Abstract
AIM: To assess whether DNA methylation patterns in chronic alcoholics are different from non-alcoholic sibling controls.
METHODS: We examined the methylation patterns in DNA samples from 25 chronic alcoholics and 22 matched siblings as controls (one per family). DNA was extracted from peripheral blood and analyzed for differences in the methylation patterns after bisulfite-conversion. We used the Illumina GoldenGate Methylation Cancer Panel I (Illumina, San Diego, CA), which probes the methylation profile at 1505 CpG sites from 807 cancer related genes. We excluded the 84 X-chromosome CpG sites and 134 autosomal CpG sites that failed to show a within sample reliability score of at least 95% for all samples, leaving 1287 autosomal CpG sites (associated with 743 autosomal genes) with reliable signals for all samples. A methylation score was calculated as the average methylation for the 1287 CpG sites examined. Differences were assessed by a two-sample t-test. We also examined the average sib pair differences in methylation scores at each of the 1287 sites. All analyses were performed using SPSS, version 9.0, P < 0.05 was considered significant.
RESULTS: Methylation levels at the 1287 CpG sites averaged 28.2% for both alcoholics and controls. The mean difference in methylation scores between alcoholic and non-alcoholic sibs by CpG site was < 1% with small inter-individual variances; and only 5 CpG sites had an average sib difference > 5%. Subgroup analysis showed that methylation scores were significantly lower for the alcoholic-dependent subjects who smoked compared to their non-smoking unaffected siblings. Specifically, among smokers who are alcoholic, global methylation indices were significantly lower than in non-alcoholic sib controls, whereas among non-smoking alcoholics, the global indices were significantly higher (P = 0.008).
CONCLUSION: Although we observed no effect of alcoholism alone on DNA methylation, there is a decrease in alcoholics who smoke, suggesting a mechanism for alcohol-tobacco synergy for carcinogenesis.
Keywords: DNA methylation, Alcohol, Epigenetics, Cancer, Carcinogenesis, Smoking, Cigarettes, Tobacco
INTRODUCTION
Epigenetics is the study of heritable differences related to changes in gene expression that are not due to differences in DNA sequences themselves. Although still in its infancy, epigenetics is expanding rapidly as a field of study. DNA methylation, one of the two main types of epigenetic inheritance, is involved in many physiological and pathophysiological conditions, including regulation of gene expression and silencing of repeat elements in the genome. Epigenetic mechanisms have been implicated in the long term memory formation by neurons and are a growing area of research in diseases such as Alzheimer’s dementia[1]. DNA methylation is thought to play important roles in many diseases, including multiple sclerosis, diabetes mellitus, schizophrenia, alcohol dependence and cancer[2-6].
It has been shown that global methylation status in peripheral blood monocytes is associated with plasma homocysteine levels in healthy individuals. The importance of homocysteine to DNA methylation status stems from the fact that homocysteine is a precursor of S-adenosyl methionine, which acts as the methyl donor when cytosine residues in the dinucleotide sequence CpG are methylated by DNA methyltransferases. Chronic alcoholics commonly have elevated homocysteine levels. Bönsch et al[7], showed associations among alcohol-associated elevated plasma homocysteine levels, global methylation levels assayed by difference in CpG methylation sensitive vs. insensitive restriction enzyme (Hpall/Mspl) digestion, and the subsequent expression of DNMT mRNAs in alcoholic patients, compared to controls. These findings support the hypothesis that ethanol exposure increases global levels of DNA methylation and suggests that changes in DNA methylation may result in changes in gene expression. Support for this hypothesis includes several reports of DNA hypermethylation associated with alcohol use at specific individual genes in peripheral blood cells[8-10]. Other studies have identified changes in methylation associated with smoking, suggesting both alcohol and smoking may contribute to changes in DNA methylation[11,12]. In all likelihood, many more genes whose levels of expression are partially controlled by the methylation status of the DNA in their promoters are yet to be discovered.
