Abstract
Overexpression of the short isoform of p53 (p44) has unexpectedly uncovered a role for p53 in the regulation of size and life span in the mouse. Hyperactivation of the insulin-like growth factor (IGF) signaling axis by p44 sets in motion a kinase cascade that clamps potentially unimpeded growth through p21Cip1. This suggests that pathways of gene activity known to regulate longevity in lower organisms are linked in mammals via p53 to mechanisms for controlling cell proliferation. Thus, appropriate expression of the short and long p53 isoforms might maintain a balance between tumor suppression and tissue regeneration, a major requisite for long mammalian life span.
Keywords: daf2, GH, growth retardation, aging, testis, Akt, Sir2
Powerful genetic experiments carried out in Caenorhabditis elegans over the past decade have very clearly shown that longevity is linked to cascades of gene activity that transduce insulin-like signals. Components of the insulin/insulin-like growth factor (IGF) signaling pathways were recently found to affect life span in the mouse as well. Whereas the life span of lower organisms such as C. elegans and Drosophila is short, mammals live a relatively long time, and cellular proliferation, which ceases in somatic tissues in adult Drosophila and C. elegans, must remain active in mammals so that dead or damaged cells that accumulate with time can be replaced. The obvious advantage that the ability of tissues to undergo regeneration confers on longevity is counterbalanced by the need for an additional level of control that can put limits on the activity of proliferating cell populations in organisms that live a long time.
We have focused on p53 as a likely candidate to integrate insulin/IGF and proliferation signals in the control of mammalian life span. Although a p53-like protein has been identified in C. elegans (CEP-1; Derry et al. 2001) and in Drosophila melanogaster (Dmp53; Jin et al. 2000; Ollmann et al. 2000), both of these p53 proteins lack regions with significant homology to the N-terminal domain of mammalian full-length p53. The function of this domain is to alter gene transcription at a number of different targets, some of which are directly involved in cell-cycle control and can cause proliferation arrest (for review, see Zhao et al. 2000). Thus, the p53 homologs in lower organisms more closely resemble a short form of p53 (DeltaN-p53) that was recently identified in mammalian cell lines and in normal cells from several different tissues (Courtois et al. 2002; Yin et al. 2002). In cells in which this short form is the only p53 protein present, the ability to transactivate target genes such as Mdm2 and p21/Cip1/Waf1 is lost (Courtois et al. 2002) and, along with it, the ability of p53 to control cellular proliferation and growth (Mowat et al. 1985). In the absence of full-length p53, DeltaN-p53 is tumorigenic (Mowat et al. 1985), whereas in the presence of full-length p53, it is growth-suppressive (Rovinski and Benchimol 1988; Lavigueur et al. 1989). The fact that this would have no functional consequences in the postmitotic cells of adult C. elegans and Drosophila suggests that the short form of p53 might represent the primitive form of the p53 protein.
In order to determine the mechanism by which the short form of p53 might control growth, in particular mammalian growth, we generated transgenic mice in which a genomic fragment coding for the short form of p53 (Lavigueur et al. 1989) is expressed in the context of full-length p53. Translation of the short isoform initiates at codon 41 in exon 4 and produces a 44-kD protein (Rovinski et al. 1987). Overexpression of p44 upsets the balance that normally exists between the full-length and short forms of p53 and leads to a phenotype of growth suppression and premature aging in mice. Growth suppression by p44 links small size, proliferation deficits, cellular senescence, and organismal aging to abnormal IGF signaling in the mouse.
Results
Shortened health and life spans
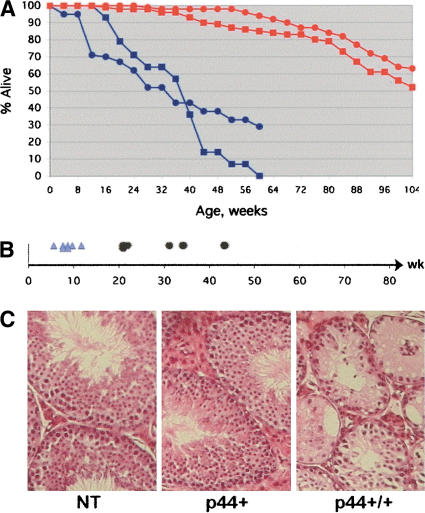
We microinjected the genomic fragment encoding p44 (a gift from Alan Bernstein, CIHR; Lavigueur et al. 1989) and were able to generate two transgenic lines (P and Q) in which the p44 transgene was stably propagated (see Materials and Methods for details). Beginning as early as 4 mo of age, p44 homozygotes in both transgenic lines showed signs of aging. As shown by the survival curve presented in Figure 1A, whereas 85% and 95% of nontransgenic (ICR) males and females, respectively, were still alive at 60 wk of age, 0% of male P+/+ and only 30% of female P+/+ mice were alive at that time. Although both males and females exhibited shortened reproductive health spans, males were more severely affected, becoming infertile as early as 5 mo of age (Fig. 1B). Loss of fertility was accompanied by a catastrophic loss of sperm-producing cells in the testis, with massive degeneration of the seminiferous epithelium leading to a “Sertoli-cell only” phenotype in many tubules (Fig. 1C).
Figure 1.
