as FHIT, APC, RB1 and CDKN2A are located [4]–[12]. In recent years, high resolution array-based CGH (aCGH) has been applied to identify target oncogenes and TSGs through defining recurrent gains and losses in various cancers. Until recently, two studies performed aCGH analysis on primary ESCC samples, revealing recurrent, high-level amplifications in 3q27.1, 7p11, 8q21.11, 8q24.21, 11q13.3, 11q22, 12q15–q21.1, 18q11.2, and 19q13.11–q13.12, and homozygous deletions in 4q34.3–q35.1 and 9p21.3 [13]; [14]. However, compared to “pure” ESCC cell lines, primary ESCC samples contain lots of normal cells which may affect aCGH results in different ways. Although several comprehensive whole genome studies on ESCC cell lines has been reported, the cell lines used are mainly originated from Japanese ESCC (TE series) and South African ESCC patients, respectively [15]–[17]. Profiling of multiple ESCC cell lines originated from different high-risk areas in Asian via aCGH will not only allow the identification of recurrent chromosomal changes in Asian ESCC, but also provide valuable insight for future studies using these cells lines as ESCC models.
In this study, we profiled 10 commonly used ESCC cell lines originated from mainland Chinese (EC1, EC18 and EC109), Hong Kong Chinese (HKESC1, HKESC2, HKESC3 and SLMT1) and Japanese (KYSE70, KYSE410 and KYSE520) patients for whole-genome DNA copy number alterations using aCGH analysis. Among identified alterations, amplification of 11q13 is the most frequent gain observed, harboring FGF19, SHANK2 and CCND1. We further found that CCND1 expression was frequently upregulated in primary ESCC tumors, and DNA amplification contributes to its overexpression, which is correlated with lymph node metastasis of primary ESCC tumors.
Results
Genomic Profiles of ESCC Cell Lines by 1-Mb aCGH
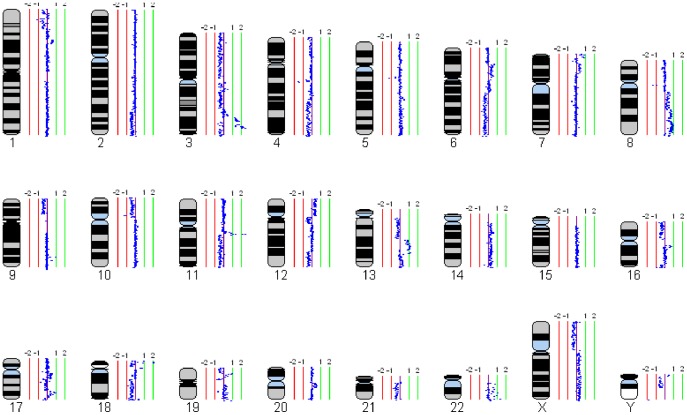
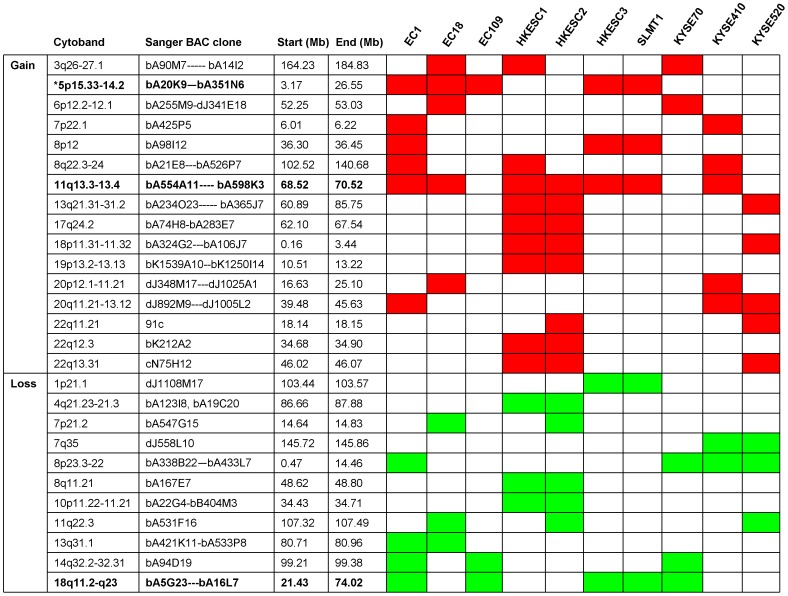
Ten ESCC cell lines were analyzed using 1-Mb aCGH (Sanger 3040-BAC/PAC clone array). Signal intensity ratios for each BAC were processed and displayed as log2 plots using SeeGH software [18]. Figure 1 shows the representative SeeGH karyograms of one ESCC cell line (EC18) analyzed, demonstrating the identification of various gains and losses. Other SeeGH karyograms of ESCC cell lines analyzed are shown in Figure S1. Figure 2 summarizes the recurrently altered regions (with log2 ratios more than 1 or less than −1). In general, chromosomal gains were more frequently detected than losses. The most frequent alterations include gain of 11q13 (70%) and complete loss of 18q11-23 (50%). Other gains occurring in three or more cell lines are 3q26-27 (40%), 5p15-14 (50%), 8p12 (30%), 8p22-24 (30%), 13q21-31 (30%), 18p11 (30%) and 20q11-13 (40%). Other losses occurring in three or more cell lines are 8p23-22 (40%), 11q22 (30%) and 14q32 (30%) (Fig. 2).
