Abstract
The tumor suppressor p53, a sensor of multiple forms of cellular stress, is regulated by post-translational mechanisms to induce cell-cycle arrest, senescence, or apoptosis. We demonstrate that endoplasmic reticulum (ER) stress inhibits p53-mediated apoptosis. The mechanism of inhibition involves the increased cytoplasmic localization of p53 due to phosphorylation at serine 315 and serine 376, which is mediated by glycogen synthase kinase-3 β (GSK-3β). ER stress induces GSK-3β binding to p53 in the nucleus and enhances the cytoplasmic localization of the tumor suppressor. Inhibition of apoptosis caused by ER stress requires GSK-3β and does not occur in cells expressing p53 with mutation(s) of serine 315 and/or serine 376 to alanine(s). As a result of the increased cytoplasmic localization, ER stress prevents p53 stabilization and p53-mediated apoptosis upon DNA damage. It is concluded that inactivation of p53 is a protective mechanism utilized by cells to adapt to ER stress.
Keywords: Endoplasmic reticulum stress, p53, glycogen synthase kinase-3β, protein phosphorylation, protein localization, apoptosis
The tumor suppressor gene p53 is frequently mutated in human cancers (Vousden and Lu 2002). Its product, the p53 protein is induced and phosphorylated by various forms of cellular stress, including γ-irradiation, UV radiation, DNA cross-linking, oxidative stress, hypoxia, and nucleotide depletion (Wahl and Carr 2001; Sharpless and DePinho 2002; Vousden and Lu 2002). Activated p53 functions as a transcription factor to regulate the expression of many different downstream genes, whose products are implicated in cell cycle arrest, DNA repair, or apoptosis (Vousden and Lu 2002). To achieve proper function, p53 is tightly regulated by means of post-translational modifications, cofactor binding, and subcellular localization. The function of p53 is tightly controlled by Mdm2, an E3 ubiquitin ligase implicated in the inactivation of the tumor suppressor by accelerating its nuclear export and degradation by the 26S proteasome (Michael and Oren 2002). Phosphorylation of p53 within its amino-terminal domain facilitates p53 stabilization by disrupting p53-Mdm2 interaction (Wahl and Carr 2001; Michael and Oren 2002) and prevents its nucleocytoplasmic export (Zhang and Xiong 2001).
Similar to nuclear DNA damage, stress conditions in other organelles are able to activate signal-transduction pathways leading to the induction of genes encoding for proteins that play key roles in damage sensing and apoptosis (Ferri and Kroemer 2001). For example, expression of mutant proteins, viral infection, energy or nutrient deprivation, extreme environmental conditions, or Ca2+ release from the lumen of the endoplasmic reticulum (ER) disrupt proper protein-folding activity in this organelle (Ferri and Kroemer 2001; Kaufman et al. 2002). This leads to the accumulation of unfolded proteins, which initiates transcriptional and translational-signaling pathways known as the unfolded protein response (UPR; Ferri and Kroemer 2001; Kaufman et al. 2002). UPR is an adaptive response that involves the up-regulation of the expression, and thus function of ER-resident chaperons that augment ER-folding capacity (Ferri and Kroemer 2001; Kaufman et al. 2002). Also, UPR induces the expression of genes engaged in ER-associated protein degradation (Travers et al. 2000) and attenuates translation by inducing the phosphorylation of the α subunit of translation initiation factor eIF2 through the activation of the pancreatic ER-resident kinase PERK (Harding et al. 2002). If these adaptive mechanisms are not sufficient to alleviate ER stress, then an apoptotic program is initiated through the activation of the JNK pathway and caspases 7, 12, and 3 (Ferri and Kroemer 2001; Harding et al. 2002; Kaufman et al. 2002).
Given the role of p53 in stress sensing and proapoptotic signaling, we were interested to investigate whether p53 responds to ER stress. Herein, we report that ER stress induced by pharmacological or physiological means signals to p53. We demonstrate that ER stress induces the destabilization of p53 protein and prevents cells from p53-dependent apoptosis. This is mediated, at least in part, through the increased cytoplasmic localization of p53 as a result of phosphorylation at serines 315 and 376. We also demonstrate that ER stress induces glycogen synthase-3 β (GSK-3β) kinase activity, which phosphorylates p53 at serine 376 in vitro and mediates p53 phosphorylation at serines 315 and 376 in vivo. Furthermore, we show that GSK-3β interacts physically with p53 in the nucleus of ER-stressed cells, promotes the cytoplasmic localization of the protein, and prevents p53-mediated apoptosis. Our findings reveal a novel mechanism utilized by cells to adapt to ER stress through the inactivation of the tumor-suppressor protein by GSK-3β.
Results
ER stress enhances the cytoplasmic localization of p53
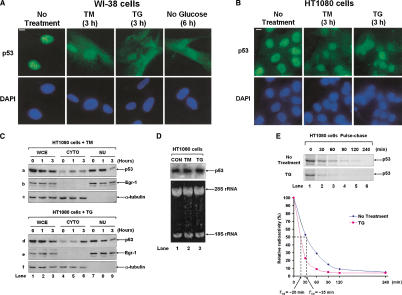
We first noticed that ER stress induces the cytoplasmic localization of p53. Specifically, human diploid WI-38 cells (Fig. 1A) or human fibrosarcoma HT1080 cells (Fig. 1B) were treated with pharmacological inducers of ER stress, such as the protein glycosylation inhibitor tunicamycin (TM), the lumenal Ca2+ mobilizing agent thapsigargin (TG), or glucose deprivation, which is another cause for ER stress (Scheuner et al. 2001). We observed that nuclear staining of endogenous wild-type p53 was decreased for both cell types with a concomitant increase in cytoplasmic staining, which was more obvious for WI-38 cells (Fig. 1A,B). To verify the cytoplasmic localization of p53 in HT1080 cells, ER-stressed cells were subjected to subcellular fractionation, and p53 levels were detected by immunoblotting (Fig. 1C). We found that the cytoplasmic levels of p53 were increased after TM (Fig. 1C, panel a, lanes 5 and 6) or TG treatment (Fig. 1C, panel d, lanes 5 and 6), which correlated with a concomitant decrease in the nuclear p53 levels (Fig. 1C, panels a and d, lanes 8 and 9). The purity of the nuclear and cytoplasmic fractions was verified by immunoblotting with antibodies against the transcription factor Egr-1 (Fig. 1C, panels b and e) and α-tubulin (Fig. 1C, panels c and f), respectively. These findings suggested that ER stress promotes the cytoplasmic localization of the tumor suppressor protein.
Figure 1.
ER stress alters the subcellular localization of p53. Immunostaining of endogenous p53. WI-38 cells (A) or HT1080 cells (B) were treated with 10 μg/mL of TM, 1 μM of TG, or incubated in glucose-free medium for the indicated time. Endogenous p53 was visualized by immunostaining with anti-p53 monoclonal antibodies (top) as described in Materials and Methods. The exposure time for green fluorescence was identical in all samples (A,B; top). The nucleus was detected by DAPI staining (A,B; bottom). (C) Subcellular fractionation of HT1080 cells. Cells were treated with either 10 μg/mL of TM or 1 μM of TG. At the indicated time points, cytoplasmic and nuclear fractions were prepared as described in Materials and Methods. Whole-cell extracts (WCE; 50 μg of protein), cytoplasmic extracts (CYTO; 40 μg of protein), or nuclear extracts (NU; 8 μg of protein) were subjected to Western blotting with anti-p53 rabbit polyclonal antibody (a,b), anti-Egr-1 rabbit polyclonal antibody (b,e), or anti-α-tubulin mouse monoclonal antibody (c,f). (D) HT1080 cells were treated with either 10 μg/mL of TM or 1 μM of TG for 2 h, total RNA (10 μg) was isolated and subjected to Northern Blotting using [32P]dCTP human p53 cDNA as probe (top). The quality of RNA and equal loading were verified by staining the denaturing gel with ethidium bromide. The position of 28S and 18S rRNA are shown (bottom). (E) HT1080 cells were labeled with [35S]methionine/cysteine for 30 min and chased with radioactive-free medium in the absence or presence of 1 μM of TG for the indicated times. Protein extracts (1 mg) were subjected to immunoprecipitation with anti-p53 rabbit polyclonal antibody followed by SDS-PAGE and autoradiography. The radioactive bands were quantified by PhosphorImager, and the plots of the relative intensity of radioactive p53 bands toward time and the half-life time values are indicated.