Accelerated aging in p44 transgenic mice. (A) Decreased life span. Survival data are presented for nontransgenic (NT; red) and p44 homozygous transgenic (blue) mice. Males, squares; females, circles. The 50% survival mark was 24 mo for wild-type mice and 8 mo for p44 homozygous mice. (B) Premature loss of fertility. Shortened reproductive period in male p44 transgenic mice. First fertile age for individual mice is indicated by light blue triangles, and last fertile age by black dots. Onset of fertility is normal, but the loss of reproductive capacity before 1 yr of age is premature in males. (C) Testicular degeneration. Histology of the testes of p44 transgenic and NT animals at 9.5 mo of age. H&E-stained sections. Note the widespread degeneration of the seminiferous epithelium and tubule atrophy in P+/+ testis compared to P+ and NT, which are normal.
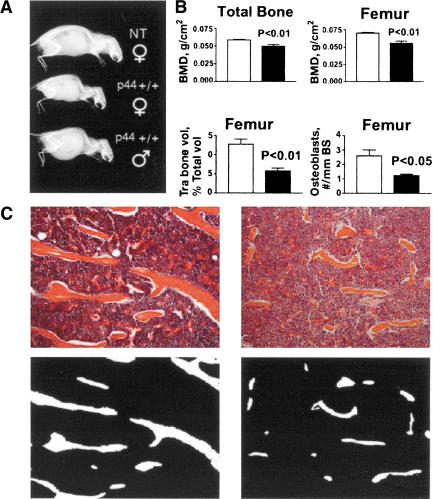
Mice overexpressing p44 also showed signs of aging in bone at relatively young ages. As illustrated by the x-rays of siblings shown in Figure 2A at 5 mo of age, both male and female homozygotes already exhibit lordokyphosis. Total body bone mineral density (excluding the skull) was significantly reduced in P+/+ mice compared to controls (Fig. 2B, upper left) as early as 4 mo of age. Bone mineral density at the distal femur (Fig. 2B, upper right) and proximal tibia (data not shown) was also significantly reduced in P+/+ mice compared to control. Histomorphometric analysis of H&E-stained sections from the femur (Fig. 2B, lower left), and tibia and lumbar spine (data not shown) of P+/+ mice revealed significant reduction in trabecular bone volume (TBV) compared to nontransgenic mice at all sites measured. Bone trabeculae were found to be much longer and thicker in nontransgenic mice (Fig. 2C, left) than in P+/+ mice (Fig. 2C, right). Reduced TBV was associated with a similar reduction in the osteoblast number at the femur in P+/+ mice (Fig. 2B, lower right). Osteoclast numbers were slightly, but not significantly, lower in the P+/+ mice compared to nontransgenic controls (data not shown). The data are consistent with a low bone turnover process in p44 mice, the typical form of osteoporosis associated with aging in humans. As sex steroid deficiency causes a high-turnover osteoporosis characterized by increased osteoclast bone destruction (Bellido et al. 1995; Erben et al. 2000), the osteoporotic phenotype is not due to hypogonadism associated with testicular degeneration of the type shown in Figure 1C above.
Figure 2.
Accelerated bone aging in p44 transgenic mice. (A) Skeletal changes. X-rays of three siblings from the Q line at 5 mo of age. Genotypes and sexes as indicated; ♀, female; ♂, male. Homozygous transgenic mice of both sexes show pronounced lordokyphosis even at this early age. (B) Changes in density and cell number. Bone mineral density (BMD; upper panel), trabecular bone volume (TBV) and osteoblast number (N.Ob/BS; lower panel) in the NT and p44+/+ transgenic mice. p44+/+ mice have lower BMD in total bone and femur (upper panel). Bone histomorphometry data of the femur showed a significant reduction both in TBV and osteoblast number in the p44+/+ mice compared to NT (lower panel). (C) Trabecular bone changes. Upper panel: A representative histological section of the NT and P44+/+ femurs, stained with H&E (original magnification, 100×). Bone trabeculae are much longer and thicker in NT mice (left) compared to P44+/+ mice (right). Lower panel: Bone trabeculae (in white) in binary images of the same histological sections showing more trabecular area in nontransgenic (left) compared to p44+/+ mice.
p44 protein levels and interaction with p53
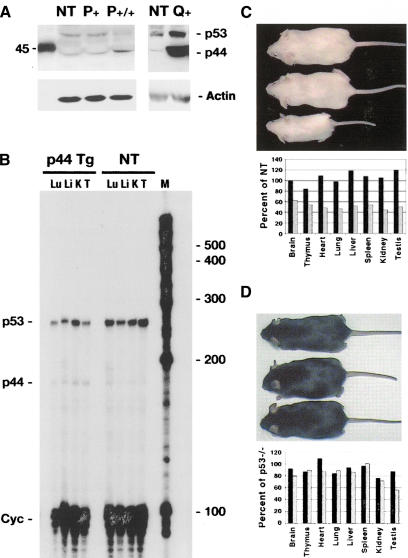
Using a polyclonal anti-p53 antiserum (CM5), both p53 and p44 can be detected on Western blots of protein from the thymus of mice in both the P and Q lines of p44 transgenic mice and in nontransgenic (ICR) mice (Fig. 3A; only relevant bands shown). The transgene gives rise to a protein that migrates as a 44-kD band on Western blots. These blots are informative in several other respects as well. First, they show the relative levels of p44 in the P and Q lines. Second, they show that an immunoreactive band that comigrates with p44 in transgenic tissue is present in nontransgenic tissue. In addition to thymus shown in Figure 3A, we have seen this band in several tissues from nontransgenic mice, including testis and heart (data not shown). In addition, using an RNAse protection assay that can distinguish between p53 and p44 transcripts, a band corresponding to the protected p44 fragment can be detected in tissue extracts from nontransgenic as well as transgenic mice, although at a very low level (Fig. 3B). The expression of both RNA and protein in nontransgenic (normal) tissue supports the idea that p44 is a naturally occurring isoform of p53. Comparable levels of the short form of p53 were reported in human cell lines (Courtois et al. 2002; Yin et al. 2002). In addition, Western blot results indicate that the level of p44 in P homozygotes is twice that of P hemizygous mice, which is approximately the same as in nontransgenic (NT) mice (Fig. 3A). Together, these results with human cell lines and NT mice support our contention that the p44 phenotype is based on a stoichiometrically relevant increase in the level of p44 protein.