Changes in DNA methylation are recognized as one of the most common forms of molecular alteration in human neoplasia[13,14]. Hypermethylation of CpG islands located in the promoter regions of tumor suppressor genes has been firmly established as a mechanism for gene inactivation in cancers[15,16]. In contrast, global hypomethylation of genomic DNA[17] and loss of IGF2 imprinting were observed in tumor cells[18] and a correlation between hypomethylation and increased gene expression was reported for many oncogenes[19,20]. In addition, monitoring global changes in DNA methylation has been used for molecular classification of cancers[21,22]. Gene hypermethylation has been correlated with clinical risk groups for neuroblastoma[23], as well as with hormone receptor status and response to tamoxifen in breast cancer[24,25]. Therefore, it may be feasible to use methylation markers to classify and predict cancer risk, different kinds or stages of cancer, cancer therapeutic outcomes and patient survival.
Alcoholism and cancer risk
About 3.6% of all cases of cancer and a similar proportion of cancer deaths are attributable to heavy consumption of alcohol. These figures are higher in selected regions of the world, in particular in Central and Eastern Europe. Among women, 60% of cancers attributable to alcohol use occur in the breast[26]. Chronic excessive alcohol consumption is the strongest risk factor for upper aerodigestive tract (UADT) cancer (oral cavity, pharynx, hypopharynx, larynx and esophagus)[27]. Chronic and heavy alcohol use also increases the risk for cancer of the liver, colon, rectum and breast[28]. Many epidemiological studies have demonstrated a correlation between chronic and heavy alcohol ingestion and the occurrence of cancer in these organs[29-31]. Because the ingestion of all types of alcoholic beverages is associated with an increased cancer risk, more likely than not, ethanol itself is the crucial compound that increases cancer risk, rather than congeners (propanol, butanol, pentanol) or other additives. The exact mechanisms of ethanol-associated carcinogenesis have remained obscure.
Multiple mechanisms are believed to be involved in alcohol-associated cancer development of the UADT, including the effect of acetaldehyde (AcH the first metabolite of ethanol oxidation), induction of cytochrome P-4502E1 leading to the generation of reactive oxygen species, and enhanced procarcinogen activation, modulation of cellular regeneration, and nutritional deficiencies. Folate deficiency, primarily the consequence of low dietary intake and destruction by AcH, is common in alcoholics and contributes to the inhibition of transmethylation, which is an important factor in the regulation of genes involved in carcinogenesis. Acetaldehyde also decreases DNA repair mechanisms and the methylation of cytosine in DNA. However, it has been shown recently that chronic alcoholics have significantly increased levels of genomic DNA methylation in peripheral blood mononuclear cells (PBMC), compared to samples from unrelated volunteer blood donors[7].
Most studies to date have examined changes in global methylation in alcohol users or methylation changes at a few candidate genes, rather than at a broader panel of specific sites. This study was designed specifically to obtain preliminary data on the methylation status in PBMC of genes known or suspected of playing a role in cancer development. The primary aim was to assess the change in global DNA methylation levels at these gene specific sites in well-characterized chronic alcoholics and to compare it to suitably matched non-alcoholic family members as controls. We also wanted to explore whether there are observable, meaningful differences in methylation patterns between the two groups at different gene loci and whether there are relationships between life time alcohol use and the degree or pattern of DNA methylation.
MATERIALS AND METHODS
We examined the methylation patterns in DNA samples from 25 chronic alcoholics and 22 of their non-alcoholic biological siblings. We utilized the resources available through the UCONN Alcohol Research Center of UCHC to help us identify suitable alcohol-dependent subjects and their non-alcohol-dependent family members to serve as controls. The kindreds studied have been well characterized and followed longitudinally. They are enrolled in the long-standing Collaborative Study on the Genetics of Alcoholism[32,33]. After IRB approval, suitable subjects were identified and informed consent for participation in this study was obtained.
The alcohol-dependent subjects were at least 21 years of age and had a history of alcohol use for at least 5 years. All subjects were interviewed using the Semi-Structured Assessment for the Genetics of Alcoholism, a reliable and valid psychiatric diagnostic instrument[34]. Alcohol-dependent subjects met the DSM-IV diagnosis of alcoholic dependence. Males were consuming at least 15 drinks per week or 5 or more standard drinks in a day and females at least 8 or more drinks per week or 4 or more standard drinks in a day within the past year. Non-alcohol-dependent biological siblings of the subjects served as controls. The controls were screened for heavy alcohol use or history of cancer by self-reported questionnaires. They were required to have had a normal physical examination and no personal history of any kind of cancer other than superficial skin cancer. We excluded any subjects with known genetic abnormalities or chronic liver diseases (other than alcohol-related liver disease) and subjects with known nutritional disorders and/or anemia, which may have served as confounding variables. The sample examined included 22 sibships comprised of 25 probands and 22 siblings (3 sibships included 2 probands).