Figure 1. Whole genome profile of an ESCC cell line (EC18) by 1-Mb aCGH.
Normalized log2 signal intensity ratios were plotted using SeeGH software. A log2 signal ratio of 0 represents equivalent copy number between the sample and the reference DNA (details in Materials and Methods). Cytoband pattern for each chromosome is shown in the left. Vertical lines denote log2 signal ratios from −2 to +2 with copy number increasing in the right and decreasing in the left. Each dark blue dot represents a single BAC clone.
Figure 2. Minimal aberrant regions identified by aCGH.
Abnormalities with ≥20% frequency in 10 ESCC cell lines are shown. * Regions with abnormalities in ≥50% cell lines are shown in bold.
Amplification and Overexpression of 11q13 Genes in ESCC Cell Lines
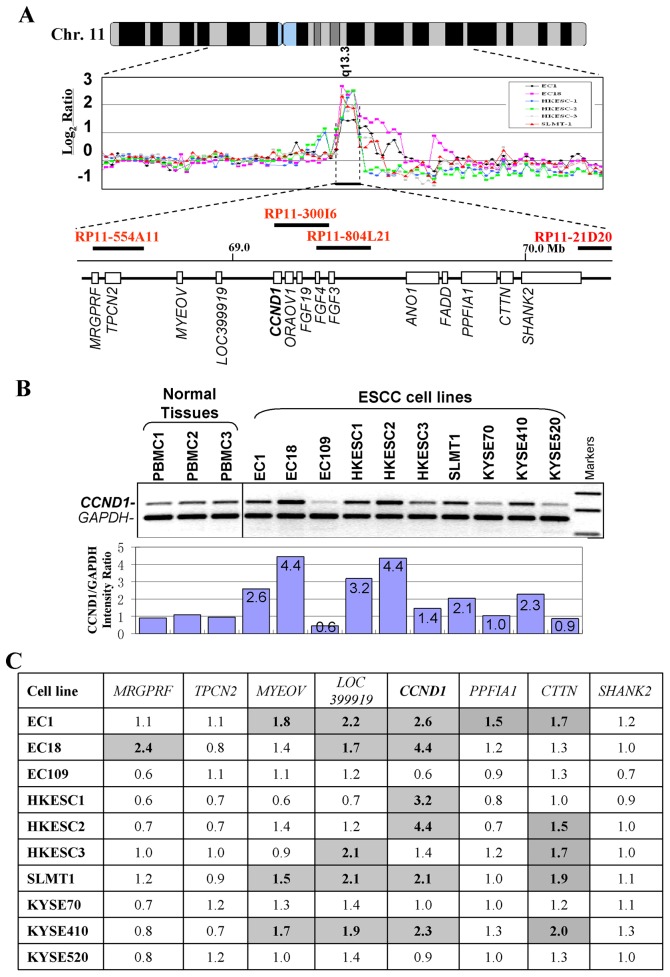
Among regions identified with recurrent alterations, amplification of 11q13.3-13.4 is the most frequent gain in ESCC (Fig. 3A), especially in those cell lines derived from Chinese patients (6/7 cell lines, 85%). Several genes with potential oncogenic functions have been identified in this locus, including CCND1 and CTTN. Thus, 11q13 was further investigated for the confirmation of gains and identification of amplified genes, by duplex genomic DNA PCR in 10 ESCC cell lines. CCND1 is mapped to the center of the 11q13 amplicon, and high-level amplification of CCND1 was confirmed in 6/10 cell lines (Fig. 3B, C), while CTTN exhibited the second highest level of amplification (in 5/10 cell lines).
Figure 3. CCND1 is located at the center of the 11q13 amplicon in ESCC. A .
, aCGH profiles of six ESCC cell lines at the CCND1 locus. Normalized log2 signal ratios were plotted. Amplifications were defined as log2 signal intensities ≥1. Horizontal lines denote log2 signal ratios from −1 to 3 with copy number increasing upwards. Each black/colorful dot represents a single BAC clone. Names of related BAC clones are also shown. Transcript map of the core 11q13 amplicon is shown in the bottom. B, Semi-quantitative duplex genomic DNA PCR analysis of CCND1 in 10 ESCC cell lines and 3 normal PBMC samples. Signal intensity ratios of CCND1/GAPDH are shown. C, Summary of gene copy number changes of several genes within the 11q13 amplicon. Numbers shown in the table are folds of copy numbers of ESCC cell lines relative to the mean values of three PBMC samples. PBMC, peripheral blood mononuclear cell.