Figure 3.
Overexpression of the short isoform of p53 in p44 transgenic mice. (A) p44 protein in transgenic and NT mice. Western blots of 100 μg thymus protein extracted from transgenic (P,Q) and NT (NT) tissues. The anti-p53 polyclonal antiserum CM5 detects both isoforms of p53, as indicated. Biotinylated markers were run alongside the tissue extracts during SDS-PAGE; the marker corresponding to 45 kD is shown. (B) p44 and p53 RNA in transgenic and NT mice. RNAse protection assay was used to detect both p53 (220-bp protected fragment) and p44 (140-bp protected fragment) RNA. Simultaneous hybridization to a probe that detects cyclophilin (Cyc) was used to control for assay conditions. Lu, lung; Li, liver; K, kidney; T, testis; M, marker RNA bands. (C) Effect of p44 on body size. Size of NT (top), hemizygous (middle), and homozygous (bottom) P mice at 3 mo of age. Organ weights of representative hemizygous (black bars) and homozygous (stippled bars) transgenic mice are plotted as a percentage of NT weights. Transgenic animals were generated on a p53+/+ background. (D) Genetic interaction of p44 and p53. Nontransgenic (top), hemizygous (middle), and homozygous (bottom) P mice generated on a p53-/- background. Organ weights of representative hemizygous (black bars) and homozygous (stippled bars) transgenic mice are plotted as a percentage of p53-/- weights.
Mice in both lines were smaller than their NT littermates, but otherwise normal in appearance. As shown in Figure 3C, fully mature homozygous mice in line P were approximately half-sized compared to hemizygous or NT littermates. This is consistent with earlier findings in both cell culture experiments and in animals that p44 is growth-suppressive (Rovinski and Benchimol 1988; Lavigueur et al. 1989; Courtois et al. 2002). As p44 was shown to suppress growth only in the presence of full-length p53, we wanted to verify that the small size of p44 homozygous mice also required the presence of full-length p53. To do this genetically, we generated p44 transgenic mice on a p53 null background. In the absence of p53, p44 had no significant effect on overall size or on the size of internal organs (Fig. 3D). In all, we generated 28 p44 transgenics on a p53 null background, and they were all normal-sized. We also observed that these animals were prone to develop tumors at the same rate as p53 null animals, in contrast to animals in both p44 lines, which have a very low incidence of tumors (data not shown). Thus, the rescue of the p44 phenotype with respect to size extends to tumor susceptibility as well. This demonstrates that the growth-suppressive effect of p44 at both the cellular and organismal levels requires p53.
Slower growth rates
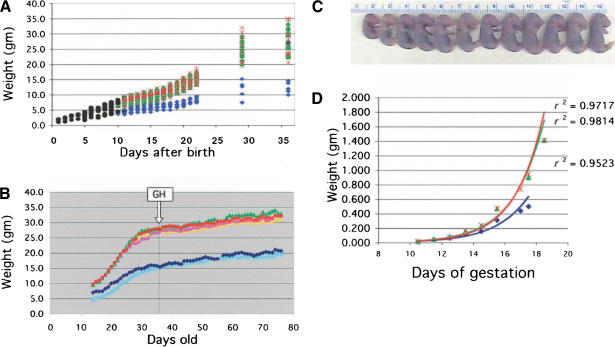
To understand the basis of the reduced size of p44 homozygous mice, we compared the postnatal growth rates of individuals with each possible genotype. The progeny of p44 intercrosses were weighed each day from birth to weaning and at weekly intervals thereafter. Homozygous p44 pups (Fig. 4A, blue symbols) had a reduced body mass relative to hemizygotes (green symbols) and NTs (red symbols) at all postnatal stages, and attained a final size of 50% of normal or less. Postnatal grow this primarily dependent on growth hormone (GH), and defects in GH or its receptor cause growth retardation in both humans and mice (Rosenfeld et al. 1994; Guevara-Aguirre et al. 1997; Lupu et al. 2001). To determine whether growth retardation in p44 homozygous mice was due to a defect in GH, we treated p44 and NT animals with recombinant rat GH once daily for 6 wk, beginning at 5 wk of age. A pharmacological dose was chosen to address the question of whether the phenotype was due to reduced pituitary GH secretion. In the wild-type this dose would be expected to replace the endogenous GH, which it would suppress, and thus would not result in an increase in body mass. There was no difference in the growth curves of animals of either genotype given GH and those given vehicle alone (Fig. 4B).
Figure 4.
