Abstract
Oncogenic ras alleles are among the most common mutations found in patients with acute myeloid leukemia (AML). Previously, the role of oncogenic ras in cancer was assessed in model systems overexpressing oncogenic ras from heterologous promoters. However, there is increasing evidence that subtle differences in gene dosage and regulation of gene expression from endogenous promoters play critical roles in cancer pathogenesis. We characterized the role of oncogenic K-ras expressed from its endogenous promoter in the hematopoietic system using a conditional allele and IFN-inducible, Cre-mediated recombination. Mice developed a completely penetrant myeloproliferative syndrome characterized by leukocytosis with normal maturation of myeloid lineage cells; myeloid hyperplasia in bone marrow; and extramedullary hematopoiesis in the spleen and liver. Flow cytometry confirmed the myeloproliferative phenotype. Genotypic and Western blot analysis demonstrated Cre-mediated excision and expression, respectively, of the oncogenic K-ras allele. Bone marrow cells formed growth factor–independent colonies in methylcellulose cultures, but the myeloproliferative disease was not transplantable into secondary recipients. Thus, oncogenic K-ras induces a myeloproliferative disorder but not AML, indicating that additional mutations are required for AML development. This model system will be useful for assessing the contribution of cooperating mutations in AML and testing ras inhibitors in vivo.
Introduction
Acute myeloid leukemia (AML) is characterized by an accumulation of immature myeloid precursors in the bone marrow and, in many cases, the peripheral blood. From analysis of recurrent chromosomal translocations in bone marrow samples from AML patients, it is clear that loss-of-function mutations in transcription factors critical for hematopoiesis are involved in leukemogenesis. These balanced reciprocal translocations often result in the expression of fusion proteins that act as dominant negative inhibitors of the normal transcription factors in hematopoietic development, such as the AML1-ETO product of the t(8;21) translocation and the PML-RARα product of the t(15;17) translocation. Loss-of-function point mutations in transcription factors involved in hematopoietic differentiation, such as C/EBPα and AML1, have also been identified in AML. Although these mutations involving hematopoietic transcription factors are necessary for AML pathogenesis, they are not sufficient (1–4). It is hypothesized that the AML phenotype requires a combination of mutations in the hematopoietic progenitor cells, which contribute to impaired differentiation and an increased proliferation and/or survival advantage (5, 6). The ras proto-oncogenes are likely candidates for proteins in which gain-of-function mutations would confer the signals for enhanced cell proliferation and survival in AML pathogenesis.
The ras proteins are a family of guanine nucleotide–binding proteins involved in cell proliferation, differentiation, and survival (7–10). The ras signaling pathways are activated by a spectrum of hematopoietic cytokine receptors in response to ligand and therefore play important roles in the proliferation and enhanced survival of hematopoietic progenitors. Members of the ras family include K-ras, N-ras, and H-ras. They are expressed ubiquitously during development and must be post-translationally modified by farnesylation and/or geranylgeranylation for membrane localization and functional activity. The ras proteins cycle between an inactive GDP-bound state and an active GTP-bound state; this cycling is mediated by GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs). In the GTP-bound state, ras activates the Raf/MAP kinase pathway, with subsequent activation of transcription factors and altered expression of genes promoting cell proliferation and differentiation. The ras proteins can also promote cell survival by activating the phosphoinositide-3-kinase (PI3K) pathway, with subsequent stimulation of AKT kinase and inactivation of the proapoptotic protein Bad. The ras proteins also activate a multitude of downstream effectors (including Ral-GEFs, AF-6, PKC-ζ, Nore1, Rin 1, RASSF1, and phospholipase C ε), whose roles in oncogenesis are incompletely understood. Functionally, the H-, N-, and K-ras isoforms have different properties with respect to activation of downstream effectors, transformation abilities, post-translational modifications, subcellular localizations, role in mouse development, and relative frequency of mutation in various human tumors (7, 11).
Oncogenic ras mutations are present in approximately 25–44% of AML patients and are among the most frequently detected genetic alterations in AML (12, 13). Activating N-ras mutations are detected in 20–25% of AML patients and K-ras mutations are seen in 10–15% of AML patients. These activating mutations most frequently involve single amino acid substitutions at ras codons 12, 13, or 61, which abrogate intrinsic ras GTPase activity and lead to constitutive ras activation.
In the absence of oncogenic ras mutations, ras may be activated through other mechanisms, such as the oncogenic activation of upstream tyrosine kinases seen in a significant proportion of AML cells, with consequent accumulation of ras-GTP. Examples of tyrosine kinase gain-of-function mutations in AML include FLT3 activation loop point mutations, FLT3 juxtamembrane region internal tandem duplications (ITDs) (14), C-KIT point mutations (15), and the tyrosine kinase fusion products of chromosomal translocations such as TEL/TRKC associated with t(12;15) (16, 17) or TEL/ABL associated with t(9;12) (18, 19). Most of these tyrosine kinase mutations confer growth factor–independent growth to hematopoietic cell lines and cause a myeloproliferative disease in mouse models (17, 20, 21).
Dysregulation of ras-GAP is another mechanism of indirect ras activation in leukemic cells. Loss-of-function mutations in neurofibromatosis 1 (NF1), a ras-GAP protein, lead to accumulation of high levels of ras-GTP (22). Children with type 1 neurofibromatosis are predisposed to malignant myeloproliferative syndromes, which include monosomy 7 syndrome and juvenile myelomonocytic leukemia (JMML) (23). Mice heterozygous for NF1 deletion are prone to develop a myeloproliferative syndrome resembling human JMML (24). NF1-deficient mouse fetal liver cells demonstrate hypersensitivity to GM-CSF stimulation and elevated levels of ras-GTP (25, 26) and confer a JMML-like myeloproliferative disease upon transplantation (25).
To date, activating ras mutations have been studied in various cell culture, bone marrow transplantation, and transgenic mice model systems expressing ras from exogenous promoters. A variety of phenotypes has been reported. CD34+ stem cells transduced with amphotrophic retrovirus expressing oncogenic N-ras had impaired erythropoietin-induced erythroid differentiation at the late erythroblast stage, reminiscent of a myelodysplastic phenotype (27, 28), and impaired G-CSF– and GM-CSF–induced granulocytic differentiation at the blast/promyelocyte stage (29). In a transplant model using bone marrow cells retrovirally transduced with oncogenic N-ras, recipient mice developed myeloproliferative disorders with long latency (107–385 days) and incomplete penetrance, suggesting that secondary mutations were necessary for the phenotype (30). Mice transplanted with bone marrow cells retrovirally transduced with viral H-ras developed peripheral myeloid leukocytosis, as well as frequent pre–T thymic lymphomas and/or pre–B cell lymphoblastic leukemia/lymphomas (31). Mice transplanted with bone marrow cells expressing oncogenic H-ras from the Moloney MuLV long terminal repeat (LTR) promoter often developed thymic lymphomas (32). Transgenic mice expressing activated N-ras from the mouse mammary tumor virus (MMTV) LTR developed lymphoblastic T cell and B cell lymphomas and poorly differentiated mammary carcinomas (33). Transgenic mice expressing viral H-ras from the MMTV promoter/enhancer developed B cell lymphoblastic lymphomas at low frequency (34).