DNA methylation analysis
DNA was prepared from peripheral blood samples using a commercial kit (Gentra PureGene, Qiagen, Valencia, CA) and 500 ng of each DNA sample was bisulfite reacted using the EZ-96 DNA methylation-gold kit from Zymo Research (Orange, CA).
We used a high-throughput single nucleotide polymorphism genotyping system[35] for DNA methylation detection, based on genotyping of bisulfite-converted genomic DNA. This technology, developed by Illumina, combines a miniaturized bead-based array platform, a high level of assay multiplexing, and scalable automation for sample handling and data processing. We used the Illumina GoldenGate Methylation Cancer Panel I (Illumina, San Diego, CA), which probes the methylation profile at 1505 CpG sites from 807 genes selected by the manufacturer, based on their relevance to carcinogenesis. This assay is reported to have a sample replicate variation of < 6%[36] and can resolve a 10% or greater methylation difference with 95% confidence.
We excluded the 84 X-chromosome CpG sites in the Illumina Cancer Panel because the methylation levels for X-chromosome sites vary greatly by sex [the X-chromosome (Lyon) inactivation in females is associated with methylation of CpG-rich islands[37]]. We also excluded from analysis 134 autosomal CpG sites that did not give an assay reliability score of at least 95% for all samples, leaving 1287 autosomal CpG sites with reliable signal for all samples. The included 1287 CpG sites were associated with 743 autosomal genes.
Statistical analysis
For each participant, we calculated a methylation score by computing the average methylation over the 1287 CpG sites examined. Differences in the mean methylation scores between the two samples were assessed by a two-sample t-test. We also examined the average sib pair differences in methylation scores at each of the 1287 sites evaluated with use of a paired t test. All analyses were performed using SPSS, version 9.0, P < 0.05 was considered a statistically significant result.
RESULTS
A total of 25 alcoholics and 22 matched controls (one control per family) were recruited for this study. The average age of probands and controls was not significantly different. Probands were more likely to be male (Fisher’s exact test, P = 0.004). Three sib pairs contained 2 probands. As anticipated, the alcohol-dependent subjects had significantly higher amounts of alcohol use, both in terms of days (frequency) and drinks (quantity) per week (Table 1). Bisulfite reacted DNA was examined at 1421 autosomal CpG sites contained on the Illumina DNA methylation chip. Analysis was limited to the 1287 probes which generated valid test signals (95% quality confidence signal) from all samples. Methylation levels at the 1287 CpG sites averaged 28.2% for all samples combined. The mean methylation score was not significantly different between the alcohol-dependent subjects and their unaffected siblings (Table 2). The mean difference in methylation scores between affected and unaffected sibs by CpG site was < 1% (Table 2) with a tight distribution, and only 5 CpG sites had an average sib difference > 5% (Figure 1). The sib difference and t-test statistic for these 5 CpG sites are listed in Table 3. Finally, as a test of the assay’s reproducibility, we performed replicate bisulfite conversion and methylation assays for DNA samples from alcoholics and non-alcoholic participants from 3 sibships. The mean difference in replicate sample methylation for the 1287 CpG sites was less than 1% (Table 2).
Table 1.
Selected demographic and alcohol-use features at baseline
| Alcohol-dependent siblings (n = 25) | Non-alcoholic siblings (n = 22) | |
| Age (yr) (SD, range) | 40.7 (9.5, 24-54) | 40.6 (11.4, 21-59) |
| Gender (M:F) | 18:7 | 4:18 |
| Current smoker | 14 | 7 |
| Race: EA, AA, NA | 14, 10, 1 | 13, 8, 1 |
| Hispanic Ethnicity | 2 | 3 |
| Past 12-mo drinking (mean ± SD) | ||
| Drinking days per week | 3.91 ± 1.95b | 0.95 ± 1.18 |
| Drinks per drinking day | 7.90 ± 4.01b | 2.68 ± 1.34 |
| Drinks per week | 34.0 ± 30.8b | 2.7 ± 3.6 |
| 1Heavy drinking days per week | 2.53 ± 2.66b | 0.18 ± 0.34 |
“Heavy drinking days” were defined as days in which men consumed more than 10 drinks and women more than 8 drinks.
t-test P < 0.001 vs non-alcoholic siblings. EA: European American; AA: African American; NA: Native American.