Growth impairment during post- and prenatal development. (A) Postnatal growth. Hemizygous p44 mice were intercrossed to produce animals of each possible genotype. Black circles, genotype unknown; blue diamonds, homozygote; green triangles, hemizygote; red cross, NT. Each symbol represents an individual animal. The pups were weighed daily until weaning, and weekly thereafter. (B) Effect of rGH on postnatal growth. GH treatment was started at week 3 of the study, as indicated by the boxed arrow, and continued for 6 wk. Animals were weighed each day and injected s.c. with either 100 ng/g body weight recombinant growth hormone (rGH) or vehicle. Light blue diamonds, P+/+-GH; dark blue diamonds, P+/+-vehicle; yellow triangles, P+-GH; green triangles, P+-vehicle; pink circles, NT=GH; red circles, NT-vehicle. P+/+, homozygous transgenic; P+, hemizygous transgenic; NT, non-transgenic. (C) Size variation between sibling embryos. Individual size variation at E17.5 in the progeny resulting from a single intercross of p44 hemizygous parents. (D) Prenatal growth. Embryo weight vs. age during the period of fetal growth for individual homozygotes (blue diamonds), hemizygotes (green triangles), and NT embryos (red crosses) resulting from p44 hemizygous intercrosses. Regression curves and their associated coefficients of determination (r2) are superimposed on the data and follow the same color scheme.
To determine whether prenatal growth rates of p44 homozygotes were also slower than normal, we compared the sizes of individual embryos resulting from intercrosses of hemizygous transgenic mice at successive times during embryogenesis. Small embryos were found at all stages tested, as shown in Figure 4C for a litter of embryos at E17.5. At each time point, embryos were weighed, then genotyped, and the results used to calculate the rates at which animals with each genotype grew (Fig. 4D). Homozygous p44 embryos were smaller at all postimplantation stages, during both the earlier period of organogenesis (E6.5 to E9.5, data not shown) and later fetal growth stages (E10.5-E19, Fig. 4D). Although hemizygotes and NT embryos grew at the same rate (green and red curves), homozygotes grew at a slower rate (blue curve). Thus, p44 affects size by slowing down the rates at which the homozygous animal grows both pre- and postnatally.
Aberrant IGF signaling
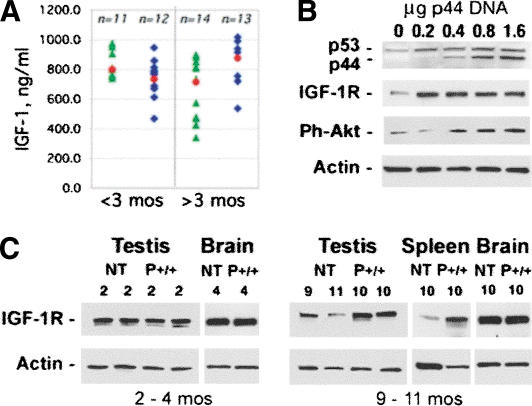
The combination of altered size and life span strongly implicated faulty signaling through the IGF pathway as the basis of the p44 homozygous phenotype. Although GH failed to rescue postnatal size deficits (Fig. 4B), it was still formally possible that circulating levels of IGF-1, which serve as an index of the effect of GH on postnatal growth, were abnormally low. However, as shown in Figure 5A, serum IGF-1 levels were actually found to be somewhat elevated in older p44+/+ animals. IGF-1 receptor levels were also found to be elevated in tissues that continue to proliferate throughout life (testis and spleen), but not in a tissue that contains mostly postmitotic cells (brain) in older P+/+ mice. In younger mice, there appear to be no differences in IGF-1 receptor (IGF-1R) levels. To show that it is p44 that is directly responsible for the elevated IGF-1R levels, we transfected NT mouse embryonic fibroblasts (MEFs) with increasing amounts of p44 DNA and measured the IGF-1R level. As shown in Figure 5C, even the smallest dose of p44 (0.2 μg) resulted in a higher protein level of the receptor compared to mock-transfected cells. Furthermore, the key molecular event in IGF-1R signaling—phosphorylation of Akt/PKB—also responded to p44 in a dose-dependent way (Fig. 5C). Western blot analysis demonstrated that the levels of the IGF-1R and of activated Akt were also several-fold higher in cells derived from P+/+ embryos than from cells derived from NT (wild-type) E12.5 embryos (Fig. 6A; compare the lanes marked “rhIGF-1”).
Figure 5.
p44 effects on IGF and its receptor. (A) Serum levels of IGF-1: Serum was prepared from retro-orbital cavity blood and subjected to competitive enzyme immunoassay to determine circulating levels of IGF in younger (<3 mo of age) and older (>3 mo of age) mice. Each blue or green symbol represents an individual animal. Blue diamonds, P+/+; green triangles, P+. The mean for each group is indicated by a red circle. (B) IGF-1 receptor expression in tissues. Western blots of 30 μg tissue extracts from young (2-4 mo) or old (9-11 mo) NT or p44 homozygous (P+/+) mice were probed with an antibody against the IGF-1 receptor (IGF-1R). Shown are representative samples of testis, brain, and spleen, with the genotypes and ages of each animal indicated above the lane with the corresponding protein extract. (C) IGF-1 receptor expression in response to increasing dose of p44 DNA. Mouse embryonic cells derived from NT mice were transfected with 0-1.6 μg pE44-1 plasmid DNA and grown for 24 h in 1% FCS. Cells were then switched to 0% for 3 h and treated for 15 min with rhIGF-1 or left untreated. Protein extracts (30 μg) were subjected to SDS-PAGE, and Western blots were probed with antibodies against the IGF-1R and phospho-Akt (Ser473). Cytoplasmic actin was used as a loading control.