As is evident from the studies described above, a consistent phenotype from oncogenic ras expression in hematopoietic cells has not been established, in part because different exogenous promoters were used to express various oncogenic ras isoforms at supraphysiological levels and often in nonphysiological tissues. The variety of malignancies that developed likely reflected the tissue specificities of the promoters used rather than the specific transforming capabilities of oncogenic ras. The phenotypes generated from overexpression of genes from exogenous reporters may be very different from the phenotypes that arise when genes are expressed from their endogenous reporters. The critical role of gene dosage in cancer is supported by data indicating that haploinsufficiency may contribute to oncogenesis (35, 36).
In this study, a conditional expression strategy was used to characterize the biological consequences of expressing oncogenic K-ras from its endogenous promoter in hematopoietic cells. Expression of oncogenic ras in the mice induced a myeloproliferative syndrome with short latency and high penetrance, similar to that observed with leukemia-associated activated receptor tyrosine kinases, and conferred growth factor–independent colony formation of primary bone marrow cells.
Methods
Mouse strains.
Lox-stop-lox (LSL)–K-ras G12D mice (on mixed 129Sv/Jae and C57BL/6 backgrounds) were crossed to Mx1-Cre transgenic mice (37) (on a BALB/c background) to obtain double-transgenic LSL–K-ras G12D+/Mx1-Cre+/– mice on a mixture of 129Sv/Jae, C57BL/6, and BALB/c genetic backgrounds. For induction of Cre expression, 4- to7-week-old mice were injected intraperitoneally with 250 μg of polyinosinic-polycytidylic acid (pI-pC; Sigma-Aldrich, St. Louis, Missouri, USA) every other day for three doses. All mice were housed in microisolator cages, were monitored daily for evidence of disease, and were sacrificed when moribund. All experiments were conducted with the ethical approval of the Harvard Medical Area Standing Committee on Animals.
Molecular and biochemical analysis.
LSL–K-ras G12D mice were genotyped by PCR amplification of genomic DNA obtained from tail tissue to detect the 500-bp wild-type K-ras and 550-bp floxed LSL–K-ras G12D products. For verification of Cre-mediated recombination, DNA from mouse bone marrow, spleen, and liver as well as from individual colonies of primary bone marrow methylcellulose cultures was amplified in PCR reactions using primers flanking the LSL cassette to detect the 285-bp wild-type K-ras and 315-bp excised lox–K-ras G12D products (38). For K-ras protein expression analysis, immunoprecipitation and Western blotting of tissue lysates from spleen, thymus, and lung were performed as described (39, 40).
Histopathology.
Mouse organs were fixed for at least 72 hours in 10% neutral buffered formalin (Sigma-Aldrich), dehydrated in alcohol, cleared in xylene, and infiltrated with paraffin. Tissue sections 4 μm in thickness from paraffin-embedded tissues blocks were placed on charged slides, deparaffinized in xylene, rehydrated through graded alcohol solutions, and stained with hematoxylin and eosin.
Flow cytometric analysis.
Spleen and bone marrow single-cell suspensions were prepared as described (41) by brief incubation in red blood cell lysis buffer (Gentra Systems, Minneapolis, Minnesota, USA) and freezing in 90% fetal bovine serum and 10% dimethyl sulfoxide. Prior to analysis, cells were washed in PBS/0.1% NaN3/0.1% BSA and were preincubated for 20 minutes on ice with supernatant from the 2.4G2 hybridoma cell line (anti-CD16/CD32; American Type Culture Collection, Rockville, Maryland, USA) to block nonspecific Fc receptor–mediated binding. Aliquots of 0.5 × 106 to 2.0 × 106 cells were stained for 20 minutes on ice with monoclonal antibodies, washed in staining buffer, and stained with secondary antibodies where necessary. Antibodies used were allophycocyanin (APC)-conjugated Gr-1 and CD4; phycoerythrin-conjugated Mac-1, Thy1.2, CD8, and CD117; biotin-conjugated CD19; and APC-conjugated streptavidin. All antibodies were purchased from Pharmingen (San Diego, California, USA), except for APC-CD4 and APC-streptavidin, which were purchased from Caltag (Burlingame, California, USA). Flow cytometric analysis was performed with a four-color FACSCalibur cytometer (Becton-Dickinson, Mountain View, California, USA). A minimum of 10,000 events was acquired and analyzed with CELLQUEST software.
For analysis of myeloid progenitors, bone marrow cells were stained as previously described (42) and analyzed with a highly modified triple-laser FACS (488-nm argon laser, 599-nm dye laser, and UV laser; FACSVantage; Becton Dickinson Immunocytometry Systems, Mountain View, California, USA). Myeloid progenitors were distinguished as Lin–Sca-1–c-Kit+CD34+FcγRII/IIIlo common myeloid progenitors (CMPs), Lin–Sca-1–c-Kit+CD34+FcγRII/IIIhi granulocyte-monocyte progenitors (GMPs), and Lin–Sca-1–c-Kit+CD34–FcγII/IIIlo megakaryocyte-erythrocyte progenitors (MEPs) (43).
Colony-forming assays.
Primary bone marrow cells from pI-pC–treated mice were cultured in methylcellulose-containing media using MethoCult GF M3434 media (StemCell Technologies, Vancouver, British Columbia, Canada) containing 50 ng/ml murine stem cell factor (SCF), 10 ng/ml murine IL-3, 10 ng/ml human IL-6, and 3 units/ml human erythropoietin (EPO), or MethoCult GF M3231 media from which SCF, IL3, IL6, and EPO were absent. For primary methylcellulose cultures, 4 × 103 to 1 × 105 cells were seeded in duplicate and harvested after 10 days. Cytospin preparations of individual colonies were stained with Wright-Giemsa solutions. Serial replating assays were performed as previously described (44) in two independent experiments with 104 cells replated in duplicate for each round of replating and colony counts performed on day 7.
Murine bone marrow transplantation.
Bone marrow cells were harvested from the femurs and tibias of pI-pC–treated LSL–K-ras G12D+/Mx1-Cre+ (KM+) mice. After red blood cell lysis, cells were washed in PBS, resuspended in Hank’s balanced salt solution (Invitrogen Life Technologies, Carlsbad, California, USA), and injected (106 cells/0.5 ml) into the lateral tail veins of sublethally irradiated (650 cGy) wild-type littermates. Mice were housed in microisolator cages and were provided with autoclaved chow and acidified water.
Results
Generation of mice expressing endogenous levels of oncogenic K-ras in the hematopoietic system.