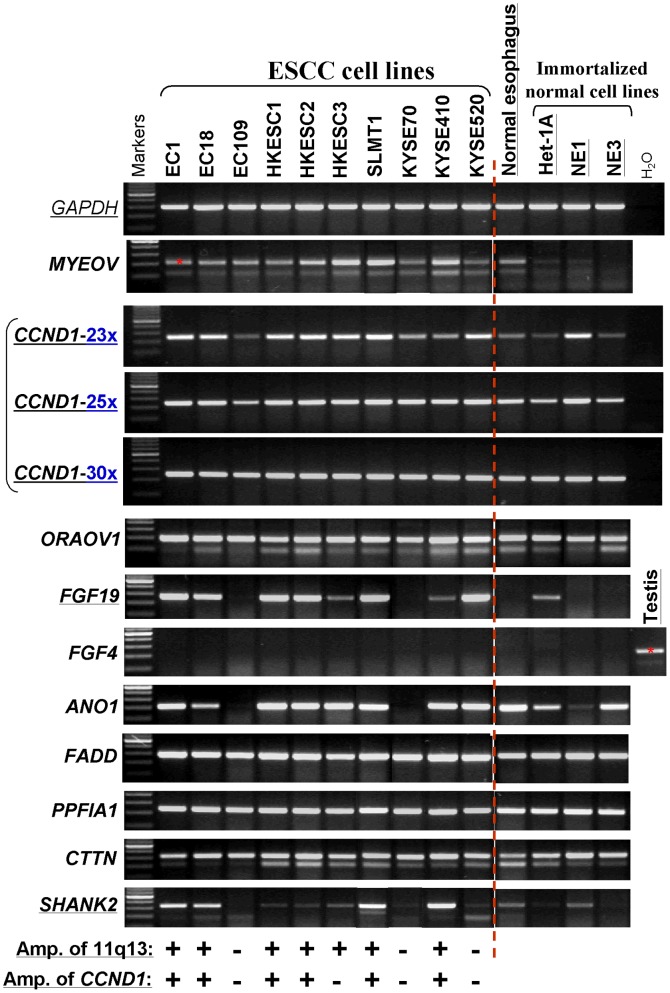
Several genes around CCND1 at 11q13 were further examined by semi-quantitative RT-PCR in ESCC cell lines. Results showed that FGF19 and SHANK2 were also remarkably overexpressed in ESCC cell lines, while only weakly or not expressed in normal esophagus or immortalized normal cells (Fig. 4), although no obvious amplification of SHANK2 was detected in these cell lines. FGF3 (data not shown) and 4 were basically not expressed in any ESCC cell line or normal esophagus. Overexpression of CCND1 and CTTN was also observed in most ESCC cell lines when compared to normal esophagus, although they were generally expressed in normal esophagus and immortalized cells (Fig. 4). Taken together, these data confirmed that 11q13 is the most frequent amplification in ESCC and delineated several genes including CCND1, CTTN, FGF19 and SHANK2, as potential critical oncogenes affected.
Figure 4. Expression levels of several 11q13 genes around CCND1 in ESCC cell lines examined by semi-quantitative RT-PCR.
The amplification status of 11q13 region by aCGH and CCND1 by multiplex DNA PCR are listed at the bottom. +, amplified; -, not amplified. 23x, 25x, 30x: RT-PCR cycles. All other genes were examined by RT-PCR with 30 cycles, with GAPDH for only 23 cycles.
CCND1 Overexpression in Primary ESCC and its Clinicopathological Association
Expression of CCND1 protein was further investigated by immunohistochemistry in ESCC tissue microarray (TMA) of 171 primary ESCC and adjacent surgical margin histologic normal esophageal tissues. Cases with >10% of tumor cells showing positive nuclear staining were scored for CCND1 overexpression. Ninety-four of the 171 cases (55%) showed CCND1 overexpression, including 42 cases of grade 1+, 35 cases of grade 2+ and 17 cases of grade 3+ (Fig. 5A). In contrast, only scattered positive cells were found in basal cells of the normal esophageal epithelia.
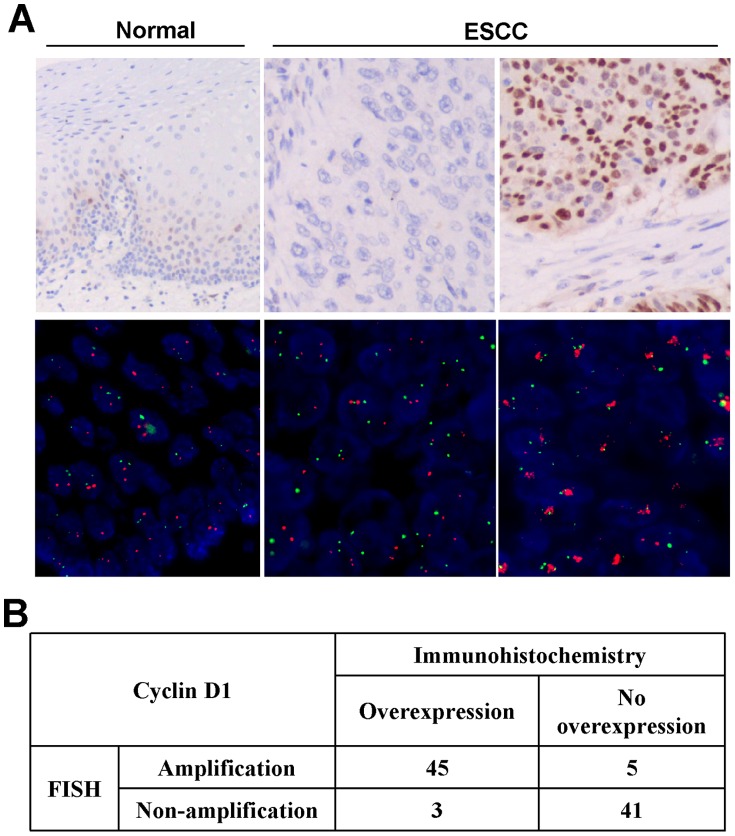
Figure 5. Representative immunohistochemical staining and FISH analysis.