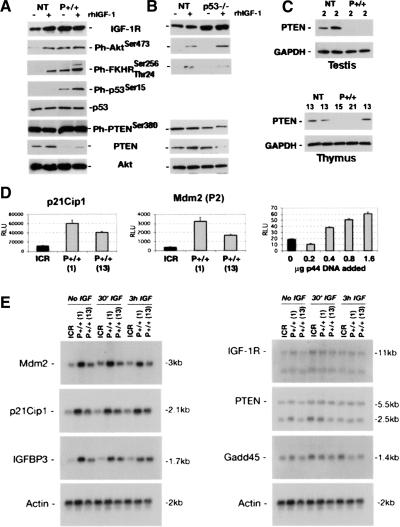
Figure 6.
Altered expression of IGF signaling molecules in p44 cells and tissues. (A) IGF-1 receptor expression in embryonic cells. Cells were grown as described in the text. NT cells respond to rhIGF-1 with an increase in receptor level. In p44 homozygous cells (P+/+), the receptor level is high even in the absence of added ligand. Increased IGF-1R activity is transduced to downstream targets, including Akt (Ph-Akt), forkhead (Ph-FKHR), and p53 itself (Ph-p53). PTEN is affected at both the total protein and phosphorylated protein levels. Total Akt levels were used as a loading control. (B) Effect of p53 null background on IGF-1 signaling. Same conditions as above, but with cells derived from p53-/- embryos. Compared to wild-type (NT), the level of phosphorylated Akt is higher in p53 null cells when IGF was added to the culture medium, which is similar to what was observed in p44 cells. In contrast, there was no hyperactivation of FKHR, which is actually lower in p53 null cells than in wild-type cells. (C) Expression of PTEN in tissues. Protein extracts (30 μg) from testis or thymus of individual mice were subjected to SDS-PAGE and Western blots probed with anti-PTEN or -GAPDH antibodies. GAPDH was used as a loading control. Ages and genotypes are indicated above each lane. The thymus extracts were all from females. (D) Luciferase reporter assays of p53 trans-activation function. Cells were transfected with the p21Cip1-luciferase reporter (p21Cip1) or Mdm2(P2)-luciferase [Mdm2(P2)] reporters and assayed for luciferase activity after 24 h in medium containing 1% FCS. MEFs from two different P+/+ embryos (labeled 1 and 13) or from NT embryos (labeled ICR) were used. In the right panel, only NT MEFs were used, and were transfected with increasing amounts of p44 DNA, as described in the Figure 5C legend. The resultant levels of p53/p44 in this experiment were assayed by Western blot and are shown in Figure 5C (uppermost panel). (E) Northern analysis of p53 target gene expression. Cells were grown in the absence of IGF-1 (No IGF), in the presence of IGF-1 during the last 30 min of culture (30′ IGF), or for 3 h with IGF-1 (3h IGF). Total RNA (9 μg) was subjected to denaturing gel electrophoresis, blotted, and hybridized to radioactive cDNA probes. Left panel: Targets demonstrating increased expression with overexpression of p44. Right panel: Targets demonstrating compromised p53 function with overexpression of p44. Rehybridization with a probe for β-actin (bottom) was used to control for any loading differences.
To begin to understand how this might affect intracellular IGF signaling pathways, we analyzed several critical downstream steps in signal transduction. Compared to wild type, embryonic cells derived from p44 homozygous embryos displayed an exaggerated forkhead response, which is evident as an increase in the level of phosphorylated FKHR (Tang et al. 1999); increased Ser15 phosphorylation; and thus activation of full-length p53 (Gu et al. 2000), and significantly reduced levels of active PTEN, which included increased phosphorylated PTEN as well as a decrease in the total amount of PTEN (Fig. 6A; Vazquez et al. 2001). This alteration in PTEN in MEFs from p44 homozygous embryos is significant, because we also observed greatly decreased PTEN levels in several tissues from adult p44 homozygous mice, as shown in Figure 6C. In testes from both of two young adult male mice and in thymi from two of three old (13-21-mo-old) female mice, there was no PTEN detectable.
As various molecular mechanisms might be responsible for these changes in IGF signaling, we performed experiments at the RNA level to determine the effect of p44 on trans-activation and trans-suppression functions of p53. The results are presented in Figure 6D,E. Using cells from two different p44 homozygous embryos (1 and 13), we found that p44 has both positive and negative effects on transcription-regulating functions of p53 depending on the specific target gene. In p44 homozygous cells, luciferase reporter genes driven by the p21/Cip1/Waf1 promoter and the P2 promoter of the Mdm2 gene (which is activated only by p53) were more active than in NT cells (Fig. 6D). In addition, transfection of NT cells with increasing doses of p44 DNA demonstrated that p44 could have both suppressive and potentiating effects on reporter gene activity, with the lowest dose actually lowering Mdm2-luciferase activity and the higher doses increasing it (Fig. 6D, right panel). Next, we examined Northern blots of RNA extracted from p44 homozygous or NT MEFs for the expression of a number of p53 target genes. These results confirmed that both Mdm2 and p21Cip1 were increased in p44 MEFs, and identified IGFBP-3 as another p53 target gene whose expression is potentiated in the presence of p44 (Fig. 6E, left panel). However, Gadd45, another p53 target, was inhibited in its response to IGF-1 in both p44 homozygous embryo-derived cell populations compared to NT MEFs (Fig. 6E, right panel). MEFs derived from individual p44 homozygous embryos consistently fell into two classes that were either more or less different from nontransgenic MEFs, as illustrated here by the MEFs from P+/+ (1) and (13), respectively. As each litter of embryos produces approximately equal numbers of more or less affected fibroblasts, and as male adult animals are consistently more severely affected than females, we suspect that this is due to a pervasive sexual dimorphism in the p44 phenotype.