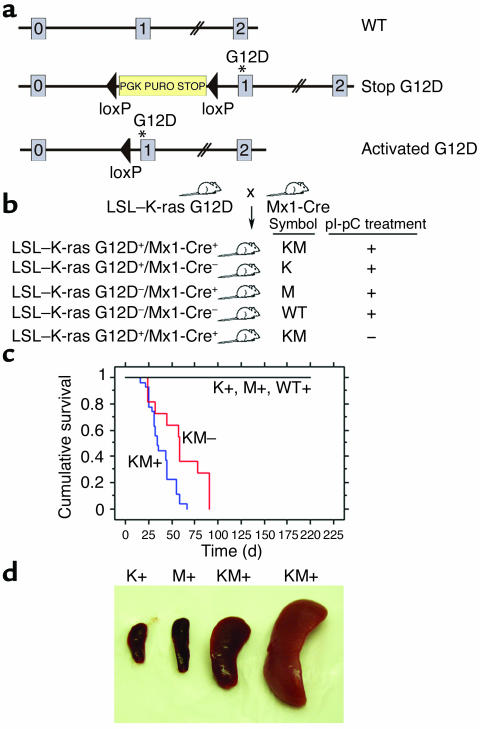
We studied LSL–K-ras G12D mice harboring a conditional oncogenic K-ras allele targeted to the endogenous K-ras locus by homologous recombination. The conditional allele contains a cassette with a transcriptional termination stop codon flanked by loxP sites (LSL cassette) located upstream of a mutation of glycine to aspartic acid in codon 12 (38) (Figure 1a). The stop cassette prevents potential deleterious consequences of expression of the oncogenic K-ras allele during development. However, Cre recombinase–mediated excision of the stop cassette in adult animals allows expression of oncogenic K-ras from the endogenous K-ras promoter sequences, thus preserving temporal, lineage-appropriate, and quantitative transcriptional regulation. For expression of oncogenic K-ras in the hematopoietic system, LSL–K-ras G12D mice were crossed to Mx1-Cre transgenic mice that express Cre from the IFN-α/β–inducible Mx1 promoter (Figure 1b) (37). Cre expression from the Mx1 promoter is induced in vivo by treating mice with IFN-α/β or pI-pC, a synthetic double-stranded RNA that induces expression of endogenous IFN. In this way, Cre expression, subsequent excision of the stop cassette, and expression of oncogenic K-ras will occur in cells expressing the IFN receptor, including hematopoietic cells.
Figure 1.
Lethal myeloproliferative disease in mice expressing a conditional oncogenic K-ras allele. (a) Schematic of wild-type (top), floxed (middle), and activated (bottom) K-ras alleles. K-ras exons 0, 1, and 2 are depicted. Gene targeting to the endogenous K-ras locus generated the floxed LSL–K-ras G12D allele (38) containing a transcriptional termination codon flanked by loxP sites upstream of a mutation of glycine to aspartic acid in codon 12 in exon 1. Excision of the stop cassette by Cre recombinase allows expression of the oncogenic K-ras allele. Asterisk indicates G12D mutation in exon 1. (b) Breeding schematic of LSL–K-ras G12D and Mx1-Cre mice, with subsequent pI-pC treatment of progeny to generate KM+, KM–, K+, M+, and WT+ littermate mice. K, LSL–K-ras G12D; M, Mx1-Cre. + or – indicates presence or absence of pI-pC treatment. (c) Kaplan-Meier comparative survival analysis of KM+, KM– and negative control mice. Cumulative survival was plotted against days after treatment with pI-pC. For KM– mice, cumulative survival was plotted against days after their littermates received pI-pC treatment. KM+ (n = 25) and KM– (n = 8) mice developed a lethal myeloproliferative disease with median latencies of 35 and 58 days, respectively. K+ (n = 11), M+ (n = 8), and WT+ (n = 10) mice were healthy during an observation period of more than 200 days. (d) Splenomegaly in mice expressing oncogenic K-ras. Spleen weights (left to right): K+, 70 mg; M+, 130 mg; KM+, 560 mg; and KM+, 2,200 mg.
Oncogenic K-ras expression in hematopoietic cells results in a myeloproliferative disorder.
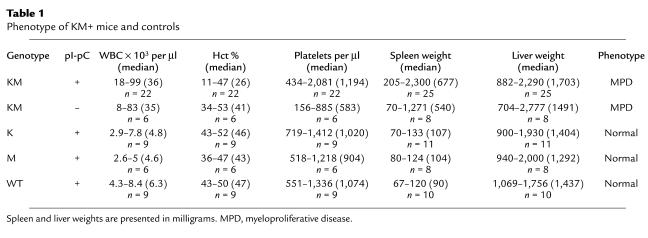
All pI-pC–treated LSL–K-ras G12D+/Mx1-Cre+ (KM+) mice developed a lethal hematopoietic disease with a median survival of 35 days (P < 0.0001 compared with negative controls by log rank test; range, 16–67 days) (Figure 1c). Of interest, all LSL–K-ras G12D+/Mx1-Cre+ mice that were not treated with pI-pC (KM– mice) also developed a similar hematopoietic disease with a longer median survival of 58 days (P = 0.0039, KM+ vs. KM– mice, by log rank test). The negative controls, single-transgenic LSL–K-ras G12D+ mice, Mx1-Cre+ mice, or wild-type littermate mice treated with pI-pC (K+, M+, or WT+ mice, respectively), showed no evidence of disease. The predominant phenotype identified in all KM+ mice examined was a myeloproliferative disorder characterized by leukocytosis, splenomegaly, and myeloid hyperplasia in the bone marrow. White blood cell (WBC) counts ranged between 18 × 103/μl and 99 × 103/μl with a median WBC count of 36 × 103/μl (Table 1). KM– mice also developed leukocytosis with median WBC counts of 35 × 103/μl. In the vast majority of cases, leukocytosis in KM+ mice was due to an increase in the granulocyte population. For example, in KM+ mice, neutrophils accounted for 15–65% of WBCs (median, 42%; n = 22), compared with K+, M+, and WT+ mice, in which neutrophils were 12–43% of WBCs (median, 19%; n = 24; P < 0.001 by Mann-Whitney test). There were no significant differences in the absolute numbers of other WBC subsets, although there was a slight increase in the percentage of atypical lymphocytes. KM+ mice were anemic, with a mean hematocrit of 27% (range, 11–48%), compared with a normal mean hematocrit of 45% in the K+, M+, and WT+ controls (P < 0.001 by Mann-Whitney test). There were no appreciable differences in the platelet counts between experimental and control mice.
Table 1.
Phenotype of KM+ mice and controls
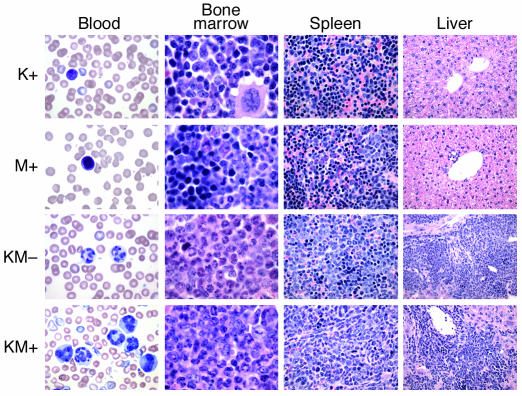
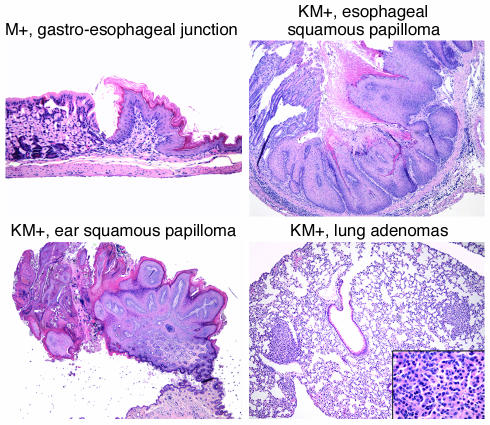
KM+ mice were emaciated, with ruffled fur and moderate-to-severe palpable splenomegaly. Spleen weights ranged between 205 and 2,300 mg, compared with normal-sized spleens (67–133 mg) in K+, M+, and WT+ negative control mice (P < 0.001; Table 1 and Figure 1d). Hematoxylin and eosin–stained sections of spleen showed marked extramedullary hematopoiesis with effacement of splenic architecture and expansion of the red pulp compartment by varying degrees of granulocytic, monocytic, erythroid, and megakaryocytic lineage cells (Figure 2). Eleven of sixteen cases examined demonstrated a predominantly granulocytic/monocytic proliferation within the spleen and a hypercellular bone marrow revealing myeloid (predominantly granulocytic) hyperplasia (Figure 2). In the remaining five of sixteen cases examined, marked extramedullary hematopoiesis in the spleen was also observed; however, this represented a striking erythroid expansion with fewer numbers of granulocytic and monocytic forms. A more subtle expansion of myeloid elements was seen in the bone marrow of these mice. In all cases examined, the liver showed significant perivascular and periportal infiltration by populations of myeloid and erythroid cells identical to those seen in the spleen (Figure 2). The overall hematopoietic phenotype was similar to the myeloproliferative diseases induced in murine bone marrow transplant assays by tyrosine kinase fusions such as BCR-ABL, TEL-PDGFRβ, TEL-TRKC, and TEL-JAK2 (17, 41, 45, 46). Interestingly, other pathological findings observed with high frequency but incomplete penetrance in KM+ mice were squamous papillomas involving the anal and vulvo-vaginal skin (12 of 19), ear (8 of 22), esophageal mucosa (14 of 17), and oral mucosa (8 of 25) (Figure 3). In addition, multiple nodules were observed in the lungs of KM+ mice (10 of 17), consisting of proliferating type II pneumocytes that stained positive for keratin (data not shown). These findings are consistent with the adenomas previously described in the lungs of LSL–K-ras G12D mice treated with intranasal instillation of adenovirus-expressing Cre (38). Finally, thymic T cell lymphoblastic lymphomas (7 of 25) and nodal lymphoid hyperplasia distinct from sinusoidal infiltrates of myeloid elements (6 of 17) were also noted.