To better understand howp53 is regulated in ER-stressed cells, we measured p53 mRNA and protein stability in HT1080 cells. Specifically, Northern blot analysis indicated that p53 mRNA levels were not affected by TM or TG treatment (Fig. 1D). On the other hand, [35S]methionine labeling and pulse-chase analysis indicated that TG treatment decreased the half-life of p53 protein (Fig. 1E), suggesting that ER stress promotes the destabilization of the tumor suppressor.
ER stress impairs p53 stabilization and p53-dependent apoptosis in response to DNA damage
Given that ER stress promotes the cytoplasmic localization and degradation of p53, we next sought to examine whether ER stress interferes with p53 stabilization in response to DNA damage. Treatment of human diploid WI-38 cells with the anticancer drug adriamycin (ADR) resulted in the induction of p53 protein (Fig. 2A, top, lanes 2 and 3), which was significantly reduced in the presence of either TM (Fig. 2A, top, lanes 4 and 5) or TG (Fig. 2A, top, lanes 6 and 7). Similar to DNA damage with ADR, induction of p53 protein was significantly impaired in ER-stressed cells subjected to γ-irradiation (γ-IR; Supplementary Fig. 1B) or exposed to ultra violet (UV) light (data not shown). Because DNA damage stabilizes p53 by inducing its phosphorylation within the amino terminus, which blocks its Mdm2-mediated degradation (Wahl and Carr 2001), we thought that p53 destabilization in ER-stressed cells might have also been due to an inhibition of p53 phosphorylation within the amino terminus domain upon DNA damage. To examine this possibility, we performed a treatment with actinomycin D (ACD), which stabilizes p53 in the absence of amino terminus phosphorylation at serine 15 and serine 20 (Ashcroft et al. 2000). We found that ER stress prevented p53 stabilization induced by ACD in WI-38 cells (Fig. 2B), indicating that phosphorylation of the amino terminus is not essential for the effects of ER stress on p53 in cells subjected to DNA damage. We also noticed that DNA damage-mediated activation of Chk2 kinase, which was manifested by a shift of protein mobility in polyacryl-amide gels (Hirano et al. 2000), was not impaired in the presence of ER stress (Supplementary Fig. 1). This data provided evidence that ER stress does not affect the ATM/ATR pathway, which induces the phosphorylation of p53 in response to DNA damage (Wahl and Carr 2001).
Figure 2.
ER stress prevents p53 stabilization and impairs p53-mediated apoptosis in response to genotoxic stress. (A) WI-38 cells were treated with 1 μM of ADR alone or with either 10 μg/mL of TM or 1 μM of TG for the indicated times. Protein extracts (50 μg) were used for immunoblotting with an anti-p53 rabbit polyclonal antibody. A nonspecific (NS) band on the same blot was used as loading control. (B) WI-38 cells were treated with 5 nM ACD in the absence or presence of either 1 μg/mL of TM or 0.1 μM of TG for the indicated times. Protein extracts (50 μg) were immunoblotted with an anti-p53 rabbit polyclonal antibody. Blot was stripped and reprobed with an anti-actin antibody. (C) HCT116 p53+/+ cells were treated with 375 μM of 5-FU in the absence or presence of either 1 μg/mL of TM or 0.1 μM of TG for the indicated times. The protein levels of p53 were detected by immunoblotting of 50 μg protein extracts with anti-p53 rabbit polyclonal antibody. A nonspecific band on the same blot was used as loading control. (D,E) HCT116 p53 +/+and HCT116 p53-/- cells were pretreated with either 0.3 μg/mL of TM or 0.03 μM of TG for 1 h, followed by treatment with 375 μM of 5-FU for 3 h. The cells were fed with fresh medium and subjected either to FACS analysis 48 h post-treatment (D) or colony formation assay (E) as described in Materials and Methods. The numbers represent the average values of colony formation quantification for each treatment from three independent experiments. Untreated (CON) cells were scored as 100%.
To further substantiate our findings, we examined the effects of ER stress on p53-mediated cell death induced by DNA damage. For this experiment, we chose the human colon carcinoma HCT116 cells, which contain wild-type p53, and the isogenic derivatives with inactivated p53 gene by a tissue culture knockout approach (Bunz et al. 1998). Because HCT116 p53+/+ cells are more susceptible than p53-/- cells to the killing effects of 5-fluorouracil (5-FU) treatment (Bunz et al. 1999), we reasoned that inhibition of p53 activation by ER stress should rescue the p53+/+ cells, but not the p53-/- cells from 5-FU-induced death. First, we confirmed that treatment of HCT116 p53+/+ cells with either TM or TG prevents p53 stabilization in response to 5-FU treatment (Fig. 2C, top). Then, we examined p53-mediated cell death either by FACS analysis (Fig. 2D) or by colony formation assays (Fig. 2E). To diminish the cytotoxic effects of TM or TG, low concentrations of the ER stress inducers were used. The optimal concentration of 5-FU for HCT116 cells was reported previously to be 375 μM (Bunz et al. 1999). FACS analysis demonstrated that a higher population of HCT116 p53+/+ cells in sub-G1 phase (∼55%), as opposed to those cells treated with ER stress plus 5-FU (∼30%; Fig. 2D). On the other hand, HCT116 p53-/- cells were resistant to the killing effects of 5-FU either in the absence or presence of ER stress. In these experiments, we noticed that treatment of HCT116 cells with the ER stress inducers alone did not induce a G1 arrest as reported previously for other cells (Brewer et al. 1999). The lack of an effect of ER stress on G1 arrest in our experiments was most likely due to low drug concentrations, as higher concentrations were able to elicit a G1 arrest response in HCT116 cells independently of p53 (data not shown). However, higher drug concentrations were also cytotoxic. Consistent with FACS analysis data, colony formation assays demonstrated that HCT116 p53+/+ cells treated with either TM or TG were protected from 5-FU-mediated suppression on clonogenic survival, as opposed to HCT116 p53-/- cells, which were considerably resistant to 5-FU regardless of ER stress (Fig. 2E). Collectively, ER stress inhibits p53-dependent cell death in response to DNA damage.
Phosphorylation-dependent cytoplasmic localization of p53 in ER-stressed cells
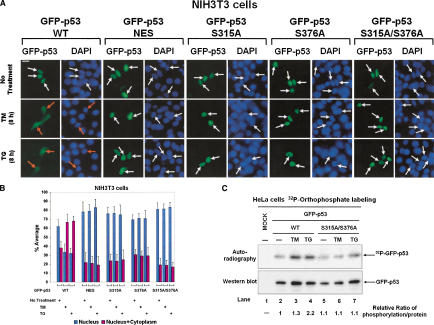
To further investigate the mechanisms of increased p53 cytoplasmic localization upon ER stress, we used a chimeric protein composed of a GFP marker fused with human wild-type p53 (GFP-p53 WT; Boyd et al. 2000). We also used a mutant of human p53 bearing the L348A and L350A substitutions within the carboxy-terminal NES (GFP-p53 NES; Boyd et al. 2000). GFP-p53 NES is defective in nucleocytoplasmic shuttling, and therefore, it is always localized in the nucleus (Stommel et al. 1999; Boyd et al. 2000). Because localization of GFP-p53 proteins was originally characterized in NIH-3T3 cells (Boyd et al. 2000), we initially used these cells to analyze the effects of ER stress on p53 localization. We noticed that the GFP-p53 was predominantly nuclear in transfected NIH-3T3 cells in the absence of treatment (Fig. 3A) as described previously (Boyd et al. 2000). On the contrary, treatment with either TM or TG increased the cytoplasmic presence of GFP-p53 WT. Importantly, GFP-p53 NES was unable to relocalize to the cytoplasm after ER stress, indicating that increased cytoplasmic localization of p53 in ER-stressed cells requires an intact NES. Quantitative analysis of the nuclear localization of the GFP fusion proteins demonstrated the cytoplasmic relocation of GFP-p53 WT, but not of GFP-p53 NES in ER-stressed NIH-3T3 cells (Fig. 3B). Similar results were obtained when GFP-p53 proteins were expressed in HT1080 cells or human colon carcinoma HCT116 cells (data not shown).
Figure 3.