Table 2.
Global methylation scores
| Alcohol-dependent siblings | Non-alcoholic siblings | |
| Global methylation index for 1287 CpG sites | ||
| Mean methylation (SD) | 0.282 (0.016) | 0.282 (0.012) |
| Median methylation (SD) | 0.082 (0.010) | 0.079 (0.009) |
| Range | 0.01-0.97 | 0.01-0.97 |
| Sib pair difference in global methylation level at each of 1287 sites (alcoholic minus non-alcoholic sibling methylation level) | ||
| Mean difference (SD) | 0.00005 (0.019) | |
| Replicate pair difference in methylation level at each of 1287 sites (3 non-alcoholic and alcoholic siblings with replicate bisulfite treatment and methylation quantification) | ||
| Mean difference (SD) | 0.0008 (0.006) | 0.001 (0.003) |
No global measures of methylation significantly differ between groups.
Figure 1.
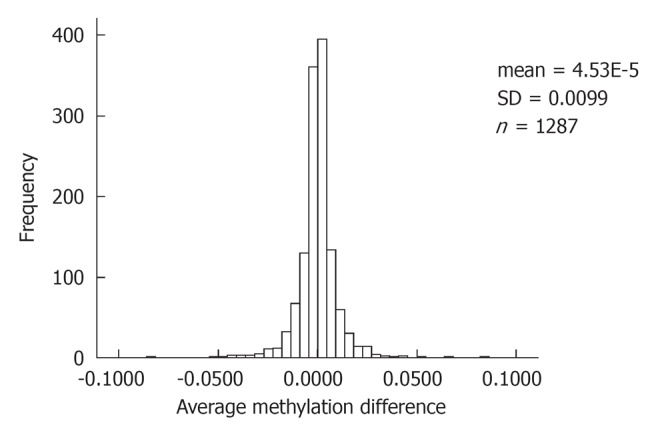
Frequency histogram of average within sib pair difference in methylation at 1287 CpG sites methylation level of alcoholic minus non-alcoholic sibling.
Table 3.
Alcoholic minus non-alcoholic sib differences in methylation scores at 5 CpG sites with average difference in methylation frequency > 0.05
| Gene symbol | Illumina CpG probe ID | Average Sib difference | Paired t-test statistic (2-tailed) | P value |
| LTA | 820 | 0.083 | 2.46 | 0.021 |
| CRK | 3392 | 0.068 | 3.18 | 0.004 |
| GSTM1 | 4902 | 0.054 | 1.82 | 0.081 |
| HPN | 4931 | -0.084 | -2.14 | 0.043 |
| MSH3 | 2787 | -0.052 | -1.35 | 0.189 |
LTA: Lymphotoxin α precursor; CRK: v-crk sarcoma virus CT10 oncogene homolog isoform b; GSTM1: Glutathione S-transferase M1 isoform 1; HPN: Hepsin (transmembrane protease, serine 1); MSH3: MutS homolog 3.
Because tobacco use may also affect methylation levels, we conducted a subgroup analysis comparing the global methylation sib pair differences for sib pairs in which neither smoked (n = 7), those in which both smoked (n = 7), and sib pairs for which the proband smoked and the control sib did not (n = 7) (in two sib pairs, the control sib but not the alcoholic sib smoked; smoking status was not available for one proband). We found that, for the two groups of sib pairs concordant for smoking status, compared with the non-concordant group, the alcohol-dependent subjects had higher average methylation levels at the 1287 sites examined (F = 284, df = 2, P < 0.001). Similarly, for non-smoking sib pairs, in 6 of 7 pairs, alcoholic subjects had a higher average methylation index. In contrast, for discordant pairs with an alcoholic smoker, in 6 of 7, the alcoholic subject had a lower average methylation index than the non-alcoholic, non-smoking sibling (χ2 = 8.2, df = 2, P = 0.017) (Table 4).
Table 4.