Figure 2.
Mice expressing oncogenic K-ras develop a myeloproliferative disease. Representative histopathology (original magnifications in parentheses) from peripheral blood (×100), bone marrow (×100), spleen (×40), and liver (×20), showing expansion of predominantly mature myeloid elements, without an increase in immature/blast forms.
Figure 3.
Mice expressing oncogenic K-ras develop esophageal squamous papillomas and lung adenomas. Shown are normal gastroesophageal junction from M+ mouse (×10) and esophageal squamous papilloma (×5), ear squamous papilloma (×2), and lung adenomas (×5; inset, ×40) from KM+ mice. Original magnifications in parentheses.
In KM– mice, extramedullary hematopoiesis in the spleen and liver was also evident. In six cases examined, one mouse demonstrated a marked myeloproliferative disease; four had mild, predominantly myeloid extramedullary hematopoiesis in the spleen and liver with mild myeloid (predominantly granulocytic) hyperplasia in the marrow; and one mouse had no hematopoietic abnormalities. In a subset of KM– mice, pulmonary adenomas (five of six), thymic lymphomas (two of six), oral squamous papillomas (two of six), and ear squamous papillomas (two of six) were also observed. No vulvo-vaginal, anal or esophageal squamous papillomas were observed in KM– mice (n = 6).
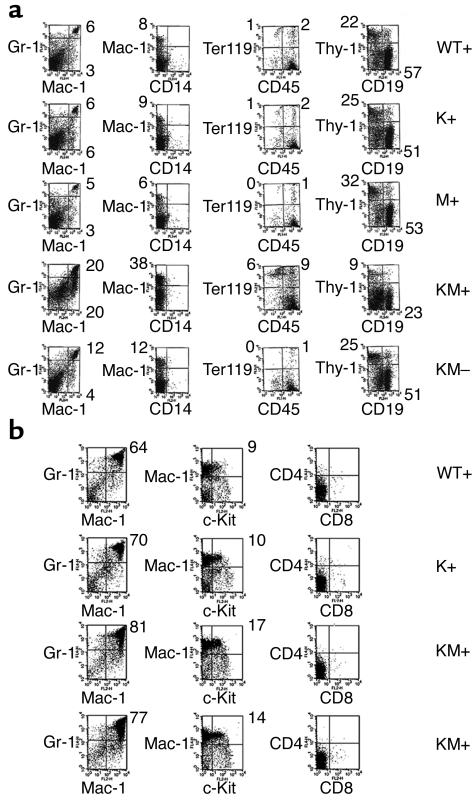
Flow cytometric analysis of spleen cells (Figure 4a) from KM+ mice further confirmed the myeloproliferative phenotype. In the spleen, 17–20% of cells were positive for the myeloid markers Gr-1 (Ly 6-G) and Mac-1, indicative of mature neutrophils, compared with 5–6% for K+, M+, and WT+ negative control mice. In untreated KM– mice, Gr-1+/Mac-1+ cells represented 12% of spleen cells. The increased percentage of myeloid cells was associated with a concomitant decrease in the percentages of CD4 and CD8 single-positive T cells and CD19+ B cells. Flow cytometric analysis of KM+ bone marrow cells also showed evidence of myelomonocytic expansion, with 77–81% Gr-1+/Mac-1+ cells, compared with 64–70% Gr-1+/Mac-1+ cells in negative control K+, M+, and WT+ mice (Figure 4b). Erythroid lineage Ter-119+ cells were present in normal percentages, except in anemic KM+ animals, which had an increased percentage of Ter-119+ cells (Figure 4a).
Figure 4.
Flow cytometric analysis of spleen and bone marrow cells. (a) Spleen cells from KM+ (n = 4), KM– (n = 2), and negative control WT+, K+, and M+ mice (n = 2 each) were stained with a combination of antibodies to Gr-1, Mac-1, CD14, CD45, Ter-119, Thy-1, and CD19. Dot plots were gated for live cells based on forward and side scatter profiles. Representative data are shown. (b) Bone marrow cells from KM+ (n = 4) and negative control mice (n = 2 each) were stained with a combination of antibodies to Gr-1, Mac-1, c-Kit, CD4, CD8, Ter-119, and CD45. Dot plots were gated for live cells based on forward and side scatter profiles. Representative data are shown. The percentages of cells in quadrants of interest are indicated.
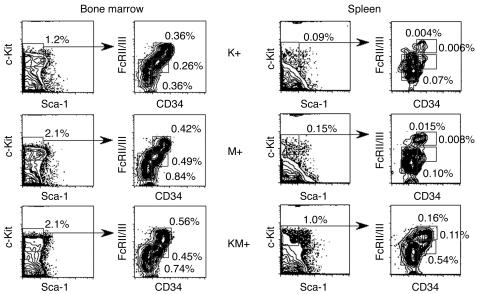
Further analysis of K+, M+, and KM+ bone marrow demonstrated no significant differences in the relative percentages of myeloid progenitor populations (Figure 5), composed of IL-7Rα–Lin–c-Kit+Sca-1– cells (43), or in the relative percentages of CMP, GMP, and MEP subpopulations (Figure 5). This result must be interpreted with caution, as it is not known whether K-ras is normally expressed in the myeloid progenitor compartment. In KM+ spleen, the relative percentages of CMP, GMP, and MEP populations increased, reflecting increased extramedullary hematopoiesis.
Figure 5.
Analysis of myeloid progenitors in bone marrow and spleen of K+, M+, and KM+ mice. Percentages of myeloid progenitors (IL-7Rα–Lin–Sca1–c-Kit+ cells) and percentages of CMPs (FcγRloCD34+), GMPs (FcγRhiCD34+), and MEPs (FcγRloCD34–) relative to whole bone marrow and spleen are indicated. Quadrants represent the respective gated populations of CMPs, GMPs, and MEPs.