Phosphorylation-dependent cytoplasmic localization of p53 in ER-stressed cell. (A) Subcellular localization of GFP-p53. Plasmid DNA (0.5 μg) containing either GFP-p53 WT or each of the indicated GFP-p53 mutants was transiently transfected into NIH-3T3 cells. Twenty-four hours later, cells were left untreated or treated with either 10 μg/mL of TM or 1 μM of TG for 8 h, and then examined for GFP-p53 fluorescence. Cell nuclei were visualized by staining with DAPI. White arrows indicate nuclear localization of GFP-p53 only, whereas orange arrows indicate either cytoplasmic or both cytoplasmic and nuclear localization of GFP-p53. (B) Quantitative analysis of GFP-p53 localization. For each condition, 1000 GFP-p53 positive and live cells were scored. Cells were classified into two groups as follows: the first with predominantly nuclear p53 (blue bars) and the second with p53 both in the nucleus and cytoplasm (red bars). Values are means ± SD from six separate experiments. (C) Induced phosphorylation of GFP-p53 by ER stress in vivo. HeLa cells were transfected with 1 μg of either GFP-p53 WT or GFP-p53 S315A/S376A plasmid DNA. Twenty-four hours later, cells were subjected to [32P]orthophosphate labeling followed by immnunoprecipitation of 500 μg of protein extracts with anti-p53 rabbit polyclonal antibody. Half of the immunoprecipitates were subjected to SDS-PAGE and autoradiography (top; 4 h exposure), the other half to immunoblotting with anti-p53 polyclonal antibody followed by ECL detection for 30 sec (bottom). The relative ratio of p53 phosphorylation was calculated by normalizing the intensity of phosphorylated p53 to the intensity of total p53 protein detected by ECL.
The carboxyl terminus of human p53 contains one bipartite nuclear localization signal (NLS) at amino acids 305-322 (i.e., K305RALPNNTSSS315PQKKKP322) and two monopartite NLSs at amino acids 369-375 and 379-384 (i.e., L369KSKKGQS376TS378RHKKLM384; Liang and Clarke 2001). These NLSs are also conserved in mouse and rat p53 (Liang and Clarke 2001). The bipartite NLS is the most active in directing p53 nuclear import, whereas the two monopartite NLSs exhibit weaker effects (Liang and Clarke 2001). Phosphorylation within these NLSs has been thought to influence the accessibility of p53 to nuclear import receptors, thus modulating the nuclear translocation of the protein (Liang and Clarke 2001). Potential targets include serine 315 within the bipartite NLS and serine 376 between the two bipartite NLSs (Liang and Clarke 2001).
To test whether phosphorylation within the NLSs plays a role in the nuclear import of p53, we generated GFP-p53 proteins with serine to alanine mutations of residue 315 (S315A), 376 (S376A), or residues 315 and 376 (S315A/S376A). NIH-3T3 cells were then transfected with each of the GFP-p53 mutant constructs, and the localization of the fusion proteins was tested by GFP fluorescence in unstressed cells as well as in cells stressed with either TM or TG. We observed that localization of S315A, S376A, or S315A/S376A mutant in the presence of ER stress remained exclusively nuclear in contrast to GFP-p53 WT, which was both nuclear and cytoplasmic (Fig. 3A,B). These data suggested that phosphorylation at serines 315 and 376 plays a role in the cytoplasmic localization of the tumor suppressor.
To verify the induction of p53 phosphorylation at serines 315 and 376 by ER stress in vivo, GFP-p53 WT or GFP-p53 S315A/S376A was transiently expressed in HeLa cells. [32P]orthophosphate labeling and immunoprecipitations demonstrated that phosphorylation of GFP-p53 WT was enhanced after treatment with either TM or TG as opposed to phosphorylation of GFP-p53 S315A/S376A, which was not affected by the ER stress inducers (Fig. 3C, cf. lanes 3,4 and 2, or lanes 6,7 and 5). These data suggested that serines 315 and 376 are phosphorylated in ER-stressed cells in vivo.
GSK-3β is activated by ER stress and mediates p53 phosphorylation at serines 315 and 376
Recent findings indicated a prolonged increase in the activity of glycogen synthase kinase (GSK)-3β in ER-stressed cells (Song et al. 2002). Because GSK-3β functionally and physically interacts with p53 in response to DNA damage (Turenne and Price 2001; Watcharasit et al. 2002), we investigated a possible role of the kinase in p53 regulation in ER-stressed cells. Because phosphorylation of GSK-3β at serine 9 negatively regulates its enzymatic activity, reduction of phosphorylation at serine 9 is used as an indicator of kinase activation (Cohen and Frame 2001). Treatment of WI-38 cells with either TM or TG caused a reduction in GSK-3β phosphorylation at serine 9 (Fig. 4A, top, lower band), whereas total GSK-3β protein levels remained stable. Phosphorylation of GSK-3α at serine 21, which is recognized by the same phosphospecific antibody, was also reduced upon ER stress (Fig. 4A, top, upper band). Similar results were obtained with NIH-3T3 cells (data not shown). Next, we examined the localization of the GSK-3β and p53 in WI-38 cells by confocal microscopy in the absence or presence of ER stress inducers (Fig. 4B). We observed that, in unstressed cells, the localization of endogenous p53 was predominantly nuclear, whereas GSK-3β localization was both nuclear and cytoplasmic. In ER-stressed cells, however, the nuclear localization of p53 was decreased, and this correlated with an increase in the cytoplasmic localization of the protein. We also noticed that in ER-stressed cells, p53 formed punctate structures of as yet unknown origin. On the other hand, in ER-stressed cells, the localization of GSK-3β was predominantly nuclear. When the images were merged, we noticed that the two proteins were colocalized in the nucleus, and their colocalization was enhanced in response to ER stress, implicating GSK-3β in regulation of p53 relocalization. Similar results were obtained with HT1080 cells (data not shown). On the basis of these data, we sought to examine whether GSK-3β physically interacts with p53 and whether this interaction is affected by ER stress. Given the down-regulation of p53 upon prolonged ER-stress, we monitored the interaction of endogenous p53 and GSK-3β in WI-38 cells upon treatment with either TM or TG for a short time (45 min; Fig. 4C). Immunoprecipitation of p53, followed by immunoblotting for GSK-3β revealed an induction of association between the two proteins in response to ER stress (Fig. 4C, top, cf. lane 2 and lanes 3 or 4). Similar results were obtained with transiently expressed epitope-tagged p53 and GSK-3β proteins in ER-stressed HeLa cells (data not shown). Taken together, these data suggested a functional interaction between GSK-3β and p53 in response to ER stress.
Figure 4.
Physical and functional interactions between p53 and GSK-3β in ER-stressed cells. (A) ER stress activates GSK-3β. WI-38 cells were treated with 10 μg/mL of TM or 1 μM of TG as indicated. Cell lysates were subjected to Western blotting with anti-phosphoserine 9 antibody of GSK-3β (top, lower bands), which also cross-reacts with phosphoserine 21 of GSK-3α (higher bands). The total levels of GSK-3β on the same blot were detected by immunoblotting (bottom). (B) ER stress promotes GSK-3β nuclear localization and interaction with p53. WI-38 cells untreated or treated with either 10 μg/mL of TM or 1 μM of TG for 2 h were subjected to immunostaining for endogenous p53 (green) or GSK-3β (red). The localization or colocalization (yellow) of both proteins was detected by laser-scanning confocol microscopy. (C) ER stress enhances the interaction between p53 and GSK-3β. WI-38 cells were treated with 10 μg/mL of TM or 1 μM of TG for 45 min. Protein extracts (400 μg) were subjected to immunoprecipitation with 1 μg of a mouse anti-p53 antibody (Ab-6, lanes 2-4). As a control, protein extracts (400 μg) from untreated WI-38 cells were immunoprecipitated with 1 μg of purified mouse IgG. Immunoprecipitates were subjected to immunoblotting with anti-GSK-3β rabbit polyclonal antibody (top) followed by immunoblotting with anti-p53 rabbit polyclonal antibody (second from top). Whole-cell extracts (WCE; 40 μg of protein) were used to detect the amount of either GSK-3β (third from top) or p53 (bottom) in the protein extracts used for p53 immunoprecipitation. (D) GSK-3β phosphorylates the carboxyl terminus of p53 in vitro. HA-tagged GSK-3β (WT or KD) was transiently expressed in HeLa cells. Protein extracts (200 μg) were used for immunoprecipitation with anti-HA rabbit polyclonal antibody followed by in vitro kinase assays with either 1 μg of GST-p53 160-318 (lanes 1,3) or 1 μg of GST-p53 319-393 (lanes 2,4) in the presence of [γ-32P]ATP. Phosphorylated GST-p53 proteins were detected by SDS-PAGE and autoradiography (top), the GST-p53 proteins by Coomassie blue staining (bottom). (E) GSK-3β phosphorylates p53 at serine 376 in vitro. A total of 1 μg of purified GST-p53 319-393 (wild type or S376A mutant) fusion protein was incubated with 30 ng of histidine-tagged GSK-3β and subjected to in vitro kinase assay in the absence or presence of 20 mM LiCl. As a control, 20 mM NaCl in lanes 2 and 4 were used. Phosphorylated GST-p53 was detected by autoradiography (top), the total GST-p53 by Coomassie blue staining (bottom). (F) Phosphorylation of p53 by GSK-3β in vivo. HeLa cells were transfected with 1 μg of either GFP-p53 WT or GFP-p53 S315A/S376A plasmid DNA in the presence of 2 μg of pcDNA/3.1, GSK-3β WT or GSK-3β KD plasmid DNA. Phosphorylation and expression levels of GFP-p53 were detected as described in Fig. 3C.
