Abstract
Part of the latest SAMPL challenge was to predict how a small fragment library of 500 commercially available compounds would bind to a protein target. In order to assess the modellers’ work, a reasonably comprehensive set of data was collected using a number of techniques. These included surface plasmon resonance, isothermal titration calorimetry, protein crystallization and protein crystallography. Using these techniques we could determine the kinetics of fragment binding, the energy of binding, how this affects the ability of the target to crystallize, and when the fragment did bind, the pose or orientation of binding. Both the final data set and all of the raw images have been made available to the community for scrutiny and further work. This overview sets out to give the parameters of the experiments done and what might be done differently for future studies.
Keywords: X-ray crystallography, Surface plasmon resonance, Isothermal titration calorimetry, Modelling, Fragment screening
Introduction
We approached this in a significantly different way than a ‘normal’ fragment screening campaign in the sense that the data set was to be complete (or as complete as physically possible). To elaborate, in a ‘normal’ fragment screening campaign, it is usual to have a fairly short timeline, so the project is set up to screen the fragments as quickly as possible using the most effective method first, and then use subsequent methods for verification and to determine the other parameters of value. For example, in our laboratory, we will typically screen the fragment set using SPR (taking about 1 week) and then only do protein crystallography on the hits from the SPR. We would only use ITC on those that were tight binders (better than 200 μM) and where we wanted verification of the binding energy. We would generally soak all fragments into pre-formed crystals and not attempt doing co-crystallization of compounds with the protein. In contrast, for the SAMPL project, it was one of the major goals of the project to have a complete data set for the modelling community to go back to and reference. For the DINGO data set, we systematically soaked every fragment of the set into the protein crystals and collected data sets for each of these complexes. In addition, co-crystallization of the target protein with fragments was undertaken as an orthogonal approach. The target chosen for the SAMPL challenge requires an inhibitor in order for crystallisation to occur, so the presence or absence of crystals with any given fragment in co-crystallisation trials is predictive of whether that fragment binds to the protein target. The SPR was done several times and dosage curves were also done several times on all those compounds that were ‘hits’.
Bovine pancreatic trypsin [1] was used as the target for several reasons. It is easily obtainable from commercial vendors; the crystallographic community has studied it rather thoroughly; it is a protease that is similar to other proteases of human health interest; and there is a body of literature that supports a prospective challenge such as SAMPL, including known positive controls that could be used for verification of our methods [2–4]. The Maybridge 500 fragment library was chosen as it was commercially available and we had tested it against some other targets and knew that it had fragments that could bind to trypsin.
After starting the project, it became apparent that our choices did have some drawbacks. Trypsin is a protease that will self-proteolyze, so is unstable over time for all of our experiments (ITC, crystallization, etc.). The Maybridge 500 fragment library has some compounds that are insoluble under the conditions we used in several of the techniques where aqueous solubility has significant advantages (e.g. ITC). And finally, in our effort to be comprehensive, trying to collect X-ray crystallographic data sets of 500 different fragments soaked into trypsin crystals required the growth of well over 3,000 ‘production’ crystals and the collection of well over 1,000 data sets at the Australian Synchrotron [5].
Methods
All SPR Experiments were performed using a Biacore T100 instrument (GE Healthcare). Trypsin was immobilized onto a CM5 chip using standard amine coupling chemistry. Benzamidine was used as a positive control to validate trypsin activity on the chip. The binding capacity of immobilized trypsin (Rmax) was increased by purifying the protein using size exclusion chromatography and immobilizing the protein in the presence of 5 mM benzamidine and up to 20 mM CaCl2. Typically in SPR experiments, a gradual decrease in analyte binding capacity (Rmax) by the immobilized protein is indicative of protein decay. CaCl2 is a structural inhibitor of trypsin [6] and its presence was observed to prolong the activity of the immobilized surface.