Oncogenic K-ras expression.
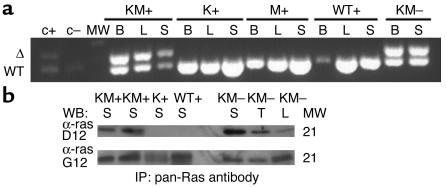
Cre-mediated excision of the stop cassette upstream of the oncogenic K-ras allele was demonstrated via PCR of genomic DNA from various tissues (Figure 6a). Only DNA from diseased KM+ and KM– tissues yielded a PCR product 30 bp larger than the wild-type allele, corresponding to excision of the transcriptional stop cassette and the presence of the oncogenic K-ras allele containing a single loxP site. Expression of oncogenic K-ras protein was demonstrated by Western blot analysis in diseased KM+ and KM– tissues but not in tissues from K+, M+, or WT+ negative control mice (Figure 6b).
Figure 6.
Cre-mediated activation of the oncogenic K-ras allele and expression of oncogenic K-ras protein in diseased KM+ and KM– tissues. (a) PCR for WT and activated (Δ) K-ras alleles demonstrates the presence of the activated K-ras allele in KM+ and KM– but not K+, M+, or WT+ tissues. B, bone marrow; L, liver; S, spleen; MW, molecular weight marker; c+, positive control DNA from an individual KM+ methycellulose colony; c–, negative control DNA from an individual K+ methylcellulose colony. (b) Oncogenic K-ras expression in diseased KM+ and KM– tissues. Tissues extracts were immunoprecipitated with a pan-ras antibody, followed by immunoblotting with polyclonal antibodies specific to wild-type ras (α-ras G12) and oncogenic ras G12D (α-ras D12). The WT ras doublet corresnomas; WB, Western blot; IP, immunoprecipitation.
Colony-forming unit activity.
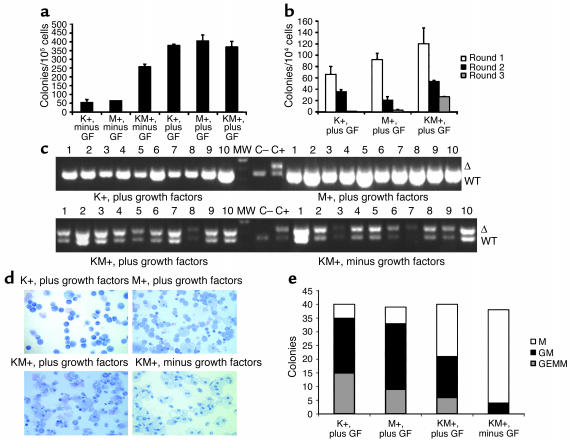
Bone marrow cells from KM+, K+, and M+ mice were plated in methylcellulose cultures in the presence or absence of growth factors (SCF, IL-3, IL-6, and EPO). All formed colonies in the presence of growth factors (Figure 7a). However, only bone marrow cells from KM+ mice readily formed colonies in a growth factor–independent manner. These colonies were smaller in size and less differentiated than colonies that grew in the presence of growth factors. In addition, colonies derived from KM+ bone marrow did not exhibit any enhanced serial replating activity (Figure 7b). Genomic DNA prepared from individual methylcellulose culture colonies was assayed for the presence of the activated K-ras allele. Only DNA from individual KM+ colonies yielded a PCR product corresponding to excision of the stop cassette and activation of the oncogenic K-ras allele (Figure 7c). The activated K-ras allele was detected in KM+ colonies that grew in the presence or absence of growth factors. In individual methylcellulose colonies, the Cre excision efficiency was 100% in KM+ bone marrow–derived cells (n = 42; Figure 7 and data not shown). The phenotype of individual colonies was determined by microscopic visualization of Wright-Giemsa–stained cytospin preparations (Figure 7, d and e). In duplicate experiments, no erythroid burst-forming unit (E-BFU) colonies were observed in K+, M+, or KM+ methylcellulose cultures harvested on day 10. Subsequent experiments with KM+ and negative control bone marrow cells demonstrated E-BFU colonies in comparable numbers when cultures were harvested on day 7 (data not shown). Colonies derived from K+, M+, or KM+ bone marrow cells grown in the presence of growth factors formed granulocyte-monocyte colony-forming unit (GM-CFU) and granulocyte-erythrocyte-monocyte-megakaryocyte colony-forming unit (GEMM-CFU) colonies with similar efficiencies. In contrast, colonies derived from KM+ bone marrow cells in the absence of growth factors were predominantly monocyte colony-forming units (M-CFUs), composed of >95% macrophages. Similar results demonstrating growth factor–independent colony formation of predominantly M-CFUs were observed in methylcellulose cultures of oncogenic K-ras bone marrow cells in a model using LSL–K-ras G12D+/Lys-Cre+ mice in which the oncogenic K-ras allele was activated in cells expressing Cre driven by the lysozyme M promoter (I.T. Chan and D.G. Gilliland, unpublished observations).
Figure 7.
Methylcellulose cultures with KM+, K+, and M+ bone marrow in the presence and absence of growth factors. (a) Number of colonies generated after methylcellulose culture of 100,000 bone marrow cells in the presence (plus) and absence (minus) of growth factors (GF: SCF, IL-3, and IL-6). Growth factor–independent colony-forming activity of KM+ bone marrow cells was demonstrated in two independent experiments. The values shown are the mean of duplicate cultures from one representative experiment. (b) Number of secondary, tertiary, and quaternary colonies generated in serial methylcellulose cultures using 104 input bone marrow cells in the presence of growth factors. Serial replating assays were performed in two independent experiments. The values shown are the means of duplicate cultures from one representative experiment. (c) PCR for WT and activated (Δ) K-ras alleles demonstrates presence of activated K-ras allele in individual methylcellulose colonies derived from KM+ bone marrow in the presence and absence of growth factors, but not from K+ or M+ colonies. The intensity of the PCR products generated from KM+ growth factor–independent colonies was less robust than that from KM+ growth factor–dependent colonies, corresponding to lesser amounts of input template genomic DNA purified from the smaller individual growth factor–independent colonies. (d) Cytospins (Wright-Giemsa stain) of individual methylcellulose colonies show GM-CFUs derived from K+, M+, and KM+ bone marrow cultured in the presence of growth factors, and M-CFUs from KM+ bone marrow cultured in the absence of growth factors. (e) Quantitation of M-CFU (M), GM-CFU (GM), and GEMM-CFU (GEMM) colonies from K+, M+, and KM+ bone marrow cultured in the presence of growth factors and KM+ bone marrow cultured in the absence of growth factors.
Transplantability.
Bone marrow cells (106) from diseased KM+ mice were transplanted into semilethally irradiated wild-type littermate secondary recipients. To date, all transplant recipients remain healthy and disease-free after 120 days (data not shown), indicating that the myeloproliferative disease was not transplantable into secondary recipients.