Next, we examined whether serines 315 and 376 are phosphorylated by GSK-3β. To this end, a GST-fusion protein encompassing either amino acids (aa) 160-318 or aa 319-393 of human p53 (Cuddihy et al. 1999) was subjected to phosphorylation with purified GSK-3β in vitro. GSK-3β did not induce the phosphorylation of aa 160-318 of p53 (Fig 4D, top, lane 3), indicating that serine 315 is not a direct target of the kinase. On the other hand, GSK-3β induced the phosphorylation of aa 319-393 (Fig 4D, top, lane 4; Fig. 4E, top, lane 2), which was inhibited by 20 mM LiCl (Klein and Melton 1996; Song et al. 2002; Fig. 4E, top, lane 1). When a GST-fusion protein of aa 319-393 of p53 with substitution of serine 376 to alanine (S376A) was used, we found that phosphorylation of the carboxyl terminus domain of p53 by GSK-3β was diminished significantly (Fig. 4E, top, lane 4). This result suggested that serine 376 is a target of GSK-3β in vitro. To test whether phosphorylation at serines 315 and 376 is mediated by GSK-3β in vivo, HeLa cells were transfected with either GFP-p53 WT or GFP-p53 S315A/S376A plasmids in the presence of GSK-3β WT or KD mutant. After [32P]orthophosphate labeling and GFP-p53 immunoprecipitation, we found that GSK-3β WT enhanced the in vivo phosphorylation of GFP-p53 WT, but not of GFP-p53 S315A/S376A (Fig. 4F, cf. lanes 3 and 6). Collectively, these data demonstrated that GSK-3β mediates the phosphorylation of p53 at serines 315 and 376.
ER stress and GSK-3β activation prevent p53-dependent apoptosis
To get an insight into the biological significance of the above findings, we examined the effects of ER stress on p53-mediated apoptosis in SAOS-2 cells, which lack endogenous p53 and are susceptible to apoptosis induced by the ectopic expression of the tumor suppressor protein (Rowan et al. 1996). We observed that a large population of cells expressing GFP-p53 WT displayed morphological changes indicative of apoptosis, such as chromatin condensation and nuclear fragmentation, as visualized by DAPI staining (Fig. 5A; apoptotic cells are indicated by arrows). When cells were treated with either TM or maintained in low glucose, we noticed a decrease in the apoptotic population of cells expressing GFP-p53 WT (Fig. 5A,B). Contrary to this, GFP-p53-mediated apoptosis was not impaired in ER-stressed SAOS-2 cells transfected with the phosphorylation mutants (Fig. 5A,B). These data indicated an inverse correlation between the cytoplasmic localization of p53 and p53-mediated apoptosis in response to ER stress. They also suggested that p53 phosphorylation at serines 315 and 376 plays a role in the inhibition of p53-mediated apoptosis in response to ER stress.
Figure 5.
ER stress impairs p53-mediated apoptosis. (A) ER stress inhibits GFP-p53-mediated apoptosis in SAOS-2 cells. Cells were transfected with 0.5 μg of EGFP (negative control), GFP-p53 WT, GFP-p53 S315A, GFP-p53 S376A, or GFP-p53 S315A/S376A plasmid DNA. Four hours post-transfection, ER stress commenced after treatments with 0.4 μg/mL of TM or by growing cells in low-glucose medium. Sixteen hours later, cells were fixed and subjected to fluorescence microscopic observation for GFP. Apoptotic cells are indicated by arrows. (B) Quantification of GFP-p53-induced apoptosis in SAOS-2 cells. GFP-positive cells from A that exhibited distinct nuclear condensation or fragmentation were scored as apoptotic. For each sample, 300 GFP-positive cells were counted. The values in the histograms represent means ± SD from three independent experiments. (C) Ectopic expression of GSK-3β inhibits p53-dependent apoptosis in SAOS-2 cells. SAOS-2 cells were transfected with either 0.5 μg of EGFP plasmid or 0.5 μg of each of the indicated types of GFP-p53 plasmid DNA together with 1 μg of pcDNA3.1, GDK-3β WT, or GSK-3β KD plasmid DNA. Twenty hours later, cells were subjected to fluorescence microscopy for GFP expression and apoptotic phenotype. Quantification of apoptotic Saos-2 cells was done as in B. The values in the histograms represent means ± SD from three independent experiments.
Given the role of GSK-3β in regulating p53 phosphorylation and localization, we next examined the effects of the kinase on p53-mediated apoptosis (Fig. 5C). Using the same methodology as in Figure 5A, we observed that induction of apoptosis in SAOS-2 cells transfected with GFP-p53 WT was inhibited by almost 30% in the presence of GSK-3β WT (Fig. 5C). Contrary to this, induction of apoptosis by GFP-p53 WT was not affected by the expression of GSK-3β KD, indicating that the enzymatic activity of the kinase plays a role in this process. Interestingly, inhibition of p53-mediated apoptosis by GSK-3β WT was not possible when the GFP-p53 phosphorylation mutant S315A, S376A, or S315A/S376A was used (Fig. 5C). These data suggested that GSK-3β inhibits p53-mediated apoptosis in a manner that is dependent on phosphorylation of the tumor suppressor protein at serines 315 and 376.
ER stress affects p53 localization and p53-mediated apoptosis through GSK-3β
To further verify the role of GSK-3β in regulation of p53 in ER-stressed cells, we used mouse embryonic fibroblasts (MEFs) from a GSK-3β knockout mouse (Hoeflich et al. 2000). Immunoblot analysis indicated that GSK-3β-/- MEFs had undetectable levels of endogenous p53 (Fig. 6A, top), most probably due to inactivation of the tumor suppressor during the immortalization process. Therefore, we examined the localization of GFP-p53 proteins in ER-stressed GSK-3β-/- MEFs reconstituted with either wild-type (WT) GSK-3β or kinase dead (KD) GSK-3β (Fig. 6B). We observed that GSK-3β WT enhanced the cytoplasmic localization of GFP-p53 WT as opposed to KD mutant, which did not affect the localization of the fusion protein (Fig. 6B). Quantification of the nuclear to cytoplasmic ratio of GFP-p53 in transfected GSK-3β-/- MEFs indicated that ER stress did not induce the cytoplasmic localization of GFP-p53 in these cells unless GSK-3β WT was present (Fig. 6C).
Figure 6.