For the fragment screening experiments, two of the four channels (flow cells 2 and 4) on the chip surface had trypsin immobilized. One trypsin surface was ‘aged’ in that it was put down 24 h prior to application of the second trypsin surface to see what the effect of this would be on binding. Our expectation, which was borne out, was that this aged protein would have less binding capacity and we should see a comparably lower response in this channel for real hits. Bovine carbonic anhydrase II (CAII) was immobilized in flow cell 3 where it served as a negative protein control. Flow cell 1 was left intact and used as a reference (blank) surface. Maybridge library fragments, previously prepared at 100 mM in neat DMSO (master stocks), were diluted into SPR running buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 0.05% (v/v) Tween-20, 1 mM CaCl2 and 5% [v/v] DMSO) to 100 μM and injected over the chip surfaces. To assess the stability and reproducibility of the assay, positive controls (benzamidine and p-amino-benzenesulfonamide) were injected several times throughout the screening experiment. Three hundred and eighty-four fragments in a 384-well plate were screened within approximately 30 h. Remaining compounds were screened later using a similar screening approach. Scrubber (http://www.biologic.com.au) was utilized for data processing and analysis. SPR signals were referenced against the blank surface and further corrected for DMSO refractive index changes (excluded volume effect). Binding data were normalized for the molecular weight of the fragments. The normalization scheme of Giannetti et al. [7] was further applied to the processed data based on the maximal binding response (Rmax) determined from fitting the control compound sensorgrams. Compounds showing undesirable SPR binding characteristics similar to those described previously [7] were removed from the screening data.
The selected top 20 hits were further analysed using dosage experiments. These were performed at 20 °C by injecting a concentration series in two-fold dilutions (C = 4–256 μM). To estimate binding affinities (equilibrium dissociation constant, KD), binding responses at equilibrium (Req) were fit to a 1:1 steady state affinity model (available within Scrubber) which utilizes a nonlinear least squares regression method to fit the Langmuir adsorption isotherm (Req = Rmax*KD/[KD + C]) to each data set. A normalized saturation response (Rmax), derived using the reference compound, was applied to the responses obtained with fragment hits that, due to solubility and chip surface artefact issues, could not be injected at or near saturating concentrations. A SPR dosage experiment for benzamidine binding to immobilized trypsin is shown in Fig. 1a. Interestingly, a marginally higher affinity was consistently estimated in the presence of CaCl2 where the KD for benzamidine binding to trypsin was measured to be ~7 μM whereas in the absence of CaCl2, KD was estimated to be approximately ~15 μM (data not shown).
Fig. 1.
a–c Normalized SPR sensorgrams showing benzamidine (a) and CC 00813 (b) binding to immobilized trypsin. Both compounds were injected as an eight-membered twofold dilution series (including ‘zero buffer blank’) with a top concentration of 160 μM for benzamidine and 256 μM for CC 00813. Diagram in c show fits of the binding responses at equilibrium (t = 50–55 s, plotted against compound concentration) to a 1:1 steady state affinity model. As CC00813 failed to reach a maximal binding response (Rmax) at the top injected concentration (256 μM), its affinity (KD = 466 μM) was estimated using Rmax values determined from a benzamidine binding fit
To further confirm our SPR and crystallography hits, isothermal titration calorimetry experiments (ITC) were performed using a MicroCal Auto-iTC200 (GE Healthcare). Trypsin solutions were freshly prepared in 50 mM Tris–HCl, 10 mM CaCl2, pH 8.0 and dialysed overnight against the same buffer at 4 °C. Prior to titration, the trypsin solution was spiked with DMSO to match the 5% (v/v) DMSO in the small molecule solution. Fragment solutions (concentration in the range 1.8–16 mM, depending on the specific inhibitor) were titrated into the stirred (1,000 r.p.m.) cell (300 μL) containing trypsin solution (0.16–1.6 mM). Data were analysed using Origin software by fitting a single-site binding isotherm that yields ΔH (enthalpy of binding) and KA (binding constant). These titration experiments only allowed for estimation of the tightest binding fragments from the SPR hit list (KD < 300 μM, Table 1). Weaker binding fragments could not be accurately measured due to the very high concentrations of both protein and compound required to generate sufficient heat that can be detected in the microcalorimeter. A more detailed description of the SPR and ITC experiments, along with the PDB coordinates of the fragment hit structures, will be published in the near future (manuscript in preparation).
Table 1.