Discussion
Expression of oncogenic K-ras from its endogenous promoter in the hematopoietic system is sufficient to induce a myeloproliferative syndrome with relatively short latency. The myeloproliferative syndrome induced by oncogenic K-ras is similar to the myeloproliferative syndromes induced by activated receptor tyrosine kinases (e.g., FLT3-ITD, BCR-ABL, TEL-PDGFRβ, and TEL-JAK2). This finding is somewhat surprising, as receptor tyrosine kinases activate multiple signaling pathways in addition to ras, such as the PI3K and JAK/STAT pathways. Indeed, disruption of PI3K/AKT or STAT signal transduction by pharmacologic or genetic strategies can abrogate transformation properties and diseases induced by receptor tyrosine kinases (46–52). As oncogenic K-ras is expressed from its endogenous promoter in this study, we may infer from these observations that oncogenic K-ras is at least as potent an allele as constitutively activated receptor tyrosine kinases that activate several signaling pathways including ras/MAP kinase. The inference is supported by epidemiological observations that activating mutations in FLT3 and ras are commonly found in AML, but rarely coexist in the same patient. The myeloproliferative disease observed in KM+ mice appears similar to the JMML-like disease in mice transplanted with NF1–/– fetal liver cells (25) and the myeloproliferative disease observed with long latency and incomplete penetrance in the oncogenic N-ras bone marrow transplant model (30).
It is interesting that the predominant phenotype induced by oncogenic K-ras is a granulocyte-rich myeloproliferative disease, as Mx1 promoter activation appears to occur very early in hematopoietic development. In a model of conditional AML1-ETO expression utilizing Mx1-Cre–mediated recombination, persistent expression of AML1-ETO occurs throughout the life of the mice, suggesting that Cre-mediated deletion of the stop cassette occurs in long-term repopulating hematopoietic progenitors (1). Mx1-Cre–induced expression of oncogenic K-ras in hematopoietic stem cells might be expected to engender not only myeloid proliferation but also megakaryocyte hyperplasia or lymphoproliferative disease. Still, neutrophil lineage cells may be more sensitive to activation of the ras/MAP kinase pathway. This hypothesis is supported by the observation that loss of NF1 function in the germline also favors development of neutrophil lineage disease. Oncogenic K-ras may contribute to lymphoid diseases, as there was evidence of lymphoid hyperplasia (enlarged lymph nodes) and lymphoid malignancy (lymphoblastic lymphoma) in the KM+ and KM– mice. Mutations in ras have not been identified in patients with myeloproliferative diseases such as polycythemia vera and essential thrombocythemia (53–56). It is uncertain whether the erythroid hyperplasia seen in some KM+ mice is the result of secondary erythroid hyperplasia in response to anemia or oncogenic K-ras is capable of directly inducing erythropoiesis, albeit ineffectively. Excision of the stop cassette in erythroid, megakaryocytic, or T- and B-lymphoid–specific Cre strains will directly address the question of whether oncogenic K-ras is capable of inducing proliferative diseases in these lineages.
Earlier cell culture, transgenic and bone marrow transplant models of H- or N-ras overexpression using heterologous promoters in the hematopoietic system yielded various phenotypes, including myeloproliferative diseases/leukemias with long latency and incomplete penetrance (30), impaired erythroid and myeloid differentiation suggestive of myelodysplasia (27–29), and pre–T thymic lymphomas and/or pre–B cell lymphoblastic leukemias/lymphomas (31–34). The variety of malignancies arising in these models likely reflects the tissue specificities and expression levels of the promoters used rather than the specific transforming properties of oncogenic ras. In KM+ mice, one copy of the oncogenic K-ras allele is expressed from its endogenous promoter, which more accurately recapitulates the genotype of human AML.
KM– mice, which were not induced with pI-pC, also developed a myeloproliferative syndrome, albeit of intermediate severity, with lower WBC counts, mild to moderate splenomegaly, and longer latency than that of KM+ mice. This is most likely due to endogenous IFN expression of sufficient levels to induce the Mx1-Cre transgene and activate oncogenic K-ras expression. Indeed, untreated AML1-ETO+/Mx1-Cre+ mice demonstrate low levels of recombination in the bone marrow, ranging from 9% to 20% when monitored for 24 weeks (1). Similarly, untreated DNA polymerase β+/Mx1-Cre+ mice demonstrate background recombination levels as high as 10% in the spleen, also attributed to endogenous IFN production (37). It is likely that endogenous IFNs in untreated KM– mice activate expression of oncogenic K-ras in a small percentage of long-term repopulating hematopoietic progenitors. Oncogenic K-ras expression confers a proliferative advantage to these progenitors; eventually, their expansion yields the myeloproliferative phenotype, although with slightly delayed latency.
KM+ and KM– mice demonstrate additional histopathological findings with incomplete penetrance. Oncogenic ras mutations are detected in squamous papillomas and squamous cell carcinomas (57) and are important for initiation and progression of these lesions. Transgenic mice expressing v-H-ras from the ζ-globin promoter develop skin papillomas with high frequency in response to trauma or chemical carcinogens (58). In rare instances, activating ras mutations have been reported in lymphomas (59). The pattern of premalignant tumors observed in KM+ and KM– mice likely reflects the patterns of IFN-induced Mx1-Cre and subsequent oncogenic K-ras expression. In studies of Mx1-Cre–mediated recombination of a floxed DNA polymerase β gene, pI-pC treatment yielded high recombination efficiencies in hematopoietic tissues (liver, 100%; spleen, 94%; and thymus, 40%) and lower levels in nonhematopoietic tissues such as heart, kidney, lung, and duodenum (37). Experiments with mice doubly transgenic for the β-galactosidase reporter ROSA26tm1Sor and Mx1-Cre demonstrate Cre expression in liver, spleen, pulmonary endothelium, kidney, and gastric epithelium (60). Papillomas and thymic lymphomas have been previously observed in 30% of adenovirus-Cre–treated oncogenic K-ras mice (40). Additional mutations are likely required to form these premalignant lesions; the low frequency of these lesions observed in KM+ and KM– mice may reflect a stochastic requirement for acquisition of additional oncogenic mutations, as well as the varying levels of IFN in those specific tissues. It is unlikely that development of these lesions depends on the penetrance of pI-pC into the various tissues, as a subset of untreated KM– mice also develop the same pattern of papillomas, lung adenomas, and thymic lymphomas. KM+ mice may be a useful model system for studying carcinogenesis in squamous cell cancers with particular utility in modeling cancer prevention strategies.
Previous studies of ras overexpression in primary cells demonstrated growth arrest and apoptosis (61–63). More recently, it has been hypothesized that thresholds of ras activity exist, such that low, intermittent signaling contributes to cellular homeostasis; modest increases promote proliferation without triggering protective tumor suppressor or growth arrest pathways; and high, sustained levels induce growth arrest and senescence (7). Consistent with this hypothesis, our results with primary bone marrow methylcellulose cultures show that oncogenic ras expression from its endogenous promoter promotes a proliferative signal that bypasses the requirement for signals from growth factors and allows colony formation independent of growth factors. Growth factor–independent colony-forming activity of bone marrow cells has also been described in TEL-JAK2 and BCR/ABL murine bone marrow transplant models (51, 52). The oncogenic K-ras–expressing colonies do not have enhanced immortalization/self-renewal properties, as assessed in serial replating assays. In distinct contrast, primary mouse embryonic fibroblasts expressing endogenous levels of oncogenic K-ras bypass proliferative senescence and are immortalized (64). These differences are consistent with the hypothesis that the effects of oncogenic K-ras are dependent on cellular context (64). Additional mutations may be required to induce an immortalization or self-renewal phenotype in the hematopoietic cells.