ER stress affects p53 localization and p53-mediated apoptosis through GSK-3β. (A) Expression of endogenous p53 in GSK-3β+/+ and GSK-3β-/- MEFs. A total of 50 μg of protein from GSK-3β+/+ and GSK-3β-/- MEFs was subjected to Western blotting with anti-p53 polyclonal antibody or anti-GSK-3β polyclonal antibody. Protein levels were normalized to α-tubulin. (B) GSK-3β mediates ER stress-induced cytoplasmic relocation of p53. GSK-3β-/- MEFs were transfected with 0.05 μg GFP-p53 WT and 0.5 μg of pcDNA/3.1, GSK-3β WT, or GSK-3β KD plasmid DNA. Sixteen hours post-transfection, cells were treated with either 10 μg/mL of TM or 1 μM of TG for 4 h, followed by DAPI staining and fluorescence microscopic observation. (C) Quantification of nuclear to cytoplasmic GFP-p53 in GSK-3β-/- MEFs was done as described previously (Zhang and Xiong 2001; see also Materials and Methods). (D) Inhibition of p53-mediated apoptosis by ER stress in GSK-3β-/- MEFs reconstituted with GSK-3β. GSK-3β-/- MEFs were transfected with 0.2 μg of GFP-p53 WT and 0.5 μg of pcDNA/3.1, GSK-3β WT, or GSK-3β KD plasmid DNA. Four hours post-transfection, cells were treated with 0.4 μg/mL of TM or maintained in low-glucose medium. Sixteen hours later, cells were subjected to fluorescence microscopic observation and quantification of apoptotic cells as in Fig. 5B. The values in the histograms represent means ± SD from three independent experiments. (E) ER stress and genotoxic stress exhibit opposite effects on p53. In cells subjected to various forms of genotoxic stress, p53 is targeted by phosphorylation mainly at serine 15 and serine 20 (serine 18 and 23 for mouse p53) through the activation of ATM and/or ATR- pathway(s). Phosphorylation at these sites promotes p53 nuclear localization and stabilization by disrupting p53/Mdm2 interaction and Mdm2-mediated p53 nucleocytoplasmic export and degradation. On the other hand, ER stress promotes the cytoplasmic localization and degradation of p53 through the activation of GSK-3β, which mediates indirectly or directly the phosphorylation of p53 at serine 315 or serine 376, respectively. In cells subjected to both ER stress and genotoxic stress, p53 stabilization is impaired by the enhanced cytoplasmic relocation of the tumor suppressor caused by ER stress. Although the nuclear-signaling pathways leading to p53 phosphorylation upon DNA damage appear not to be affected by ER stress (as judged by the Chk2 activity levels in Supplementary Fig. 1), decrease in nuclear p53 by ER stress results in diminished activation capacity of the tumor suppressor in response to genotoxic stress.
To further substantiate the anti-apoptotic role of GSK-3β, we examined GFP-p53-mediated apoptosis in GSK-3β-/- MEFs subjected to ER stress (Fig. 6D). First, we noticed that GSK-3β-/- MEFs were susceptible to GFP-p53-mediated apoptosis in the absence of ER stress, whereas GFP alone was unable to induce apoptosis. When GSK-3β-/- MEFs were reconstituted with GSK-3β WT, we found that the cells were less susceptible to apoptosis by GFP-p53 WT, and this susceptibility was further decreased in the presence of either TM treatment or low glucose (Fig. 6D). On the other hand, GSK-3β KD did not affect the sensitivity of GSK-3β-/- MEFs to GFP-p53-induced apoptosis. Collectively, the above data show that cytoplasmic relocation of p53 and inhibition of p53-mediated apoptosis in ER-stressed cells proceeds through GSK-3β.
Discussion
Herein, we show that pharmacological (i.e., TM, TG) or physiological inducers (i.e., glucose deprivation) of ER stress enhance p53 cytoplasmic localization, and prevent p53-mediated apoptosis (see model in Fig. 6E). Regulation of p53 in ER-stressed cells is mediated, at least in part, by the site-specific phosphorylation at serines 315 and 376. Serine 315 lies within the bipartite NLS of p53 (residues 305-322), which was shown previously to be targeted by cdc2/cyclinB and cdk2/cyclinA (Bischoff et al. 1990; Wang and Prives 1995). Serine 315 phosphorylation was reported previously to enhance p53 binding to DNA in vitro (Bischoff et al. 1990; Wang and Prives 1995) and contribute to p53 transcriptional activation in response to DNA damage (Blaydes et al. 2001). However, other studies suggested no role for serine 315 phosphorylation in regulation of p53 stability and activity in DNA-damaged cells (Hengstermann et al. 1998; Ashcroft et al. 2000). Furthermore, p53 activation by phosphorylation at serine 315 is largely dependent on phosphorylation at other sites (Appella and Anderson 2001). Contrary to genotoxic stress, our findings show that serine 315 phosphorylation is induced by ER stress and negatively regulates p53, indicating that various forms of stress utilize phosphorylation at serine 315 to differentially regulate p53. Serine 376 was initially characterized as a potential target of the CDK7-cycH-p36 complex of transcription factor IIH (Lu et al. 1997). In unstressed cells, constitutive phosphorylation at serine 376 was suggested to contribute to p53 degradation (Chernov et al. 2001), whereas in cells subjected to γ-irradiation, dephosphorylation of serine 376 facilitates p53 binding to sequence-specific DNA by recruiting 14-3-3 protein (Waterman et al. 1998). Thus, it appears that serine 376 phosphorylation negatively regulates p53, consistent with our findings that phosphorylation at this site promotes p53 cytoplasmic localization and prevents p53-mediated apoptosis in ER-stressed cells.
We also show that phosphorylation at both serine 315 and 376 in ER-stressed cells is mediated by GSK-3β. GSK-3 is a multifunctional serine/threonine kinase initially described to play a key role in glycogen metabolism (Cohen and Frame 2001; Doble and Woodgett 2003). The kinase is a key component of Wnt signaling, which controls the entire body pattern during embryonic development (Cohen and Frame 2001; Doble and Woodgett 2003). It is well established that GSK-3 is involved in a wide range of cellular processes, including the phosphorylation of transcription and translation factors, regulation of cell cycle, and proliferation (Cohen and Frame 2001; Doble and Woodgett 2003). Here, we show that ER stress induces GSK-3β activity and binding to p53 (Fig. 4A,C). Consistent with the biochemical data, confocal microscopy data indicated that ER stress induces the nuclear localization of GSK-3β and interaction with p53 (Fig. 4B). Interestingly, DNA damage induced by camptothecin was shown to promote a direct interaction between GSK-3β and p53 in the nucleus (Watcharasit et al. 2002), indicating that the kinase is able to control p53 activity in various forms of stress. It is likely that p53 phosphorylation by GSK-3β occurs in the nucleus and facilitates the nucleocytoplasmic export and degradation of the tumor suppressor. Although GSK-3β mediates the phosphorylation of both serine 315 and serine 376 in vivo, serine 376 is the only one of the two sites that is directly phosphorylated in vitro. However, serine 376 may not be the only residue phosphorylated by GSK-3β, as mutation of serine 376 to alanine did not completely abolish the phosphorylation of the GST-p53 by the kinase in vitro (Fig. 4E). Therefore, phosphorylation of p53 by GSK-3β at various sites may also contribute to regulation of the tumor suppressor in ER-stressed cells. At a first glance, serine 376 phosphorylation by GSK-3β is an unusual finding, as efficient phosphorylation of many of GSK-3β substrates requires the presence of another phosphorylated residue optimally located four amino acids carboxy-terminal to the site of GSK-3β phosphorylation (Cohen and Frame 2001). As such, serine 376 does not conform to the general rule of substrate specificity of this kinase. In analogy to p53, however, GSK-3β was shown previously to phosphorylate cyclin D1 at threonine 286 and mediate the nuclear export and proteolysis of this cyclin (Diehl et al. 1998; Alt et al. 2000). Interestingly, similar to serine 376 of p53, threonine 286 of cyclin D1 does not meet the criteria of a bona fide substrate of the kinase (Cohen and Frame 2001). Thus, both p53 and cyclin D1 may be part of the general limitation in identifying the physiological substrates for GSK-3β (Cohen and Frame 2001). Given that GSK-3β requires numerous levels of regulation to confer signal-dependent specificity (Doble and Woodgett 2003), it is also likely that direct phosphorylation of p53 at serines 315 and 376 is mediated by downstream kinases, whose function is dependent on GSK-3β.
With regard to the biological significance of our findings, we demonstrate that ER stress impairs p53-dependent apoptosis. First, we demonstrated that ER stress prevents the stabilization of p53 in response to various forms of DNA damage and prevents p53-dependent cell death by 5-FU (Fig. 2D,E). These data are consistent with previous observations that TG treatment antagonizes drug-induced nuclear accumulation of p53 (Kaneko and Tsukamoto 1995). Second, we showed that that GSK-3β plays an essential role in the inhibition of p53-mediated apoptosis (Figs. 5C and 6D). Previous findings provided some evidence for a proapoptotic role of GSK-3β in pheochromocytoma (PC12) cells and Rat-1 fibroblasts through the inhibition of phosphatidyl-inositol-3 (PI-3) kinase pathway (Bijur et al. 2000). In addition, overexpression of GSK-3β in neurons can increase apoptosis (Hetman et al. 2000; Cross et al 2001). However, in contrast to these proapoptotic effects of the kinase, GSK-3β-/- mice die in utero due to apoptosis of hepatocytes between E13.5 and E14.5 (Hoeflich et al. 2000). This is due to the inability to elicit an antiapoptotic response to tumor necrosis factor-α (TNF-α), because GSK-3β-/- MEFs fail to activate NF-κB (Hoeflich et al. 2000). Thus, GSK-3β appears to modulate apoptosis in a tissue- and stimuli-specific fashion (Frame and Cohen 2001).