Values given in the columns for SPR and ITC are micromolar; NA means not attempted; values for the co-crystallization are the number of crystals seen out of the number of successful drops set up (in some cases the drop was not set down properly by the robotics); for fragment density, yes means that there was clean and clear density for the fragment, no means that there was no fragment density or that it wasn’t clear
| Maybridge # | Mol wt. | SPR affinity (μM) | ITC affinity (μM) | Co-crystals found | Soaked fragment density | Co-crystal fragment density | 2D |
|---|---|---|---|---|---|---|---|
| CC 33513 | 242.1 | 24 | 33.9 | 94 of 94 | Yes | Yes | 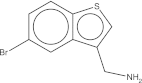
|
| CC 12313 | 199.7 | 31 | 43.1 | 96 of 96 | Yes | Yes | 
|
| CC 38513 | 176.3 | 71 | 180.7 | 91 of 95 | Yes | Yes | 
|
| CC 00413 | 215.7 | 136 | 157 | 96 of 96 | NA | Yes | 
|
| CC 11513 | 196.3 | 153 | 163.2 | 26 of 34 | Yes | Yes | 
|
| CC 21913 | 172.2 | 236 (old) 40 (new) | 97.5 | 41 of 96 | Yes | Yes | 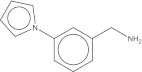
|
| CC 35913 | 205.3 | 271 | 185.9 | 54 of 91 | No | Yes | 
|
| CC 32913 | 173.2 | 400 | NA | 9 of 96 | Yes | NA | 
|
| CC 00813 | 185.7 | 466 | NA | 12 of 96 | Yes | No | 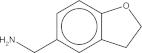
|
All of the crystallization experiments were performed at the Collaborative Crystallisation Centre (C3) at CSIRO in Melbourne, Australia. Drops were set up in two subwell sitting drop plates (SD-2, IDEX Corp) using a Phoenix robot (Art Robbins Industries) with 50 μL of crystallant in the reservoir and droplets consisting of 300 nL of the reservoir and 195 nL of the protein sample and 5 nL of seed stock [8]. Only one of the two crystallisation subwells was utilised for the initial crystallisation. A robotic procedure using a Mosquito robot (TTP) was developed to place a mixture of fragment and a cryoprotectant onto the both the crystallisation droplet and the unused subwell in the sitting drop plates [9]. The second subwell was used as part of the 2 step soaking procedure to make sure the fragments had a chance to displace the benzylamine in the crystals. After allowing the fragments to soak into the crystals for 24–48 h, the crystals were transferred manually to the fresh fragment/cryoprotectant solution in the second subwell and allowed to soak an additional 24–48 h. Crystals were gently removed using mylar loops (MiTeGen) mounted in copper pins (Crystal Positioning Systems, USA) and cryo-cooled in liquid nitrogen and placed in a 96 hole cassette that was kept submerged in liquid nitrogen until the individual pin with the crystal of interest was placed in the X-ray beam at the Australian Synchrotron. At least two crystals were harvested for each of the soaks and data sets were attempted for both in all cases. 181 frames of data, each one a 1° oscillation with 1–3 s of exposure, were taken for each crystal. All of the data sets were initially processed using a script called Jigsaw [5] (available upon request) that uses the following crystallographic programs to automatically index, scale, do molecular replacement, an initial round of refinement and then try to place a ligand in the excess density of the active site (when present): XDS [10], Pointless (CCP4) [11], SCALA (CCP4) [11], Phaser (CCP4) [11], Refmac (CCP4) [11], Flynn (OpenEye) [12]. Coot was used to visualize the model and electron density as well as manually rebuild the model where there were changes [13].
Discussion
This was a project which could not have been attempted without a lot of recent (and expensive) tools: for example, automation in crystallogenesis, X-ray data collection and computing. It is notable that the technology for one of the major techniques used in this project, surface plasmon resonance, only became available in the early 1990s. In all, to assemble the experimental underpinnings for this project took five domain experts close to 2 years, and required equipment that was millions of dollars to purchase and run. This is excluding the cost of the Australian Synchrotron, where the equivalent of about a month of continuous beamtime was required to collect the X-ray diffraction data for this challenge. If we were to attempt this same amount of data collection on a standard X-ray home source it would take closer to a decade of continuous beamtime to collect the same amount of data. Similarly, about 200 96-well crystallisation plates were set up during the course of this experiment; by hand, assembling that many experiments would take close to 3 working months, and that is without even taking a peek at the experimental results once they were set up.
The enormity of the project is quite obvious to most experimentalists, and explains why this type of challenge has not been taken on previously; the modelling community has been relying on retrospective analyses in part because the prospective data are so expensive to obtain. These experimental data are not perfect: there is ‘real world’ noise in the data—machines break, chemicals degrade, data get misplaced (despite best efforts) and then the reality is that data from different biophysical techniques cannot be cleanly compared to each other. The use of amine coupling techniques to prepare SPR chips precludes the use of Tris buffers to attach the protein to the chip. The requirement for cryoprotection of protein crystals results in protein structures with blobs of extra density which are from the ethylene glycol cryoprotectant rather than any fragment. There are numerous examples where the details of experimental setup are where the difficulties lie.