Global methylation score sib-pair differences for non-smokers vs sibship with alcoholic tobacco user (7 sib pairs)
|
Sib pair concordance for smoking status |
|||
|
Concordant |
Discordant |
||
| Both non-smokers | Both smokers | Proband smokes | |
| Mean (SD) sib pair difference in methylation at each site (alcoholic minus non-alcoholic sibling methylation level) | +0.006 (0.018) | +0.010 (0.020) | -0.009 (0.025) |
Mean sib pair difference for 1287 markers, ANOVA: F = 284 (df = 2), P < 0.0001. Among concordant non-smoking sib pairs, for 6 of 7 pairs alcoholic subject had higher methylation index among concordant. Five of 7 smoking sib pairs alcoholic subjects had higher methylation index. Among discordant pairs with an alcoholic smoker, 6 of 7 alcoholic subjects had a lower methylation index than non-alcoholic siblings.
DISCUSSION
The major findings of this study are two-fold: (1) Contrary to our a priori major hypothesis, there was no difference in average CpG methylation scores between alcohol-dependent subjects and non-alcoholic siblings; and (2) However, in a secondary analysis, we did find a small but significant decrease in PBMC methylation scores in the alcoholic subjects who smoked, when compared to their non-alcohol dependent siblings who did not smoke (Table 4). Thus, despite heavy, chronic and ongoing alcohol use in the alcohol-dependent probands, we found no effect on average methylation of the DNA of PBMCs for a set of 1287 CpG sites associated with 743 genes implicated in carcinogenesis. This is in contrast to results reported by Bönsch et al[7] who have shown a global CpG DNA hypermethylation in chronic alcoholics. However, in previous work, results among alcoholics were compared to a random, unrelated non-alcoholic control population and genes particularly relevant to cancer development were not studied. Gender and race have recently been reported to influence global genomic methylation in peripheral blood[38], emphasizing the importance of carefully matched controls in studies of this type. We believe that our family controls are a unique strength of our results.
Others have shown that global leukocyte DNA hypomethylation is associated with the risk of developing breast cancer[39]. In a mouse model of cutaneous carcinogenesis, it has been shown that the degree of DNA hypomethylation of genomic DNA increases as lesions progress from a benign to invasive cancers[40]. The discordant results can be explained by the fact that hypomethylation is most relevant when it occurs in the coding regions of the genes. In contrast to prior global CpG methylation analysis with respect to heavy and chronic alcohol use, our study found no meaningful change in levels of methylation at specific CpG sites of potential relevance to cancer-related genes, when results were compared to those of non-alcoholic siblings.
The combination of alcohol and tobacco use is known to be synergistic in markedly increasing the risk of development of malignancies of the UADT, especially squamous cell carcinomas of esophagus, lung and oropharynx[41-44]. Our finding of increased CpG methylation among alcoholics vs. non-alcoholic siblings for those 14 sib pairs concordant for smoking status, corrected for the status of their sibs (Table 4), is thus of much interest. If confirmed in larger number of subjects and in several other samples, it will suggest that factors other than hypomethylation of DNA accounts for the well established synergism of alcohol and tobacco in the pathogenesis of cancer of UADT.
Our study had several limitations. Perhaps most important is the small sample size, which, due to limitations in time and funding, was only about half as large as we had hoped. Secondly, this is not a genome-wide study, but rather examines only a select group of candidate genes, albeit genes pre-selected for their known relevance to cancer development. Nonetheless, the genes examined may not be as important in early stage carcinogenesis and/or may be affected by other epigenetic factors such as histone modifications. Another unavoidable limitation was that most alcoholics were men, whereas most non-alcoholic siblings were women. Thus, although matched genetically by family, alcoholic subjects and controls were not closely matched by gender.
A major strength of this study is the inclusion of biological siblings unaffected by alcoholism as controls. Also, the tumor genes included on the Illumina Cancer Methylation Assay chip have been well characterized previously as related to cancers of the UADT. We excluded from analysis the CpG sites related to the X and Y chromosomes that could have had a confounding effect on our results. This is supported by a recent study by Zhang et al[38] showing significantly lower global genomic DNA methylation in females. It is thought that X chromosome inactivation in women may diminish the capacity for methylating autosomal loci[45].