. A, Top panels, immunohistochemical staining for CCND1. Left, scattered positivity of CCND1, especially in the basal layer, is seen in normal esophageal epithelium (original magnification, ×200). Middle, a case of ESCC is negative for CCND1 expression (original magnification, ×200). Right, diffuse and strong nuclear staining for CCND1 in this case of ESCC (original magnification, ×200). Bottom panels, FISH analysis. Green signals refer to reference probe of chr 11 centromere while red signals are target probe for CCND1. Left, unamplified in normal esophageal epithelium, Middle, an unamplified ESCC case. Right, an amplified ESCC case. B. Results of CCND1 amplification and expression levels in 94 paraffin-embedded primary ESCC.
We then tested the correlation between CCND1 overexpression and clinicopathological features, including tumor size, grade, lymph node metastasis, and patient age. CCND1 overexpression was significantly associated with lymph node metastasis (p = 0.006) (Table 1), but not with pT, grade, or patient age (p>0.05, respectively). In the cases with CCND1 overexpression, there was no significant difference regarding lymph node metastasis, between CCND1 high-expression groups (grade 2+ and 3+) and the low-expression group (grade 1+) (p = 0.37).
Table 1. Overexpression of CCND1 was associated with lymph node metastasis.
| CCND1 expression | p value | |||
| Positive | Negative | |||
| Differentiation | Well | 18 | 11 | 0.62* |
| Moderate | 48 | 39 | ||
| Poor | 28 | 27 | ||
| Lymph Node Metastasis | No | 35 | 45 | 0.006* |
| Yes | 59 | 32 | ||
CCND1 Overexpression Resulting from Gene Amplification but Not Activated β-catenin Signaling
To confirm the gene amplification of CCND1 in primary ESCC and investigate the association with its overexpression, we applied fluorescence in situ hybridization (FISH) analysis with commercial CCND1 probe together with CEP 11 probe in 94 cases. CCND1 was amplified in 50 cases (53.2%) and not-amplified in 44 cases (46.8%) (Fig. 5B). Furthermore, gene amplification by FISH and protein overexpression by immunohistochemistry for CCND1 showed 91.5% (κ = 0.69) concordance.
Immunohistochemically, nuclear staining for β-catenin was positive in only 4/94 cases (4.3%). All β-catenin positive cases were negative for CCND1 by immunohistochemistry.
Discussion
ESCC is the sixth most fatal cancer worldwide and accounts for 90% cases of all esophageal cancers in China [19]. Although the incidence of ESCC is low in Western countries, this tumor is common in Asia, especially in some regions in China like Henan province and Shantou city [20]; [21]. Like other cancers, environmental and genetic factors contribute to ESCC pathogenesis. Tobacco, alcohol, hot drink/food, dietary deficiency and achalasia have been considered as major environmental risk factors for ESCC, as revealed by epidemiology studies [21]; [22]. Meanwhile, cytogenetic and molecular genetic analyses demonstrated multiple genetic abnormalities in ESCC, including chromosomal losses and gains. Elucidation of these aberrations will lead to better understanding of ESCC pathogenesis, and further develop therapeutics and biomarkers for the prediction of metastasis and prognosis. In this study, we carried out a comprehensive investigation of genomic abnormalities in ESCC by high-resolution array-based CGH, to delineate the minimal chromosomal regions of amplifications and deletions. The results provide detailed pictures of multiple genetic lesions in ESCC genomes, and also give hints for further identification of critical oncogenes and TSGs in this malignancy.
Consistent with previous findings [4]–[14], recurrent gains including 11q13 (CCND1, EMS1), 3q26 (EIF5A2), 21q22 (ETS2 ) and 8q24 (MYC), and losses including 3p, 5q, 9p, 13q, 18q and 21q with target genes such as FHIT, APC, RB1 and CDKN2A, were identified in ESCC cell lines in this study. Among amplicons detected, 11q13.3-13.4 is the most frequently amplified chromosomal region. Two genes, CCND1 and CTTN located in this region, were identified with high-level amplifications in multiple ESCC cell lines. While investigated at the mRNA level, we found several genes, including CCND1 CTTN, FGF19 and SHANK2, were frequently overexpressed in ESCC cell lines. Recently, Sawey et al reported that CCND1 and FGF19 were two driving oncogenes in hepatocellular carcinoma [23]. In this study, we focused on CCND1. Our study confirmed frequent amplification (53.2%) and concordant protein over-expression (51.1%) of CCND1 in primary ESCC. Moreover, a positive correlation between CCND1 amplification/overexpression and lymph node metastasis in ESCC was observed, indicating that CCND1 may serve as a prognostic marker for ESCC, in line with other studies ([24]–[27]. In addition to amplification, CCND1 overexpression may be driven by transcriptional regulation, such as the Wnt signaling pathway. Beta-catenin is a key member of the Wnt signaling pathway, which has been suggested involved in esophageal cancer initiation and progression [28]. In this study, only 4.3% (4 in 94) of ESCC cases were identified with beta-catenin nuclear expression. The results indicate that the genomic gain of 11q13 in ESCC is the primary mechanism resulting in CCND1 amplification and overexpression. This mechanism has also been reported in other tumor types such as head and neck carcinoma [29], pituitary tumors [30], and breast cancer [31].
CTTN is an actin-associated scaffolding protein, binds and activates actin-related protein complex (Arp2/3) and thus regulates the branched actin networks in the formation of dynamic cortical actin associated structures. CCND1 and CTTN are frequently co-amplified in cancers [32]–[34]. Previously, CTTN has been identified as a bona fide oncogene located at 11q13 involved in ESCC carcinogenesis, contribute to the metastasis of various caners including ESCC, breast, hepatocellular, and head and neck squamous cell carcinomas [32]–[34]. FGF19, a fibroblast growth factor, together with CCND1, was recently reported to a major driver oncogene in hepatocellular carcinoma [23]. The involvement of FGF19 and other novel oncogenes identified in ESCC pathogenesis and the related molecule mechanisms needs to be further investigated.