Trans-suppression was also affected by p44. The 11-kb transcript representing the full-length IGF-1 receptor was higher in the p44+/+ cells in the absence of IGF, which parallels what we found at the protein level. Furthermore, the addition of IGF resulted in loss of trans-suppression in wild-type cells, but had no effect on p44+/+ cells (the three lanes marked “30′ IGF”), again supporting the idea that p44 interferes with normal p53 activity. Finally, the major 5.5-kb transcript of PTEN appears to be little affected by p44. Collectively, these results demonstrate that p44 overexpression has both dominant and dominant-negative effects on the function of full-length p53. To determine which effects were due to a loss of p53 function, we analyzed IGF signaling in MEFs derived from E12.5 embryos lacking p53 (p53-/-) and compared the levels of individual protein components to the levels seen in p44+/+ cells. As shown in Figure 6B, loss of p53 was associated with increased IGF-1R, hyperactivated Akt, and a reduced level of PTEN. However, in contrast to overexpression of p44, loss of p53 expression resulted in a diminished FKHR response and a decrease rather than increase in the inactivated form of PTEN. Together with the RNA expression data, these results clearly demonstrate that removing the N-terminal domain of the p53 protein does not have a simple loss- or gain-of-function effect on p53 activity. Increased trans-activation of p21, Mdm2, and IGFBP-3 demonstrate enhancement of p53 function as a consequence of p44 overexpression, whereas the effects on Gadd45 and IGF-1R reveal diminished p53 function. The possibility of generating tetramers in which anywhere from one to all four p53 monomers are replaced by a p53 isoform in which only one of several important functional domains of the p53 protein is missing, undoubtedly contributes to this complexity.
Reduced cellular proliferation with increased cellular senescence
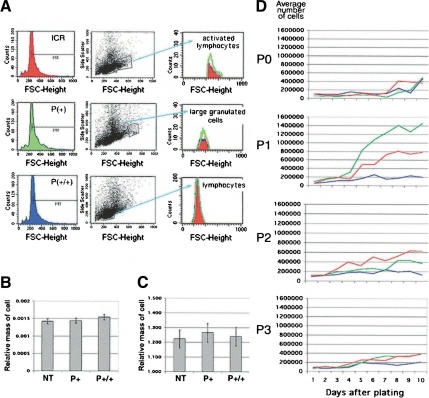
As IGF signaling can affect both cell size and cell number in lower organisms, we performed experiments to determine the underlying basis of the size deficits in p44 homozygous mice. We tested two alternate hypotheses that could account for the growth suppression caused by p44 on a p53(+/+) background: an inability of cells to grow (gain in mass) or to proliferate (increase in number). To determine if deficits in cell growth account for the small size of p44 homozygotes, we compared the sizes of cells in the testis, liver, and spleen from animals with one, two, or no copies of the p44 transgene. First, we measured the relative size (forward scatter, FSC) of splenocytes by cytofluorometry. Splenocytes of all three genotypes were similar in size when analyzed either as total splenocytes or as selected subpopulations (Fig. 7A). Next, we counted cells in histological sections of testis and liver from animals of different genotypes and found no difference in the relative masses of spermatocytes (Fig. 7B) or cells in the liver (Fig. 7C). Thus, the small size of p44 homozygous animals does not appear to be due to smaller-sized cells.
Figure 7.
p44 effects on cell size and cell number. (A) Lymphocytes. Relative sizes of gated subpopulations of a total of 20,000 spleen cells determined in forward scatter mode by FACS. Histograms of cell counts in populations of activated lymphocytes (top), large granulated cells (middle), and immature lymphocytes (bottom) are color-coded by genotype: red, NT splenocytes; green, hemizygous splenocytes; blue, homozygous splenocytes. (B) Testis. Relative mass of spermatocytes at 3 wk of age, as determined by image analysis. See Materials and Methods for details. (C) Liver. Relative area of cells from adult liver, as determined by image analysis. See Materials and Methods for details. (D) Embryonic cell proliferation. Assays were performed at successive passages with cells derived from E12.5-14 embryos. Shown is a plot of average cell counts over the 10-d period for cultures derived from NT (red), hemizygous (green), and homozygous (blue) embryos at P0, P1, P2, and P3. P0 were primary cells that had not been cultured for any length of time prior to the assay. Shown is one set of assays; identical results were obtained with cells from at least two different embryos of each genotype. Hemizygous cells consistently proliferated at a higher rate than NT cells at P1. See the discussion of the IGF-R in the text for an explanation.