Like activated receptor tyrosine kinases in myeloproliferative diseases, oncogenic ras is implicated in AML pathogenesis by virtue of its presence in a high percentage of AML samples. Still, oncogenic ras is not sufficient to induce AML, as oncogenic K-ras induced myeloproliferative disease is not transplantable into secondary recipients. Additional mutations are likely required. We favor a model in which the AML phenotype requires at least two cooperating mutations: one that promotes proliferation and enhanced survival (such as oncogenic ras or an activated receptor tyrosine kinase), and one associated with impaired differentiation and an immortalization phenotype (such as a loss-of-function mutation in a hematopoietic transcription factor, as has been reported for AML1-ETO; ref. 1). It will be important to test this hypothesis by analyzing oncogenic K-ras mice in the context of additional mutations associated with impaired hematopoietic differentiation, such as AML1-ETO (1) or PML-RARα (2). Ultimately, these oncogenic K-ras/Mx1-Cre models will be useful for testing novel therapeutic agents, such as farnesyltransferase and geranylgeranyltransferase inhibitors, which target ras (65, 66), and other agents such as ERK inhibitors, which target the ras/MAP kinase pathway.
Oncogenic ras mutations have been detected in other hematologic malignancies such as multiple myeloma, myelodysplasia, chronic myelomonocytic leukemia, and acute lymphoblastic leukemia (12, 13). As noted above, crosses of LSL–K-ras G12D mice with other transgenic mice strains expressing Cre from hematopoietic-specific promoters, such as Lys-Cre, Vav-Cre, CD19-Cre, and Lck-Cre, may yield new insights to the contributions of oncogenic K-ras in myeloproliferative and lymphoproliferative diseases as well as AML.
Many downstream effectors are activated by ras, including the Raf/MEK/ERK pathway, the PI3K/AKT pathway, and RalGEFs. It would be interesting to determine which of these pathways is required for oncogenic ras-mediated myeloproliferative disease. Selective mutation of the ras effector domain (residues 32–40) can disrupt binding and activation of individual effector pathways (67–70). Generation of additional mice harboring various conditional oncogenic K-ras mutations activating only single effector pathways for subsequent crosses to Mx1-Cre mice will yield additional insights into the pathogenesis of ras-induced myeloproliferative diseases with implications for the development of molecularly targeted therapies.
Finally, data are gradually accumulating regarding differences between H-, N-, and K-ras. The phenotypes of H-ras, N-ras, and K-ras knockout mice indicate clear distinctions in function and/or expression between family members. H-ras and N-ras knockout mice are viable with no overt abnormalities (71, 72). In contrast, K-ras knockout mice die at embryonic day 12.5 from severe anemia secondary to defects in the fetal liver microenvironment (39, 73). In addition, ras isoforms differ with respect to post-translational modifications, subcellular localization, and transformation properties (7, 11). Furthermore, N-ras mutations occur nearly exclusively in hematologic malignancies, whereas oncogenic K-ras and H-ras mutations are detected more often in solid tumors. In this Mx1-Cre mouse model, it is possible that the phenotypes from oncogenic N- or H-ras expression may be different from that of oncogenic K-ras. It will be informative to study the analogous conditional oncogenic H-ras and oncogenic N-ras alleles to understand differences in these ras isoforms and their contributions to oncogenesis.
Acknowledgments
We gratefully acknowledge administrative assistance from Alexis Bywater, and helpful discussions with members of the Gilliland lab. We also thank Benjamin Braun, Kevin Shannon, and Benjamin Neel for careful review of the manuscript. This work was supported in part by NIH grants DK51564 and CA66996 (D.G. Gilliland), a National Research Service Award for Postdoctoral Trainees (I.T. Chan) and the Leukemia and Lymphoma Society (D.G. Gilliland and I.T. Chan). D.G. Gilliland is an Associate Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: acute myeloid leukemia (AML); GTPase-activating protein (GAP); guanine nucleotide exchange factor (GEF); phosphoinositide-3-kinase (PI3K); internal tandem duplication (ITD); neurofibromatosis 1 (NF1); juvenile myelomonocytic leukemia (JMML); long terminal repeat (LTR); mouse mammary tumor virus (MMTV); lox-stop-lox (LSL); polyinosinic-polycytidylic acid (pI-pC); allophycocyanin (APC); common myeloid progenitor (CMP); granulocyte-monocyte progenitor (GMP); megakaryocyte-erythrocyte progenitor (MEP); stem cell factor (SCF); erythropoietin (EPO); pI-pC–treated LSL–K-ras G12D+/Mx1-Cre+ (KM+); LSL–K-ras G12D+/Mx1-Cre+ not treated with pI-pC (KM–); LSL–K-ras G12D+ treated with pI-pC (K+); Mx1-Cre+ treated with pI-pC (M+); wild-type littermate treated with pI-pC (WT+); white blood cell (WBC); erythroid burst-forming unit (E-BFU); monocyte colony-forming unit (M-CFU); granulocyte-erythrocyte-monocyte-megakaryocyte colony-forming unit (GEMM-CFU).
References
- 1.Higuchi M, et al. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 2.Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RARα under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 3.He LZ, et al. Acute leukemia with promyelocytic features in PML/RARα transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown D, et al. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu. Rev. Genomics Hum. Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 6.Gilliland DG. Hematologic malignancies. Curr. Opin. Hematol. 2001;8:189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Hingorani SR, Tuveson DA. Ras redux: rethinking how and where Ras acts. Curr. Opin. Genet. Dev. 2003;13:6–13. doi: 10.1016/s0959-437x(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Sagi D. A Ras by any other name. Mol. Cell. Biol. 2001;21:1441–1443. doi: 10.1128/MCB.21.5.1441-1443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheele JS, Ripple D, Lubbert M. The role of ras and other low molecular weight guanine nucleotide (GTP)-binding proteins during hematopoietic cell differentiation. Cell. Mol. Life Sci. 2000;57:1950–1963. doi: 10.1007/PL00000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain’t over ‘til it’s over’. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 11.Ellis CA, Clark G. The importance of being K-Ras. Cell. Signal. 2000;12:425–434. doi: 10.1016/s0898-6568(00)00084-x. [DOI] [PubMed] [Google Scholar]
- 12.Beaupre DM, Kurzrock R. RAS and leukemia: from basic mechanisms to gene-directed therapy. J. Clin. Oncol. 1999;17:1071–1079. doi: 10.1200/JCO.1999.17.3.1071. [DOI] [PubMed] [Google Scholar]
- 13.Reuter CW, Morgan MA, Bergmann L. Targeting the Ras signaling pathway: a rational, mechanism-based treatment for hematologic malignancies? Blood. 2000;96:1655–1669. [PubMed] [Google Scholar]
- 14.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 15.Beghini A, et al. C-kit mutations in core binding factor leukemias. Blood. 2000;95:726–727. [PubMed] [Google Scholar]
- 16.Eguchi M, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25) Blood. 1999;93:1355–1363. [PubMed] [Google Scholar]
- 17.Liu Q, et al. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. EMBO J. 2000;19:1827–1838. doi: 10.1093/emboj/19.8.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papadopoulos P, Ridge SA, Boucher CA, Stocking C, Wiedemann LM. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. [PubMed] [Google Scholar]
- 19.Golub TR, Barker GF, Stegmaier K, Gilliland DG. Involvement of the TEL gene in hematologic malignancy by diverse molecular genetic mechanisms. Curr. Top. Microbiol. Immunol. 1996;211:279–288. doi: 10.1007/978-3-642-85232-9_28. [DOI] [PubMed] [Google Scholar]