Among other frequent chromosomal gains, 3q26-27 is a novel amplicon detected in ESCC. Well-known oncogenes residing in this region include EVI1, EIF5A2 and PIK3CA, which have been reported to exhibit potential oncogenic functions. In addition, TRIO and CTNND2 at 5p, MYC at 8q22, KLF5 and POU4F1 at 13q and NKX2.2 at 20p were also considered to play oncogenic roles in other tumors [35]; [36] and might contribute to ESCC carcinogenesis. Regions of high-level deletions containing known or candidate TSGs were also detected, including 18q11-23 containing SMAD2, SMAD4 and DCC (EC1, EC109, HKESC3, SLMT1 and KYSE70), 8p22 containing DLC1 (EC1, KYSE70, 410 and 520) and a region on 14q32 containing DLK1 and MEG3 (EC1, EC109 and KYSE70). Other high-level deletions containing candidate TSGs include 4q21.23-21.3 (MAPK10, PTPN13 and ARGAP24), 7p21.2 (DGKB), 7q35 (CNTNAP2), 8q11 (CEBPD), 10p11 (PARD3), 13q31.1 (SPRY2) and 16q22-23 (ATBF1). Most of these deleted regions of ESCC have also been reported in one or more other cancer types [37]; [38]. Importantly, genetic/epigenetic disruptions or altered expression levels of some TSG candidates mentioned above, including SMAD2, SMAD4, DCC, DLC1 and PARD3, have been reported in ESCC tumors and cell lines, indicating that aCGH study using multiple tumor cell lines could facilitate the identification of critical cancer genes in human tumors. [39]–[42].
In conclusion, multiple minimal regions of deletions and amplicons were detected in 10 commonly used ESCC cell lines in this study, and several novel oncogenes located in the most frequently amplified region 11q13, including FGF19, SHANK2 and CCND1, have been identified. This study provides crucial data for further identification and characterization of critical oncogenes and TSGs involved in ESCC pathogenesis.
Materials and Methods
Cell Lines
Ten ESCC cell lines (EC1, EC18, EC109, HKESC1, HKESC2, HKESC3 and SLMT1 are from Chinese patients, while KYSE70, KYSE410 and KYSE520 are from Japanese patients) and three immortalized normal esophageal epithelial cell lines (Het-1A, NE1 and NE3) were used in the study [43]. Cell lines were maintained in RPMI-1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), cultured in 5% CO2 at 37°C.
Tumor Samples
A total of 171 formalin-fixed paraffin-embedded (FFPE) Chinese ESCC samples (from 2007 to 2008) and adjacent surgical margin histological normal esophageal tissues were obtained, after receiving patients’ written informed consents and Institutional Review Board (IRB) approval from the Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, China. All patients were previously untreated (i.e., with no chemotherapy or radiotherapy), with resectable primary tumors. The mean age of patients was 58 years (range 33–78), and male to female ratio was 4.2∶ 1 (138∶ 33). Hematoxylin and eosin (HE)-stained sections were reviewed to confirm the diagnosis and define tumor areas. Tumor stage (pT) and grade were defined according to the current WHO classification of tumors.
Array CGH Analysis
High-molecular-weight chromosomal DNA was isolated from cell lines using Qiagen kit (Qiagen, Hilgen, Germany). The purity and molecular weight of DNA were examined on agarose gels. 1-Mb resolution whole-genome arrays with 3040 BAC/PAC clones were provided by Sanger Institute, UK (http://www.sanger.ac.uk/Projects/Microarrays/) [44]. Clones details are listed in Ensembl (http://www.ensembl.org/Homo_sapiens/index.html).
Array-CGH was performed with slight modifications [43]–[45]. Briefly, sample DNA (600 ng) was labeled with Cy5-dCTP (Amersham Pharmacia), whereas reference DNA of normal peripheral blood mononuclear cells (PBMCs) from healthy Chinese donors with Cy3-dCTP using the BioPrime Array CGH Genomic Labeling System (Invitrogen, Carlsbad, CA). Unincorporated nucleotides were removed using Purification Module supplied in the labeling system. Labeled samples and normal DNA (50 µl each) were mixed and ethanol precipitated together with 67.5 µl of human Cot-1 DNA (Invitrogen). Then, the mixed DNA were resuspended in 30 µl of hybridization buffer, denatured and prehybridized in a humidity chamber inside a hybridization oven at 5 rpm for 2 hrs at 37°C. The prehybridization mixture was prepared as follows: 80 µl of Herring Sperm DNA (10 mg/ml, Sigma) and 67.5 µl of human Cot-1 DNA (Invitrogen) were precipitated, resuspended in 120 µl of hybridization buffer and denatured for 10 min at 72°C. After prehybridization, the prehybridized probe was added onto the slide, and incubated at 5 rpm for 48 hours at 37°C. Slides were rinsed and washed three times, air-dried and stored at 4°C.