Looking at the differences in the techniques, we see that the pH and buffer was different for each: SPR used 50 mM Hepes pH 7.4, 150 mM NaCl, 0.05% Tween P20, 20 mM CaCl2 + 5% DMSO for the fragment; ITC used 50 mM Tris pH 8.0, 10 mM CaCl2 + 5% DMSO for the fragment; and crystallization used 22.5% w/v PEG 3350, 0.18 M (NH4)2SO4, 0.12 M NaSCN, 0.09 M Bis–Tris pH 5.5, 0.01 M Tris pH 8.5, which gave a final pH of 5.8. DMSO was used in all cases as the fragments were solubilized in neat DMSO at the start. It should be noted that the crystals obtained for soaking were in space group P212121, whereas most of the crystal structures determined for the co-crystallization with fragments were found in P312. This is due to the fact that when DMSO is present during the crystallization process, the space group tends to fall into the trigonal space group. There may also be some influence due to a pH change- the co-crystal experiments were done at pH 7.0 instead of pH 5.8. We have typically found that SPR is a reliable method for estimating binding constants of fragments up to KD = 250–500 μM, but beyond this level the error associated with the measurements can become significant. In particular, the insolubility of fragments in SPR compatible buffers and at high fragment concentrations, can cause chip surface interaction artefacts, and this prevents fragment injections at or near the saturating concentrations required for accurate affinity estimations. As discussed previously, by applying a normalization scheme based on the saturation response from a positive control it is possible to estimate affinities without achieving saturation (Fig. 1b, c). Using this approach we attempted to estimate KD up to 1 mM values, but as can be seen in Table 1, the correspondence between the SPR and crystallography methods breaks down beyond 300 μM and we saw no fragments in the crystal structures beyond the 500 μM barrier.
Although we were limited by the solubility and weak binding of the fragments in the ITC experiments, the correlation between the SPR and ITC is relatively good (see Table 1). For most of the SPR hits better than 300 μM, we have multiple X-ray data sets to confirm the position of the ligand found in the binding site. All of the ligands found to date sit in the same binding site as the benzamidine and benzylamine controls and all have a primary amine that binds to the Asp189 residue of trypsin (see Fig. 2). Trypsin is a rather rigid molecule and besides a few rotomer changes of side chains, there are no large loop or domain movements upon binding these fragments.
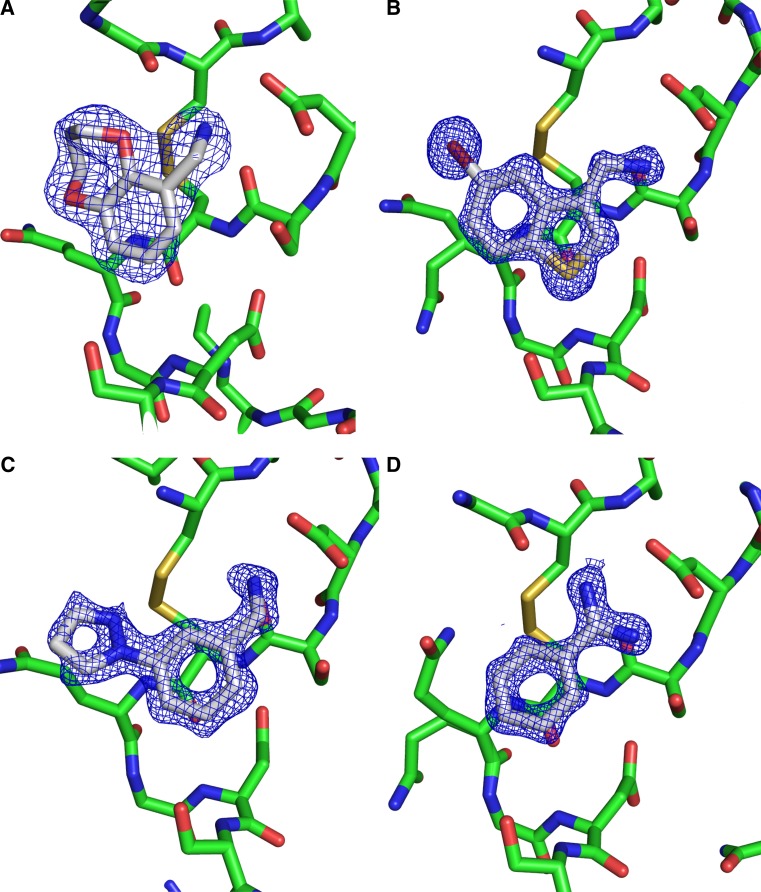
Fig. 2.