- 20.Kelly LM, et al. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 21.Million RP, Aster J, Gilliland DG, Van Etten RA. The Tel-Abl (ETV6-Abl) tyrosine kinase, product of complex (9;12) translocations in human leukemia, induces distinct myeloproliferative disease in mice. Blood. 2002;99:4568–4577. doi: 10.1182/blood-2001-12-0244. [DOI] [PubMed] [Google Scholar]
- 22.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 23.Shannon KM, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N. Engl. J. Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 24.Jacks T, et al. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 25.Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat. Genet. 1996;12:137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 26.Bollag G, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat. Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 27.Darley RL, Hoy TG, Baines P, Padua RA, Burnett AK. Mutant N-RAS induces erythroid lineage dysplasia in human CD34+ cells. J. Exp. Med. 1997;185:1337–1347. doi: 10.1084/jem.185.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darley RL, et al. Protein kinase C mediates mutant N-Ras-induced developmental abnormalities in normal human erythroid cells. Blood. 2002;100:4185–4192. doi: 10.1182/blood-2002-05-1358. [DOI] [PubMed] [Google Scholar]
- 29.Darley RL, Hoy TG, Robinson L, Burnett AK. Mutant N-Ras blocks granulocytic differentiation of human CD34+ cells. Br. J. Haematol. 1998;102:289. [Google Scholar]
- 30.MacKenzie KL, Dolnikov A, Millington M, Shounan Y, Symonds G. Mutant N-ras induces myeloproliferative disorders and apoptosis in bone marrow repopulated mice. Blood. 1999;93:2043–2056. [PubMed] [Google Scholar]
- 31.Hawley RG, Fong AZ, Ngan BY, Hawley TS. Hematopoietic transforming potential of activated ras in chimeric mice. Oncogene. 1995;11:1113–1123. [PubMed] [Google Scholar]
- 32.Dunbar CE, Crosier PS, Nienhuis AW. Introduction of an activated RAS oncogene into murine bone marrow lymphoid progenitors via retroviral gene transfer results in thymic lymphomas. Oncogene Res. 1991;6:39–51. [PubMed] [Google Scholar]
- 33.Mangues R, Symmans WF, Lu S, Schwartz S, Pellicer A. Activated N-ras oncogene and N-ras proto-oncogene act through the same pathway for in vivo tumorigenesis. Oncogene. 1996;13:1053–1063. [PubMed] [Google Scholar]
- 34.Sinn E, et al. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 35.Song WJ, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 36.Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15:2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 38.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 41.Schwaller J, et al. Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. EMBO J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto T, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 43.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 44.Deguchi K, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 45.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 46.Tomasson MH, et al. Fatal myeloproliferation, induced in mice by TEL/PDGFβR expression, depends on PDGFβR tyrosines 579/581. J. Clin. Invest. 2000;105:423–432. doi: 10.1172/JCI8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen MH, Ho JM, Beattie BK, Barber DL. TEL-JAK2 mediates constitutive activation of the phosphatidylinositol 3′-kinase/protein kinase B signaling pathway. J. Biol. Chem. 2001;276:32704–32713. doi: 10.1074/jbc.M103100200. [DOI] [PubMed] [Google Scholar]
- 48.Neshat MS, Raitano AB, Wang HG, Reed JC, Sawyers CL. The survival function of the Bcr-Abl oncogene is mediated by Bad-dependent and -independent pathways: roles for phosphatidylinositol 3-kinase and Raf. Mol. Cell. Biol. 2000;20:1179–1186. doi: 10.1128/mcb.20.4.1179-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dierov J, Xu Q, Dierova R, Carroll M. TEL/platelet-derived growth factor receptor beta activates phosphatidylinositol 3 (PI3) kinase and requires PI3 kinase to regulate the cell cycle. Blood. 2002;99:1758–1765. doi: 10.1182/blood.v99.5.1758. [DOI] [PubMed] [Google Scholar]
- 50.Sternberg DW, et al. The TEL/PDGFβR fusion in chronic myelomonocytic leukemia signals through STAT5-dependent and STAT5-independent pathways. Blood. 2001;98:3390–3397. doi: 10.1182/blood.v98.12.3390. [DOI] [PubMed] [Google Scholar]
- 51.Schwaller J, et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 52.Sattler M, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell. 2002;1:479–492. doi: 10.1016/s1535-6108(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 53.Tsurumi S, et al. N-ras and p53 gene mutations in Japanese patients with myeloproliferative disorders. Am. J. Hematol. 2002;71:131–133. doi: 10.1002/ajh.10188. [DOI] [PubMed] [Google Scholar]
- 54.Mavrogianni D, et al. Leukemogenic risk of hydroxyurea therapy as a single agent in polycythemia vera and essential thrombocythemia: N- and K-ras mutations and microsatellite instability in chromosomes 5 and 7 in 69 patients. Int. J. Hematol. 2002;75:394–400. doi: 10.1007/BF02982131. [DOI] [PubMed] [Google Scholar]
- 55.Gaidano G, Guerrasio A, Serra A, Rege-Cambrin G, Saglio G. Molecular mechanisms of tumor progression in chronic myeloproliferative disorders. Leukemia. 1994;8:S27–S29. [PubMed] [Google Scholar]
- 56.Gaidano G, et al. Genetic lesions associated with blastic transformation of polycythemia vera and essential thrombocythemia. Genes Chromosomes Cancer. 1997;19:250–255. [PubMed] [Google Scholar]
- 57.Yuspa SH. The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis. J. Dermatol. Sci. 1998;17:1–7. doi: 10.1016/s0923-1811(97)00071-6. [DOI] [PubMed] [Google Scholar]
- 58.Leder A, Kuo A, Cardiff RD, Sinn E, Leder P. v-Ha-ras transgene abrogates the initiation step in mouse skin tumorigenesis: effects of phorbol esters and retinoic acid. Proc. Natl. Acad. Sci. U. S. A. 1990;87:9178–9182. doi: 10.1073/pnas.87.23.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nedergaard T, Guldberg P, Ralfkiaer E, Zeuthen J. A one-step DGGE scanning method for detection of mutations in the K-, N-, and H-ras oncogenes: mutations at codons 12, 13 and 61 are rare in B-cell non-Hodgkin’s lymphoma. Int. J. Cancer. 1997;71:364–369. doi: 10.1002/(sici)1097-0215(19970502)71:3<364::aid-ijc10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 60.Schneider A, Zhang Y, Guan Y, Davis LS, Breyer MD. Differential, inducible gene targeting in renal epithelia, vascular endothelium, and viscera of Mx1Cre mice. Am. J. Physiol. Renal Physiol. 2003;284:F411–F417. doi: 10.1152/ajprenal.00235.2002. [DOI] [PubMed] [Google Scholar]
- 61.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 62.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 63.Ferbeyre G, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol. Cell. Biol. 2002;22:3497–3508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 65.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 66.Ahmadian MR. Prospects for anti-ras drugs. Br. J. Haematol. 2002;116:511–518. doi: 10.1046/j.0007-1048.2001.03314.x. [DOI] [PubMed] [Google Scholar]
- 67.White MA, et al. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 68.Khosravi-Far R, et al. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol. Cell. Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodriguez-Viciana P, et al. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 70.Hamad NM, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1709–1713. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ise K, et al. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene. 2000;19:2951–2956. doi: 10.1038/sj.onc.1203600. [DOI] [PubMed] [Google Scholar]
- 73.Koera K, et al. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]