Hybridized slides were scanned using an Axon 4000B scanner (Axon Instruments Inc, Union City, CA) and analyzed with the GenePixPro 4.0 image analysis software where the spots were defined and median fluorescence intensities were calculated. The background subtracted fluorescence intensities were imported into a custom-designed Microsoft Excel spreadsheet template. A particular spot was excluded if the duplicate spots have a difference of >10%. The mean values of duplicate spots were presented in graphical output in the form of mean log2 (Cy5/Cy3) ratio against distance along each individual chromosome (Mb). Chromosome copy number changes were scored as hemizygous loss if the log2 ratio was ranging from −0.2 to −0.7, and homozygous deletion if >−0.7, or genomic gain for log2 ratio of 0.2 to 0.5, and amplification if >0.5. Copy number changes seen on the sex chromosomes were excluded in the analysis.
Multiplex Genomic PCR
Multiplex PCR permitted a semi-quantitative assessment of DNA amplification/loss. GAPDH gene was selected as an internal control. We amplified CCND1 and the internal control simultaneously. For CCND1, a pair of primers (forward: 5′-tgctgcgaagtggaaaccat and reverse: 5′-caacaagttgcagggaagtc) generated a PCR product of 227 bp. For GAPDH, a pair of primers (forward: 5′-gcctcactccttttgcagac and reverse: 5′-gatgaccttgcccacagcct) generated a PCR product of 157 bp. PCR reactions were performed in a final volume of 20 µl containing 200 mM deoxynucleotide triphosphates, 2.5 mM MgCl2, 50 ng DNA, 0.5 mM each oligonucleotide and 0.5 U AmpliTaq GOLD (Applied Biosystems, Foster City, CA). Reaction conditions were as follows: an initial denature at 95°C for 10 min followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s and a final extension at 72°C for 10 min. PCR products were separated on 2% agarose gels and visualized. All reactions were carried out at least twice in independent experiments.
Semi-quantitative Reverse Transcription PCR
Total RNA were extracted from cell pellets using TRI Reagent (Molecular Research Centre, Cincinnati, OH). Reverse transcription PCR (RT-PCR) was performed as described [43], using GAPDH as a control. The primers for all genes will be provided upon request. The PCR program utilized an initial denaturation at 95°C for 10 min, followed by 30 cycles of reaction (94°C for 30 s, 55°C for 30 s and 72°C for 30 s) (or 23, 25 cycles for CCND1), with a final extension at 72°C for 10 min.
ESCC Tissue Microarray (TMA)
For each case, both tumor and normal tissues were duplicated with a diameter of 1 mm on a glass slide. Before sample acquisition, HE-stained FFPE slide of each case was observed under a microscope and the locations of typically characteristic morphology of ESCC and surrounding normal tissues were circled. Samples were taken from the circled locations in the paraffin block using the Beecher Instruments Tissue Arrayer (Silver Springs, MD). For each block, two 1 mm cores were punched from the circled regions in the donor block and arrayed on the recipient block to ensure the representation of the samples, and avoid missing information due to a loss of tissue cores. A total of 171 ESCC specimens, 54 specimens of corresponding normal mucosa were arrayed on two recipient blocks. Tissue microarray sections (4 µm) were cut 24 h before immunohistochemistry.
Automated Immunohistochemistry
Immunohistochemistry was performed with a Ventana Benchmark XT autostainer (Ventana, Tuscon, USA), using monoclonal rabbit anti-human CCND1/cyclin D1 (clone SP4) and monoclonal mouse anti-human β-catenin (Clone β-Catenin-1) from DAKO (Glostrup, Denmark) with Ventana Ultraview kits (Ventana, Tucson, USA). Slides were incubated for 24 min at 37°C with primary antibodies. Diaminobenzidine or 3-amino-9-ethylcarbazole was used as chromogens and slides were counterstained with haematoxylin before mounting. Both chromogens used on regular full sections before TMA testing gave concordant results in terms of both surfaces and intensities of immunostaining. Negative controls were created by omission of primary antibody and replacement with phosphate buffered saline (PBS).
The CCND1 and β-catenin expression levels were determined semi-quantitatively using a four-tiered scoring system, based on the positive nuclear staining fraction of tumor cells (grade 0 = 0–10%; grade 1+ = 11–25%; grade 2+ = 26–50%; grade 3+ = 51–100%). A score of 0 was considered negative while a score of 1+, 2+ or 3+ was considered positive. For β-catenin, staining in cytoplasm may have been present but was not included in the determination of positivity.
Fluorescence in Situ Hybridization (FISH)
For FISH analysis of CCND1 gene amplification, two direct-labeled probes were used, Vysis CCND1/CEP11 FISH Probe Kit including LSI CCND1 (11q13) SpectrumOrange against CCND1 (11q13) and CEP11 SpectrumGreen (Vysis/Abbott, IL, USA) against the centromere of chromosome 11. FISH analysis was done according to “LSI Locus Specific Identifier DNA probes” from Vysis. The CCND1 gene to chromosome 11 centromere ratio was measured in at least 60 nuclei from tumor cells and an average score was taken. Observing more than two copies of CCND1 for each chromosome 11 was considered to be a positive sign for CCND1 gene amplification.
Statistical Analysis
Statistical analysis was performed using SPSS version 12.0. The association between FISH and IHC results and the clinico-pathological variables are performed using the χ2 -test. For all tests, a p-value of <0.05 was considered to be statistically significant.