a CC 00413 bound to trypsin in a co-crystallization experiment. Data to 1.90 Å, space group P312. Asp189 is seen in the upper right of the figure. The protein carbon atoms are coloured green whereas the carbon atoms of the fragment are coloured in gray. A 2Fo-Fc electron density map is shown as a blue mesh. b CC 33513 soaked into trypsin crystals (space group P212121), resolution 1.4 Å. For clarity, the atom attached to the benzene ring and colored a deep red, is Br. c CC 32913 soaked into the trypsin crystals (space group P212121, resolution 1.4 Å). d Model and electron density for benzamidine, one of the two control compounds used in this experiment. All figures are in approximately the same orientation with Asp189 in the upper right hand corner of the figures (about 2 o’clock)
We conclude from looking at our experimental results that the rigidity of the target limited the hit rate of fragment binding, and that an experienced protease expert would have looked at the fragment library and picked out the likely binders simply by choosing molecules that look somewhat akin to well known protease inhibitors such as benzamidine. This would have probably taken an afternoon, rather than the 2 years to collect the experimental results! However, despite the ‘obviousness’ of the results in retrospect, there was no modeling technique that found or ranked all the experimental hits correctly, showing clearly the value of this work—without guides to let us know when an approach/method isn’t working, that method cannot advance. We are glad that the experiments have opened up new directions for modeling development, and in future years (when the memory of this data collection has faded) we may be able to do this again to see how far modeling has progressed.
Acknowledgments
We thank Kim Branson and Anthony Nicholls for the opportunity to contribute to the SAMPL challenge; to Matt Geballe and Vijay Pande for organizing the recent SAMPL meeting; our managers (Tim O’Meara, Tim Adams and Paul Savage) for supporting us; and most importantly the Australian Synchrotron for giving us the beam time to collect all of the data sets.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Kunitz M, Northrop JH. Isolation of protein crystals possessing tryptic activity. Science. 1931;73:262–263. doi: 10.1126/science.73.1888.262. [DOI] [PubMed] [Google Scholar]
- 2.Rauh D, et al. Trypsin mutants for structure-based drug design: expression, refolding and crystallisation. J Biol Chem. 2002;383:1309–1314. doi: 10.1515/BC.2002.148. [DOI] [PubMed] [Google Scholar]
- 3.Markwardt F, Landmann H, Walsmann P. Comparative studies on the inhibition of trypsin, plasmin, and thrombin by derivatives of benzylamine and benzamidine. Eur J Biochem. 1968;6:502–506. doi: 10.1111/j.1432-1033.1968.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 4.Stubbs MT, Huber R, Bode W. Crystal structures of factor Xa specific inhibitors in complex with trypsin: structural grounds for inhibition of factor Xa and selectivity against thrombin. FEBS Lett. 1995;375:103–107. doi: 10.1016/0014-5793(95)01190-P. [DOI] [PubMed] [Google Scholar]
- 5.Newman J, Fazio VJ, Caradoc-Davies TT, Branson K, Peat TS. Practical aspects of the SAMPL challenge: providing an extensive experimental data set for the modeling community. J Biomol Screen. 2009;14:1245–1250. doi: 10.1177/1087057109348220. [DOI] [PubMed] [Google Scholar]
- 6.McDonald MR, Kunitz M. The effect of calcium and other ions on the autocatalytic formation of trypsin from trypsinogen. J Gen Physiol. 1941;25:53–73. doi: 10.1085/jgp.25.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giannetti AM, Koch BD, Browner MF. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J Med Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 8.Luft JR, DeTitta GT. A method to produce microseed stock for use in the crystallization of biological macromolecules. Acta Crystallogr. 1999;D55:988–993. doi: 10.1107/s0907444999002085. [DOI] [PubMed] [Google Scholar]
- 9.Newman J, Pham TM, Peat TS. Phoenito experiments: combining the strengths of commercial crystallization automation. Acta Crystallogr. 2008;F64:991–996. doi: 10.1107/S1744309108029667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. doi: 10.1107/S0021889893005588. [DOI] [Google Scholar]
- 11.The CCP4 suite: programs for protein crystallography Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 12.Wlodek S, Skillman AG, Nicholls A. Automated ligand placement and refinement with a combined force field and shape potential. Acta Crystallogr. 2006;D62:741–749. doi: 10.1107/S0907444906016076. [DOI] [PubMed] [Google Scholar]
- 13.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]