The physiological relevance of our findings may lie within the biology of ER and its sensitivity to alterations that disrupt its functions. Induction of UPR is a protective mechanism utilized by all cells to adapt to ER stress. At the molecular level, this response is presented by the induced synthesis of specific sets of ER resident proteins known as glucose-regulated proteins (Grps) or ER chaperones (Lee 2001). In normal cells, induction of Grps is associated with tissue preservation or organ protection against ER stress (Lee 2001). In neoplastic cells, however, the antiapoptotic function of Grps is implicated in immune resistance, cancer progression, and drug resistance (Lee 2001). Therefore, inhibition of the apoptotic functions of p53 by ER stress represents an additional control mechanism used by cells to adapt to ER stress. If adaptation is not possible, then ER-stressed cells undergo apoptosis through the activation of evolutionary conserved pathways (Ferri and Kroemer 2001; Kaufman et al. 2002), which proceed independently of p53.
ER stress has been linked to human diseases that are related to aberrant protein conformation including cystic fibrosis, Alzheimer's, and prion disease (Aridor and Balch 1999). A recent study demonstrated that fibroblasts from Alzheimer's disease patients exhibit a defective p53-dependent gene transcription and are resistant to p53-mediated death upon DNA damage induced by oxidative injury (Uberti et al. 2002). Furthermore, cytoplasmic retention of p53 is also a well-documented mechanism to inactivate p53 function in certain pathological conditions. For instance, constitutive cytoplasmic localization of wild-type p53 has been reported in solid tumors such as colorectal adenocarcinoma (Sun et al. 1992; Bosari et al. 1994), neuroblastomas (Moll et al. 1995, 1996), retinoblastoma (Schlamp et al. 1997), hepatocellular carcinoma (Ueda et al. 1995), and breast cancer (Moll et al. 1992; Stenmark-Askmalm et al. 1994), and is associated with tumor metastasis and poor prognosis. Because ER stress can be induced by microenvironments of solid tumors (Lee 2001), a state of chronic ER stress in these tumors might lead to the constitutive cytoplasmic localization of p53. Although it is not clear whether this phenotype is directly linked to malignancy, it suggests that defects in nuclear accumulation of p53 could contribute to tumor progression and unresponsiveness to anti-cancer therapies in patients. Thus, improved understanding of how exactly ER stress induces p53 cytoplasmic retention may facilitate the design of novel anti-cancer strategies that can reverse this process and render p53 functional once again by allowing it to accumulate in the nucleus.
Materials and methods
Cell culture and treatments
NIH-3T3, HeLa, and HT1080 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) plus 10% calf serum (Invitrogen) and antibiotics. Human diploid primary WI-38 cells were grown in Minimum Essential medium Eagle (MEM) containing 10% fetal bovine serum (FBS, Invitrogen) and antibiotics. HCT116 human colorectal carcinoma cells (p53+/+ and p53-/-) were maintained in McCoy's 5A medium (Cellgro) containing 10% FBS and antibiotics. SAOS-2 cells, as well as spontaneously immortalized GSK-3β+/+ and GSK-3β-/- MEFs, were cultured in DMEM plus 10% FBS and antibiotics. 70Z/3 B lymphoma cells containing wild-type p53 (Aloni-Grinstein et al. 1993) were cultured in RPMI-1640 medium supplemented with 10% FBS plus 50 μM β-mercaptoethanol (Sigma).
ER stress was induced with the indicated concentrations of tunicamycin (TM) or thapsigargin (TG), or incubation in DMEM with no or low glucose (1500 mg/L D-glucose).
DNA constructs and transfection
Wild type HA-tagged GSK-3β or the kinase-dead (KD) GSK-3β cDNA in pcDNA/3.1 vector was described (Stambolic and Woodgett 1994). Wild-type and the NES mutant (L348A/L350A) of p53 in the pEGFP/N1 vector were described elsewhere (Boyd et al. 2000). The GFP-p53-S315A, -S376A mutants were generated using the QuickChange XL method (Stratagene) with the following sets of primers:
-
S315A,
5′-GCCCAACAACACCAGCTCCGCCCCCCAGCCAAAG
AAGAAAC-3′ (forward),
5′-GTTTCTTCTTTGGCTGGGGGGCGGAGCTGGTGTT
GTTGGGC-3′ (reverse);
-
S376A,
5′-GTCCAAAAAGGGTCAGGCCACCTCCCGCCATAAA
AAAC-3′ (forward),
5′-GTTTTTTATGGCGGGAGGTGGCCTGACCCTTTTT
GGAC-3′ (reverse).
The generation of the double S315A/S376A mutation was made with the S376A set of primers (see above) using the GFP-p53 S315A plasmid as template. All mutations were verified by DNA sequencing.
For transient transfections, cells were transfected with the appropriate DNA using the Lipofectamine Plus reagent (Invitrogen) as recommended by the manufacturer.
Western blotting
To prepare whole-cell extracts, cellular proteins were extracted in ice-cold lysis buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol (DTT), 100 mM NaF, 20 mM β-glycerophosphate, 50 mM Na3VO4, 1 mM PMSF, 4 μg/mL aprotinin, 4 μg/mL pepstatin A, and 4 μg/mL leupeptin. After incubation on ice for 15 min, the lysates were cleared by centrifugation at 18,000g for 15 min. Proteins were separated by SDS-PAGE, transferred onto PVDF membranes, subjected to Western blotting using the standard protocol (Sambrook et al. 1989), and visualized by the chemiluminescent detection method (ECL; Amersham) according to specification. Antibodies against p53 (FL-393) and Erg-1 (SC-588) were purchased from Santa Cruz Inc. Monoclonal antibody to actin, and α-tubulin were from ICN and Sigma, respectively. Ab-1 anti-p53 mouse monoclonal antibody was from Oncogene Science. Rabbit polyclonal anti-HA antibody was from Transduction Laboratories. Rabbit anti-Chk2 antibody was described previously (Matsuoka et al. 1998). Rabbit polyclonal antibodies to GSK-3β and phosphoserine 9 of GSK-3β were from Cell Signaling. All antibodies were used at dilutions recommended by the manufacturer.
Subcellular fractionation
Cytoplasmic and nuclear extracts were prepared as described previously (Nakamura et al. 2000) with some modification. Briefly, the cells were washed twice with ice-cold PBS, and transferred to 1.5-mL eppendorf tubes. The cells were then centrifuged at 300g for 4 min at 4°C. After centrifugation, the pellet was resuspended in 400 μL of cold buffer A [10 mM HEPESKOH (pH 7.5), 10 mM KCl, 1 mM DTT, 1 mM PMSF, 4 μg/mL aprotonin, 4 μg/mL pepstatin A, and 4 μg/mL leupeptin]. After incubation on ice for 15 min, 12.5 μL of 10% Nonidet P-40 was added, the mixture was vortexed briefly, and incubated on ice for 10 min. The nuclei were pelleted by centrifugation at 1500g for 5 min at 4°C, whereas the supernatant (cytoplasmic extracts) was recovered by centrifugation at 13,000g for 15 min. Nuclei were washed with 1 mL of ice-cold buffer A twice, and then resuspended in 50 μL of ice-cold buffer B [20 mM HEPES-KOH (pH 7.5), 0.4 M NaCl, 1 mM DTT, 1 mM PMSF, 4 μg/mL aprotonin, 4 μg/mL pepstatin A, and 4 μg/mL leupeptin], followed by incubation on ice for 30 min. The mixture was then centrifuged at 18,000g for 5 min, and the supernatant was collected as a nuclear extract.