Supporting Information
Whole genome profiles of 10 ESCC cell lines by 1-Mb aCGH. Normalized log2 signal intensity ratios were plotted using SeeGH software. A log2 signal ratio of 0 represents equivalent copy number between the sample and the reference DNA (details in Materials and Methods). Cytoband pattern for each chromosome is to the left for each plot. Vertical lines denote log2 signal ratios from −2 to +2 with copy number increases to the right and decreases to the left. Each black dot represents a single BAC clone.
(TIF)
Acknowledgments
We thank Dr Tzer Jing Seng for the help of array-CGH, Profs Gopesh Srivastava and George Tsao (University of Hong Kong) for some ESCC and normal esophageal epithelial cell lines, and DSMZ (German Collection of Microorganisms & Cell Cultures) for KYSE cell lines (Shimada et al., Cancer 69: 277–284, 1992).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grants from the National Natural Science Foundation of China (#30801344, #30928012) and the Beijing Nova Program (No. 2009A70). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer. 1993;19:594–606. doi: 10.1002/ijc.2910540413. [DOI] [PubMed] [Google Scholar]
- 2.Pera M, Manterola C, Vidal O, Grande L. Epidemiology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 3.Lambert R, Hainaut P. The multidisciplinary management of gastrointestinal cancer Epidemiology of oesophagogastric cancer. Best Pract Res Clin Gastroenterol. 2007;21:921–945. doi: 10.1016/j.bpg.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Law FB, Chen YW, Wong KY, Ying J, Tao Q, et al. Identification of a novel tumor transforming gene GAEC1 at 7q22 which encodes a nuclear protein and is frequently amplified and overexpressed in esophageal squamous cell carcinoma. Oncogene. 2007;26:7490–7498. doi: 10.1038/sj.onc.1210390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo ML, Shen XM, Zhang Y, Wei F, Xu X, et al. Amplification and overexpression of CTTN (EMS1) contribute to the metastasis of esophageal squamous cell carcinoma by promoting cell migration and anoikis resistance. Cancer Res. 2006;66:11690–11699. doi: 10.1158/0008-5472.CAN-06-1484. [DOI] [PubMed] [Google Scholar]
- 6.Wang XX, Liu R, Jin SQ, Fan FY, Zhan QM. Overexpression of Aurora-A kinase promotes tumor cell proliferation and inhibits apoptosis in esophageal squamous cell carcinoma cell line. Cell Res. 2006;16:356–366. doi: 10.1038/sj.cr.7310046. [DOI] [PubMed] [Google Scholar]
- 7.Arai H, Ueno T, Tangoku A, Yoshino S, Abe T, et al. Detection of amplified oncogenes by genome DNA microarrays in human primary esophageal squamous cell carcinoma: comparison with conventional comparative genomic hybridization analysis. Cancer Genet Cytogenet. 2003;146:16–21. doi: 10.1016/s0165-4608(03)00106-7. [DOI] [PubMed] [Google Scholar]
- 8.Miller CT, Aggarwal S, Lin TK, Dagenais SL, Contreras JI, et al. Amplification and overexpression of the dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (DYRK2) gene in esophageal and lung adenocarcinomas. Cancer Res. 2003;63:4136–4143. [PubMed] [Google Scholar]
- 9.Janssen JW, Imoto I, Inoue J, Shimada Y, Ueda M, et al. MYEOV, a gene at 11q13 is coamplified with CCND1 but epigenetically inactivated in a subset of esophageal squamous cell carcinomas. J Hum Gene. 2002;47:460–464. doi: 10.1007/s100380200065. [DOI] [PubMed] [Google Scholar]
- 10.Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, et al. Identification of a novel gene GASC1 within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 2002;60:4735–4739. [PubMed] [Google Scholar]
- 11.Chiang PW, Beer DG, Wei WL, Orringer MB, Kurnit DM, et al. Detection of erbB-2 amplifications in tumors and sera from esophageal carcinoma patients. Clin Cancer Res. 1999;5:1381–1386. [PubMed] [Google Scholar]
- 12.Jiang W, Kahn SM, Tomita N, Zhang YJ, Lu SH, et al. Amplification and expression of the human cyclin D gene in esophageal cancer. Cancer Res. 1992;52:2980–2983. [PubMed] [Google Scholar]
- 13.Carneiro A, Isinger A, Karlsson A, Johansson J, Jonsson G, et al. Prognostic impact of array-based genomic profiles in esophageal squamous cell cancer. BMC Cancer. 2008;8:98–104. doi: 10.1186/1471-2407-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi ZZ, Liang JW, Zhan T, Wang BS, Lin DC, et al. Genomic alterations with impact on survival in esophageal squamous cell carcinoma identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2011;50:518–526. doi: 10.1002/gcc.20875. [DOI] [PubMed] [Google Scholar]
- 15.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Gene. 41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuris S, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown J, Bothma H, Veale R, Willem P. Genomic imbalances in esophageal carcinoma cell lines involve Wnt pathway genes. World J Gastroenterol. 2011;17:2909–2923. doi: 10.3748/wjg.v17.i24.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi B, DeLeeuw RJ, Coe BP, MacAulay C, Lam WL. SeeGH–a software tool for visualization of whole genome array comparative genomic hybridization data. BMC Bioinformatics. 2004;5:13–13. doi: 10.1186/1471-2105-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 20.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Thompson SK, Ruszkiewicz AR, Jamieson GG, Esterman A, Watson DI, et al. Improving the accuracy of TNM staging in esophageal cancer: a pathological review of resected specimens. Ann Surg Oncol. 2008;15:3447–3458. doi: 10.1245/s10434-008-0155-0. [DOI] [PubMed] [Google Scholar]