In summary, our study did not reveal any significant differences in the average methylation score between alcoholic and non-alcoholic siblings associated with 743 genes implicated in carcinogenesis. However, subgroup analysis did show a significantly decreased methylation of genes important in cancer development among alcoholics who smoked, compared to their non-alcoholic siblings who did not smoke. This finding needs confirmation in larger independent samples. It would also be prudent to consider a priori the combined effect of alcohol and smoking when planning future studies examining the effects of alcohol on DNA methylation.
COMMENTS
Background
DNA methylation is thought to play an important role in cancer development. Chronic and heavy alcohol has long been associated with a variety of cancers and has recently been associated with increased DNA methylation levels.
Research frontiers
The authors planned this study to assess whether DNA methylation patterns in chronic alcoholics are different from non-alcoholic siblings who served as controls for comparison.
Innovations and breakthroughs
The major findings of this study are two-fold: (1) Contrary to our belief, there was no difference in average CpG methylation scores between alcohol-dependent subjects and non-alcoholic siblings; and (2) However, in a secondary analysis, we did find a small but significant decrease in methylation scores of DNA from peripheral blood mononuclear cells in the alcoholic subjects who smoked, when compared to their non-alcohol dependent siblings who did not smoke. Thus, despite heavy, chronic and ongoing alcohol use in the alcohol-dependent subjects, we found no effect on average methylation for the set of 743 genes examined, which have previously been implicated in carcinogenesis. This is in contrast to results reported by Bönsch et al who reported a global DNA hypermethylation in chronic alcoholics, albeit not adjusted for results from controls from the same families.
Applications
Subgroup analysis did show significantly decreased methylation of genes important in cancer development among alcoholics who smoked, compared to their non-alcoholic siblings who did not smoke. This finding needs confirmation in larger independent samples. It would also be prudent to consider a priori the combined effect of alcohol and smoking when planning future studies examining the effects of alcohol on DNA methylation.
Terminology
DNA Methylation: It refers to the addition of a methyl group to the DNA at specific locations, namely, the cytosine residues of CpG dimers. DNA methylation is thought to regulate a number of cellular processes in the human body and also to influence the development of cancer when it occurs at specific sites.
Peer review
The study was well planned and conducted. The conclusions drawn are supported by the results. The study however is limited by its limited sample size and the fact that it examines only a select group of genes that have been associated to cancer development. A major strength of this study is the use of siblings as controls to adjust for any differences in the DNA methylation status that may be due to inherent genetic factors that differ among different kindreds.
Footnotes
Supported by (in part) Grants from The American College of Gastroenterology (to Bonkovsky HL and Thapar M); NIH/NIAAA P60-AA003510 (to Hesselbrock V and Covault J); NIH/NIAAA U10-008401 (to Hesselbrock V); NCRR M01RR006192; NIH/NIDDK 5R01 DK 38825 (to Bonkovsky HL)
Peer reviewers: Ke-Bin Liu, Assistant Professor, Department of Biochemistry and Molecular Biology, School of Medicine, Medical College of Georgia, Augusta, GA 30912, United States; Fahd Al-Mulla, PhD, Associate Professor, Department of Molecular Pathology, Kuwait University, Faculty of Medicine, Safat 13110, Kuwait; Ying-Yan Yu, MD, PhD, Professor, School of Medicine, Shanghai Jiaotong University, Shanghai 200025, China
S- Editor Wang JL L- Editor Roemmele A E- Editor Zheng XM
References
- 1.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 2.Ballestar E, Esteller M. The impact of chromatin in human cancer: linking DNA methylation to gene silencing. Carcinogenesis. 2002;23:1103–1109. doi: 10.1093/carcin/23.7.1103. [DOI] [PubMed] [Google Scholar]
- 3.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. The coming of age of DNA methylation in medicine in the genomics and postgenomics era. Clin Immunol. 2002;103:213–216. doi: 10.1006/clim.2002.5216. [DOI] [PubMed] [Google Scholar]
- 6.Singh SM, McDonald P, Murphy B, O’Reilly R. Incidental neurodevelopmental episodes in the etiology of schizophrenia: an expanded model involving epigenetics and development. Clin Genet. 2004;65:435–440. doi: 10.1111/j.1399-0004.2004.00269.x. [DOI] [PubMed] [Google Scholar]