We next examined the proliferative capacity of cells derived from embryos of the same three genotypes and cultured at successive passages in vitro (Fig. 7D). Cells derived from hemizygous or NT embryos demonstrated a striking ability to undergo a burst of proliferation at P1 in vitro, after which the number of cells produced in each culture declined with successive passages. In contrast, cells from p44 homozygous embryos failed to undergo this proliferative burst at any passage, and instead produced about four cell doublings per passage irrespective of the age of the culture. This difference in cell number was not due to a higher rate of cell death in homozygous cells, as the fraction of dead cells in the total was the same for all genotypes (data not shown).
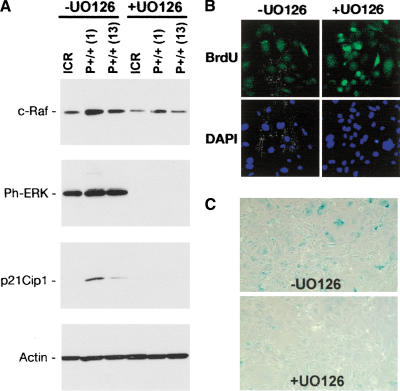
To understand the molecular nature of the proliferative defect in p44 homozygous cells, we analyzed the expression of components of a MAP kinase cascade that is known to be capable of overriding proliferative signals mediated by Akt in growth factor-stimulated cells (for review, see Roovers and Assoian 2000). The target of that cascade, ERK, has been shown to exert differential effects on cell-cycle progression via p21Cip1 depending on the amplitude and duration of the signal from the receptor. In fibroblasts, sustained ERK activation leads to induction of p21 (Woods et al. 1997) and G1 cell-cycle arrest (Meloche et al. 1992). The results of our analysis are shown in Figure 8A for NT (ICR) MEFs and MEFs from two different P+/+ embryos, P+/+ (1) and P+/+ (13), grown in the presence or absence of the MEK inhibitor UO126. The signal is transduced as a cascade of phosphorylation events on Ras, Raf, MEK, and finally, ERK, and blocking with UO126 stops the signal before ERK is activated. In the absence of UO126, the pathway is active, as evidenced by the phosphorylated state of ERK, which we detected using the pTEpY antibody (a gift from Tom Sturgill, Univ. of Virginia). In both P+/+ MEFs, but not in wild-type MEFs, this is accompanied by expression of p21Cip1. When the pathway is blocked, as evidenced by the absence of ERK phosphorylation, there is no p21Cip1 detectable. These results are consistent with a model in which sustained IGF-1R activation in p44 homozygous cells leads to sustained ERK activation, p21 induction, and cell-cycle arrest. We cannot completely exclude the possibility that MAP kinases other than MEK1/2 are involved in the observed p21 upregulation by p44 and might be similarly inhibited by UO126. Activated p38MAPK, for example, can stabilize p53 by phosphorylation of Ser15 (Bulavin et al. 1999) and thereby lead to trans-activation of the p21/Cip1/Waf1 gene by p53. However, MEK inhibitors have so far only been shown to lead to increased p38MAPK activation by kinase crosstalk (Xiao et al. 2002). We would therefore rather expect an increase in, rather than loss of, p21 expression after U0126 block of MEK1/2. The complete abrogation of p21 seen in our experiments, in fact, argues against a direct involvement of p38MAPK.
Figure 8.
Effect of ERK activation on proliferation and senescence. (A) Ras-Raf-MEK-ERK effects in MEFs. Western blots of protein extracts (30 μg) from cells grown in the absence (-) or presence (+) of UO126, a pharmacological blocker of the ERK kinases, MEK1/2. ICR, cells from NT embryo; P+/+ (1) and P+/+ (13), cells from two different p44 homozygous embryos. (B) Recovery of proliferation in p44 homozygous MEFs by pharmacological blockade of ERK. Cells were grown for 48 h in 10 mM UO126 and pulsed for 1 h with BrdU. After fixation, cells were stained with antibody G3G4 against BrdU (green, top panels) to identify nuclei synthesizing DNA and with DAPI (blue, bottom panels) to identify all cell nuclei. Original magnification, 40×. Images were captured digitally and manipulated using Adobe Photoshop, v.7. (C) Reduced cellular senescence in p44 homozygous MEFs by pharmacological blockade of ERK. Cells were subjected to SA-β-gal assay following culture in the absence or presence of UO126, as described above.
To determine whether blockade of MEK by UO126 could reverse the proliferation defect, cells were labeled with BrdU for a short time before fixation. With UO126 applied continuously in the medium for 72 h, many more P+/+ cells were found to be synthesizing DNA than when the MEK-inhibitor was absent, as evidenced by the increased BrdU-associated fluorescence in the +UO126 panel (Fig. 8B). Furthermore, blocking the MAP kinase pathway in presenescent cells also resulted in many fewer senescent cells after 72 h compared to cultures grown in the absence of UO126, as determined by senescence-activated β-gal assay (Dimri et al. 1995) on P2 cells (Fig. 8C). This is evidenced by a decrease in the number of blue-stained cells in the panel marked +UO126 in Figure 8C. These results strongly suggest that proliferation is clamped by the cell-cycle inhibitor, p21Cip1, and can return to more normal levels when the pathway that activates p21 expression is turned off.