Metabolic labeling and pulse-chase assays
Metabolic labeling with [32P]orthophosphate was performed as described previously (Wong et al. 1997). 35S-protein labeling and pulse-chase assays were performed as described (Spotts et al. 1997) with few modifications. Briefly, cells were plated at density of 1 × 106 per 10-cm dish, and maintained for 24 h. Medium was removed and cells were incubated with 4 mL of labeling medium consisting of L-methionine/L-cysteine-free DMEM (Invitrogen) supplemented with 5% dialyzed FBS (GIBCO) for 2 h. Then, the medium was refreshed with 4 mL of labeling medium containing 100 μCi of [35S]methionine/cysteine (ICN). After labeling for 30 min, the labeling medium was removed, cells were washed twice with PBS, and chased with DMEM plus 10% dialyzed FBS supplemented with 0.2% of L-methionine and 0.2% of L-cysteine, with or without the ER stress inducer. Protein extracts (1 mg) were subjected to immunoprecipitation with anti-p53 polyclonal antibody and protein G-agarose (Sigma) followed by SDS-PAGE and autoradiography. Quantification of the bands was performed by the NIH Image 1.54 software.
GSK-3β phosphorylation assays in vitro
Human GSK-3β purified from baculovirus-infected Sf21 cells was purchased from Upstate Biotechnology Inc. GST-p53 proteins were purified as described previously (Cuddihy et al. 1999). The phosphorylation assays were performed with 1 μg of purified GST-p53 protein in the absence or presence of 20 mM LiCl as described previously (Klein and Melton 1996; Song et al. 2002). Alternatively, HA-tagged GSK-3β (WT or KD) transiently expressed in HeLa cells was immunoprecipitated with anti-HA antibody followed by in vitro kinase assays with purified GST-p53 proteins.
Colony formation assays and FACS analysis
For colony formation, HCT116 cells (3 × 104/10-cm plate) were treated with 0.3 μg/mL of TM or 0.03 μM of TG plus 375 μM of 5-FU for 4 h. The culture medium was replenished, and cells were maintained at 37°C for 12 d with medium changing every other day. Grown colonies were fixed with 3.7% formaldehyde and stained with crystal violet. Plates were scanned and quantified by the NIH Image software. For FACS analysis, HCT116 cells (2 × 105/10-cm plate) were treated with TM or TG plus 5-FU as above for 4 h. The medium was replenished and cells were cultured for 48 h. Then, the cells were harvested, fixed in 70% ethanol, stained with propidium iodide (PI), and subjected to FACS analysis as described (Huang et al. 2003).
Immunofluorescence studies
Cells grown on 22-mm coverslip (Fisher) were treated as indicated, washed with PBS, fixed with 1% formaldehyde for 15 min at room temperature, and then blocked with 5% BSA, PBS, 0.1% TX-100 for 1 h. To detect p53, cells were stained with a mixture of 1:200 diluted Ab-2 and Ab-6 anti-p53 monoclonal antibodies (both from Oncogene Science). To detect GSK-3β, cells were stained with a 1:100 diluted polyclonal antibody (Cell Signaling). Cells were incubated with primary antibodies for 16 h at 4°C, washed with PBS, and incubated for 1 h with Alexa Fluro-488-conjugated secondary antibody or Alexa Fluro-546-conjugated secondary antibody (both from Molecular Probes). To visualize the nucleus, cells were counterstained with 1 μg/mL 4, 6-diamidino-2-phenylindole (DAPI, Sigma). After mounting, cells were analyzed with a laser-scanning confocol microscope (LSM 410, software v3.80, Carl Zeiss) or Olympus BX-51 fluorescence microscope.
Analysis of GFP-p53 expression and induction of apoptosis
Subcellular localization of GFP-p53 proteins was performed as described previously (Boyd et al. 2000). GFP-positive and live cells were scored and classified into two groups; the first group with fluorescence predominantly in the nucleus and the second with fluorescence in both nucleus and cytoplasm or, in few cases (<10%), with cytoplasmic fluorescence only. Quantification of the ratio of nuclear to cytoplasmic GFP-p53 in GSK-3β-/- MEFs was performed as described (Zhang and Xiong 2001). Briefly, total green fluorescence in the nucleus and whole cell were quantified by NIH Image from 40 randomly selected GFP-positive and live cells. The data was expressed as the ratio of nuclear to the cytoplasmic green fluorescence (total cellular fluorescence minus nuclear fluorescence).
For GFP-p53-induced apoptosis, cells seeded on coverslips were transfected with the indicated amounts of plasmid DNA for 16-24 h, at which point the cells were fixed, stained with 1 μg/mL 4, 6-diamidino-2-phenylindole (DAPI), and examined under a fluorescence microscope. GFP-positive cells with distinct nuclear condensation or fragmentation were scored as apoptotic.
Acknowledgments
We thank D. Moraitis for technical assistance; B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, MD) for the HCT116 cell lines; S. Boyd and T. Jacks (Massachusetts Institute of Technology, Cambridge, MA) for the GFP-p53 WT and GFP-p53 NES constructs; J.Woodgett (Ontario Cancer Institute, Toronto, ON, Canda) for HA-GSK-3β WT and KD plasmids as well as GSK-3β knockout MEFs; S. Elledge (Baylor College of Medicine, Houston, TX) for anti-Chk2 antibody; and H. Dilhuydy (Université de Montreal, Montreal, QC, Canada) for assistance in confocal microscopy. This work was supported by a grant from the National Cancer Institute of Canada (NCIC) to A.E.K. A.E.K. is recipient of a Scientist's Award from the Canadian Institutes for Health Research (CIHR).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1165804.
Supplemental material is available at http://www.genesdev.org.
References
- Aloni-Grinstein R., Zan-Bar, I., Alboum, I., Goldfinger, N., and Rotter, V. 1993. Wild type p53 functions as a control protein in the differentiation pathway of the B-cell lineage. Oncogene 8: 3297-3305. [PubMed] [Google Scholar]
- Alt J.R., Cleveland, J.L., Hannink, M., and Diehl, J.A. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes & Dev. 14: 3102-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E. and Anderson, C.W. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268: 2764-2772. [DOI] [PubMed] [Google Scholar]
- Aridor M. and Balch, W.E. 1999. Integration of endoplasmic reticulum signaling in health and disease. Nat. Med. 5: 745-751. [DOI] [PubMed] [Google Scholar]
- Ashcroft M., Taya, Y., and Vousden, K.H. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20: 3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijur G.N., De Sarno, P., and Jope, R.S. 2000. Glycogen synthase kinase-3β facilitates staurosporine- and heat shock-induced apoptosis. Protection by lithium. J. Biol. Chem. 275: 7583-7590. [DOI] [PubMed] [Google Scholar]
- Bischoff J.R., Friedman, P.N., Marshak, D.R., Prives, C., and Beach, D. 1990. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc. Natl. Acad. Sci. 87: 4766-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydes J.P., Luciani, M.G., Pospisilova, S., Ball, H.M., Vojtesek, B., and Hupp, T.R. 2001. Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. J. Biol. Chem. 276: 4699-4708. [DOI] [PubMed] [Google Scholar]
- Bosari S., Viale, G., Bossi, P., Maggioni, M., Coggi, G., Murray, J.J., and Lee, A.K. 1994. Cytoplasmic accumulation of p53 protein: An independent prognostic indicator in colorectal adenocarcinomas. J. Natl. Cancer Inst. 86: 681-687. [DOI] [PubMed] [Google Scholar]
- Boyd S.D., Tsai, K.Y., and Jacks, T. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell. Biol. 2: 563-568. [DOI] [PubMed] [Google Scholar]
- Brewer J.W., Hendershot, L.M., Sherr, C.J., and Diehl, J.A. 1999. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. 96: 8505-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J.P., Sedivy, J.M., Kinzler, K.W., and Vogelstein, B. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497-1501. [DOI] [PubMed] [Google Scholar]
- Bunz F., Hwang, P.M., Torrance, C., Waldman, T., Zhang, Y., Dillehay, L., Williams, J., Lengauer, C., Kinzler, K.W., and Vogelstein, B. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 104: 263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov M.V., Bean, L.J., Lerner, N., and Stark, G.R. 2001. Regulation of ubiquitination and degradation of p53 in unstressed cells through C-terminal phosphorylation. J. Biol. Chem. 276: 31819-31824. [DOI] [PubMed] [Google Scholar]
- Cohen P. and Frame, S. 2001. The renaissance of GSK3. Nat. Rev. Mol. Cell. Biol. 2: 769-776. [DOI] [PubMed] [Google Scholar]