- 7.Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113:1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- 8.Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, Bönsch D. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30:587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- 9.Bönsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- 10.Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res. 2009;43:388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, Philibert RA. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philibert RA, Beach SR, Gunter TD, Brody GH, Madan A, Gerrard M. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:619–628. doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33 Suppl:238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 14.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, Baylin SB, Herman JG. Hypermethylation-associated Inactivation of the Cellular Retinol-Binding-Protein 1 Gene in Human Cancer. Cancer Res. 2002;62:5902–5905. [PubMed] [Google Scholar]
- 16.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 18.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 20.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 21.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 22.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 23.Alaminos M, Davalos V, Cheung NK, Gerald WL, Esteller M. Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. J Natl Cancer Inst. 2004;96:1208–1219. doi: 10.1093/jnci/djh224. [DOI] [PubMed] [Google Scholar]
- 24.Martens JW, Nimmrich I, Koenig T, Look MP, Harbeck N, Model F, Kluth A, Bolt-de Vries J, Sieuwerts AM, Portengen H, et al. Association of DNA methylation of phosphoserine aminotransferase with response to endocrine therapy in patients with recurrent breast cancer. Cancer Res. 2005;65:4101–4117. doi: 10.1158/0008-5472.CAN-05-0064. [DOI] [PubMed] [Google Scholar]
- 25.Widschwendter M, Siegmund KD, Müller HM, Fiegl H, Marth C, Müller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 26.Boffetta P, Hashibe M, La Vecchia C, Zatonski W, Rehm J. The burden of cancer attributable to alcohol drinking. Int J Cancer. 2006;119:884–887. doi: 10.1002/ijc.21903. [DOI] [PubMed] [Google Scholar]
- 27.Pöschl G, Seitz HK. Alcohol and cancer. Alcohol Alcohol. 2004;39:155–165. doi: 10.1093/alcalc/agh057. [DOI] [PubMed] [Google Scholar]
- 28.Seitz HK, Maurer B, Stickel F. Alcohol consumption and cancer of the gastrointestinal tract. Dig Dis. 2005;23:297–303. doi: 10.1159/000090177. [DOI] [PubMed] [Google Scholar]
- 29.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Cogliano V. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8:292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 30.Maier H, Dietz A, Gewelke U, Seitz HK, Heller WD. [Tobacco- and alcohol-associated cancer risk of the upper respiratory and digestive tract] Laryngorhinootologie. 1990;69:505–511. doi: 10.1055/s-2007-998241. [DOI] [PubMed] [Google Scholar]
- 31.Tuyns AJ, Péquignot G, Jensen OM. [Nutrition, alcohol and oesophageal cancer (author’s transl)] Bull Cancer. 1978;65:58–64. [PubMed] [Google Scholar]
- 32.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 34.Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 35.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 36.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norris DP, Brockdorff N, Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- 38.Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, Nesline MK, Ambrosone CB, Karpf AR. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraga MF, Herranz M, Espada J, Ballestar E, Paz MF, Ropero S, Erkek E, Bozdogan O, Peinado H, Niveleau A, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004;64:5527–5534. doi: 10.1158/0008-5472.CAN-03-4061. [DOI] [PubMed] [Google Scholar]
- 41.Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, Fernandez L, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777–789. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 42.Peters ES, McClean MD, Marsit CJ, Luckett B, Kelsey KT. Glutathione S-transferase polymorphisms and the synergy of alcohol and tobacco in oral, pharyngeal, and laryngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:2196–2202. doi: 10.1158/1055-9965.EPI-06-0503. [DOI] [PubMed] [Google Scholar]
- 43.Talamini R, Bosetti C, La Vecchia C, Dal Maso L, Levi F, Bidoli E, Negri E, Pasche C, Vaccarella S, Barzan L, et al. Combined effect of tobacco and alcohol on laryngeal cancer risk: a case-control study. Cancer Causes Control. 2002;13:957–964. doi: 10.1023/a:1021944123914. [DOI] [PubMed] [Google Scholar]
- 44.Flanders WD, Rothman KJ. Interaction of Alcohol and Tobacco in Laryngeal-Cancer. Am J Epidemiol. 1982;115:371–379. doi: 10.1093/oxfordjournals.aje.a113315. [DOI] [PubMed] [Google Scholar]
- 45.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]