- 22.Allen JW, Richardson JD, Edwards MJ. Squamous cell carcinoma of the esophagus: a review and update. Surg Oncol. 1997;6:193–200. doi: 10.1016/s0960-7404(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 23.Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagasawa S, Onda M, Sasajima K, Makino H, Yamashita K, et al. Cyclin D1 overexpression as a prognostic factor in patients with esophageal carcinoma. J Surg Oncol. 2001;78:208–214. doi: 10.1002/jso.1152. [DOI] [PubMed] [Google Scholar]
- 25.Kunisaki C, Shimada H, Akiyama H, Nomura M, Matsuda G, et al. Prognostic factors in esophageal cancer. Hepatogastroenterology. 2004;51:736–740. [PubMed] [Google Scholar]
- 26.Liu GY, Luo Q, Xiong B, Pan C, Yin P, et al. Tissue array for Tp53, C-myc, CCND1 gene over-expression in different tumors. World J Gastroenterol. 2008;14:7199–7207. doi: 10.3748/wjg.14.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin DC, Du XL, Wang MR. Protein alterations in ESCC and clinical implications:a review. Dis Esophagus. 2009;22:9–20. doi: 10.1111/j.1442-2050.2008.00845.x. [DOI] [PubMed] [Google Scholar]
- 28.Peng H, Zhong XY, Liu KP, Li SM. Expression and significance of adenomatous polyposis coli beta-catenin E-cadherin and cyclin D1 in esophageal squamous cell carcinoma assessed by tissue microarray. Ai Zheng. 2009;28:38–41. [PubMed] [Google Scholar]
- 29.Izzo JG, Papadimitrakopoulou VA, Li XQ, Ibarguen H, Lee JS, et al. Dysregulated cyclin D1 expression early in head and neck tumorigenesis: in vivo evidence for an association with subsequent gene amplification. Oncogene. 1998;17:2313–2322. doi: 10.1038/sj.onc.1202153. [DOI] [PubMed] [Google Scholar]
- 30.Hibberts NA, Simpson DJ, Bicknell JE, Broome JC, Hoban PR, et al. Analysis of cyclin D1 (CCND1) allelic imbalance and overexpression in sporadic human pituitary tumors. Clin Cancer Res. 1999;5:2133–2139. [PubMed] [Google Scholar]
- 31.Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- 32.Rodrigo JP, Garcia-Carracedo D, Garcia LA, Menendez S, Allonca E, et al. Distinctive clinicopathological associations of amplification of the cortactin gene at 11q13 in head and neck squamous cell carcinomas. J Pathol. 2009;217:516–523. doi: 10.1002/path.2462. [DOI] [PubMed] [Google Scholar]
- 33.Luo ML, Wang MR. CTTN (EMS1): an oncogene contributing to the metastasis of esophageal squamous cell carcinoma. Cell Res. 2007;17:298–300. doi: 10.1038/cr.2007.17. [DOI] [PubMed] [Google Scholar]
- 34.Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi G, et al. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol. 2004;41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Zheng M, Simon R, Mirlacher M, Maurer R, Gasser T, et al. TRIO amplification and abundant mRNA expression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Am J Pathol. 2004;165:63–69. doi: 10.1016/S0002-9440(10)63275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nandan MO, McConnell BB, Ghaleb AM, Patel NV, Robine S, et al. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying J, Li H, Cui Y, Wong AH, Langford C, et al. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20:1173–1175. doi: 10.1038/sj.leu.2404193. [DOI] [PubMed] [Google Scholar]
- 38.Sun X, Frierson HF, Chen C, Li C, Ran Q, et al. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat Genet. 2005;37:407–412. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- 39.Park HL, Kim MS, Yamashita K, Westra W, Carvalho AL, et al. DCC promoter hypermethylation in esophageal squamous cell carcinoma. Int J Cancer. 2008;122:2498–2502. doi: 10.1002/ijc.23434. [DOI] [PubMed] [Google Scholar]
- 40.Chattopadhyay I, Singh A, Phukan R, Paukayastha J, Kataki A, et al. Genome-wide analysis of chromosomal alterations in patients with esophageal squamous cell carcinoma exposed to tobacco and betel quid from high-risk area in India. Mutat Res. 2010;696:130–138. doi: 10.1016/j.mrgentox.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Maesawa C, Tamura G, Nishizuka S, Iwaya T, Ogasawara S, et al. MAD-related genes on 18q21.1, Smad2 and Smad4, are altered infrequently in esophageal squamous cell carcinoma. Jpn J Cancer Res. 1997;88:340–343. doi: 10.1111/j.1349-7006.1997.tb00386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zen K, Yasui K, Gen Y, Dohi O, Wakabayashi N, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–2918. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- 43.Ying J, Li H, Seng TJ, Langford C, Srivastava G, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 44.Hurst CD, Fiegler H, Carr P, Wiiliams S, Carter NP, et al. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23:2250–2263. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- 45.Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole genome profiles of 10 ESCC cell lines by 1-Mb aCGH. Normalized log2 signal intensity ratios were plotted using SeeGH software. A log2 signal ratio of 0 represents equivalent copy number between the sample and the reference DNA (details in Materials and Methods). Cytoband pattern for each chromosome is to the left for each plot. Vertical lines denote log2 signal ratios from −2 to +2 with copy number increases to the right and decreases to the left. Each black dot represents a single BAC clone.
(TIF)