- Cross D.A., Culbert, A.A., Chalmers, K.A., Facci, L., Skaper, S.D., and Reith, A.D. 2001. Selective small-molecule inhibitors of glycogen synthase kinase-3 activity protect primary neurones from death. J. Neurochem. 77: 94-102. [DOI] [PubMed] [Google Scholar]
- Cuddihy A.R., Wong, A.H., Tam, N.W., Li, S., and Koromilas, A.E. 1999. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene 18: 2690-2702. [DOI] [PubMed] [Google Scholar]
- Diehl J.A., Cheng, M., Roussel, M.F., and Sherr, C.J. 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes & Dev. 12: 3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble B.W. and Woodgett, J.R. 2003. GSK-3: Tricks of the trade for a multi-tasking kinase. J. Cell. Sci. 116: 1175-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri K.F. and Kroemer, G. 2001. Organelle-specific initiation of cell death pathways. Nat. Cell. Biol. 3: E255-E263. [DOI] [PubMed] [Google Scholar]
- Frame S. and Cohen, P. 2001. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359: 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Calfon, M., Urano, F., Novoa, I., and Ron, D. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell. Dev. Biol. 18: 575-599. [DOI] [PubMed] [Google Scholar]
- Hengstermann A., Whitaker, N.J., Zimmer, D., Zentgraf, H., and Scheffner, M. 1998. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene 17: 2933-2941. [DOI] [PubMed] [Google Scholar]
- Hetman M., Cavanaugh, J.E., Kimelman, D., and Xia, Z. 2000. Role of glycogen synthase kinase-3β in neuronal apoptosis induced by trophic withdrawal. J. Neurosci. 20: 2567-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao A., Kong, Y.Y., Matsuoka, S., Wakeham, A., Ruland, J., Yoshida, H., Liu, D., Elledge, S.J., and Mak, T.W. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287: 1824-1827. [DOI] [PubMed] [Google Scholar]
- Hoeflich K.P., Luo, J., Rubie, E.A., Tsao, M.S., Jin, O., and Woodgett, J.R. 2000. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature 406: 86-90. [DOI] [PubMed] [Google Scholar]
- Huang S., Qu, L.K., Cuddihy, A.R., Ragheb, R., Taya, Y., and Koromilas, A.E. 2003. Protein kinase inhibitor 2-aminopurine overrides multiple genotoxic stress-induced cellular pathways to promote cell survival. Oncogene 22: 3721-3733. [DOI] [PubMed] [Google Scholar]
- Kaneko Y. and Tsukamoto, A. 1995. Apoptosis and nuclear levels of p53 protein and proliferating cell nuclear antigen in human hepatoma cells cultured with tumor promoters. Cancer Lett. 91: 11-17. [DOI] [PubMed] [Google Scholar]
- Kaufman R.J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C.Y., and Arnold, S.M. 2002. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell. Biol. 3: 411-421. [DOI] [PubMed] [Google Scholar]
- Klein P.S. and Melton, D.A. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. 93: 8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S. 2001. The glucose-regulated proteins: Stress induction and clinical applications. Trends Biochem. Sci. 26: 504-510. [DOI] [PubMed] [Google Scholar]
- Liang S.H. and Clarke, M.F. 2001. Regulation of p53 localization. Eur. J. Biochem. 268: 2779-2783. [DOI] [PubMed] [Google Scholar]
- Lu H., Fisher, R.P., Bailey, P., and Levine, A.J. 1997. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol. Cell. Biol. 17: 5923-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Huang, M., and Elledge, S.J. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893-1897. [DOI] [PubMed] [Google Scholar]
- Michael D. and Oren, M. 2002. The p53 and Mdm2 families in cancer. Curr. Opin. Genet. Dev. 12: 53-59. [DOI] [PubMed] [Google Scholar]
- Moll U.M., Riou, G., and Levine, A.J. 1992. Two distinct mechanisms alter p53 in breast cancer: Mutation and nuclear exclusion. Proc. Natl. Acad. Sci. 89: 7262-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll U.M., LaQuaglia, M., Benard, J., and Riou, G. 1995. Wild-type p53 protein undergoes cytoplasmic sequestration in un-differentiated neuroblastomas but not in differentiated tumors. Proc. Natl. Acad. Sci. 92: 4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll U.M., Ostermeyer, A.G., Haladay, R., Winkfield, B., Frazier, M., and Zambetti, G. 1996. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol. Cell. Biol. 16: 1126-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Roth, J.A., and Mukhopadhyay, T. 2000. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol. 20: 9391-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S., Ludwig, R.L., Haupt, Y., Bates, S., Lu, X., Oren, M., and Vousden, K.H. 1996. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO J. 15: 827-838. [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scheuner D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R.J. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7: 1165-1176. [DOI] [PubMed] [Google Scholar]
- Schlamp C.L., Poulsen, G.L., Nork, T.M., and Nickells, R.W. 1997. Nuclear exclusion of wild-type p53 in immortalized human retinoblastoma cells. J. Natl. Cancer Inst. 89: 1530-1536. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E. and DePinho, R.A. 2002. p53: Good cop/bad cop. Cell 110: 9-12. [DOI] [PubMed] [Google Scholar]
- Song L., De Sarno, P., and Jope, R.S. 2002. Central role of glycogen synthase kinase-3β in endoplasmic reticulum stress-induced caspase-3 activation. J. Biol. Chem. 277: 44701-44708. [DOI] [PubMed] [Google Scholar]
- Spotts G.D., Patel, S.V., Xiao, Q., and Hann, S.R. 1997. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 17: 1459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V. and Woodgett, J.R. 1994. Mitogen inactivation of glycogen synthase kinase-3 β in intact cells via serine 9 phosphorylation. Biochem. J. 303: 701-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark-Askmalm M., Stal, O., Sullivan, S., Ferraud, L., Sun, X.F., Carstensen, J., and Nordenskjold, B. 1994. Cellular accumulation of p53 protein: An independent prognostic factor in stage II breast cancer. Eur. J. Cancer 30A: 175-180. [DOI] [PubMed] [Google Scholar]
- Stommel J.M., Marchenko, N.D., Jimenez, G.S., Moll, U.M., Hope, T.J., and Wahl, G.M. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18: 1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.F., Carstensen, J.M., Zhang, H., Stal, O., Wingren, S., Hatschek, T., and Nordenskjold, B. 1992. Prognostic significance of cytoplasmic p53 oncoprotein in colorectal adeno-carcinoma. Lancet 340: 1369-1373. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249-258. [DOI] [PubMed] [Google Scholar]
- Turenne G.A. and Price, B.D. 2001. Glycogen synthase kinase3 β phosphorylates serine 33 of p53 and activates p53's transcriptional activity. BMC Cell Biol. 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti D., Carsana, T., Bernardi, E., Rodella, L., Grigolato, P., Lanni, C., Racchi, M., Govoni, S., and Memo, M. 2002. Selective impairment of p53-mediated cell death in fibroblasts from sporadic Alzheimer's disease patients. J. Cell. Sci. 115: 3131-3138. [DOI] [PubMed] [Google Scholar]
- Ueda H., Ullrich, S.J., Gangemi, J.D., Kappel, C.A., Ngo, L., Feitelson, M.A., and Jay, G. 1995. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat. Genet. 9: 41-47. [DOI] [PubMed] [Google Scholar]
- Vousden K.H. and Lu, X. 2002. Live or let die: The cell's response to p53. Nat. Rev. Cancer 2: 594-604. [DOI] [PubMed] [Google Scholar]
- Wahl G.M. and Carr, A.M. 2001. The evolution of diverse biological responses to DNA damage: Insights from yeast and p53. Nat. Cell. Biol. 3: E277-E286. [DOI] [PubMed] [Google Scholar]
- Wang Y. and Prives, C. 1995. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature 376: 88-91. [DOI] [PubMed] [Google Scholar]
- Watcharasit P., Bijur, G.N., Zmijewski, J.W., Song, L., Zmijewska, A., Chen, X., Johnson, G.V., and Jope, R.S. 2002. Direct, activating interaction between glycogen synthase kinase-3β and p53 after DNA damage. Proc. Natl. Acad. Sci. 99: 7951-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman M.J., Stavridi, E.S., Waterman, J.L., and Halazonetis, T.D. 1998. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19: 175-178. [DOI] [PubMed] [Google Scholar]
- Wong A.H., Tam, N.W., Yang, Y.L., Cuddihy, A.R., Li, S., Kirchhoff, S., Hauser, H., Decker, T., and Koromilas, A.E. 1997. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 16: 1291-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Xiong, Y. 2001. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292: 1910-1915. [DOI] [PubMed] [Google Scholar]