Discussion
Life span manipulations in the nematode C. elegans have shown that loss-of-function mutations in genes that code for insulin/IGF signaling proteins, such as insulin/IGF-R (daf-2), PI3K (age-1), and Akt/PKB (akt; for review, see Guarente and Kenyon 2000) significantly prolong life span and fertility. In Drosophila melanogaster, some allelic combinations of the INS/IGF-R produce flies with extended life span (Clancy et al. 2001; Tatar et al. 2001). The Ames and Snell GH-deficient mice have significantly longer life spans and exhibit significant delays in biological senescence compared to wild-type mice (Flurkey et al. 2001), and recent studies show that reduced levels of the IGF-1 (Holzenberger et al. 2003) and insulin (Bluher et al. 2003) receptors also significantly increase life span in the mouse. Thus, the IGF axis is linked to longevity in animals representing vast evolutionary distances, up to and including mammals.
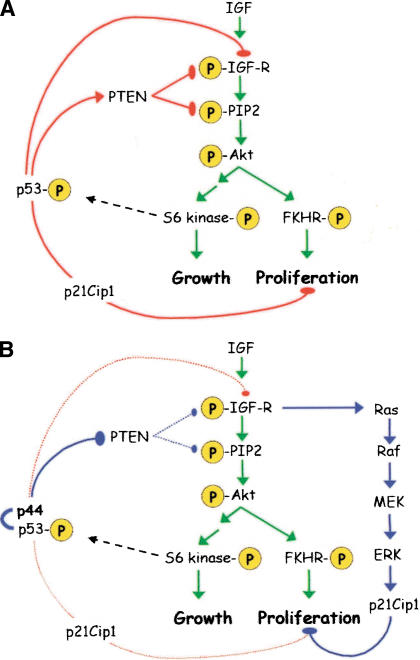
We have unexpectedly uncovered a role for the short isoform of p53 in the regulation of the IGF axis in the mouse. Overexpression of the short form of p53 and disruption of the normal ratio of p44 to p53 have effects on both size and life span that can be linked to changes in the intracellular signaling cascades initiated by IGF. As depicted in Figure 9, p53 exerts control over IGF signaling at several key points. First and foremost, p53 controls the level of the IGF-1 receptor (Werner et al. 1996). Secondly, p53 controls both the level and activity of the dual lipid-protein-phosphatase, PTEN (Schuster et al. 2001; Stambolic et al. 2001). PTEN modulates the IGF signal transduced to Akt, mainly through dephosphorylation of phosphatidyl-inositol triphosphate (Fig. 9A). The level of PTEN is controlled by direct trans-activation of the PTEN promoter by p53 (Stambolic et al. 2001). Although trans-activation of PTEN is enhanced only slightly by the presence of p44 (see Fig. 6E above), the level of the protein actually goes down in MEFs (Fig. 6A) and in tissues from p44+/+ mice (Fig. 6C). In addition to this apparent protein instability, phosphorylation at the site modified by casein kinase II (CKII; Torres and Pulido 2001; Vazquez et al. 2001; Miller et al. 2002) is dramatically increased (Fig. 6A). Collectively, these results can be interpreted as interference by p44 with protein-protein interactions between full-length p53 and CKII, which are known to be inhibitory (Schuster et al. 2001), rather than with protein-DNA interactions during assembly of the trans-activation complex on the PTEN promoter. Increased phosphorylation of PTEN by CKII leads not only to its inactivation, but also to its stabilization, which seems to be responsible for the apparent accumulation of phospho-PTEN and loss of degradation-sensitive PTEN in cells of p44 mice. Because phosphorylation blocks its recruitment into complexes at the cell membrane and inhibits its phosphatase activity (Vazquez et al. 2001), p44 interference effectively results in functional inactivation of PTEN.
Figure 9.
A model for the regulation of IGF signaling by p53. (A) Hierarchy of IGF control in normal cells. Major phosphorylation events that occur upon ligand binding are indicated by the “P”s circled in yellow and are connected by green arrows. The pathway bifurcates at Akt/PKB into an FKHR-dependent proliferation branch (right) and a TOR/S6 kinase-dependent anabolic branch (left). p53 exerts several layers of control over the first steps of the cascade (solid red arrows), but also has a fail-safe mechanism of control via p21 late in the cascade (dotted red arrow). (B) Disruption of IGF control by p44. The interaction of p44 with p53 results in permanent activation of the IGF-R, simultaneously triggering growth via the IGF pathway (green arrows) and cell-cycle arrest via Ras (blue arrows).
Control of the IGF axis by p53 is exerted at the earliest steps in the cascade, modulating effectors of both growth and proliferation further downstream. Although we have not identified the exact mechanism by which the Ras-Raf-MEK-ERK pathway switches from one promoting proliferation to one inducing cell-cycle arrest (Fig. 9B), we have shown that blocking this pathway pharmacologically can prevent replicative senescence and/or restore the ability of presenescent cells to proliferate. Pharmacological reversal of replicative senescence by blocking the expression of p21Cip1 is similar to a result with presenescent human fibroblasts in which senescence could be reversed in cells expressing p21, but not in cells expressing p16 (Beausejour et al. 2003). This strengthens our argument that hyperactivation of the IGF signaling pathway plays a causal role in the phenotype of p44 transgenic mice by setting in motion a “fail-safe” pathway to clamp downstream pathways that would otherwise lead to uninhibited growth. This also helps to resolve the paradox of small size with hyperactivity of an axis that otherwise would be expected to enhance growth. The outcome of affecting a major pathway that controls both growth and proliferation is perhaps best illustrated by contrasting the phenotype of p44 mice in which p53 function is disturbed with mice in which proliferation alone is affected. Hypomorphic alleles of Myc cause comparable reductions in overall size (Trumpp et al. 2001), but have no effect on longevity (Andreas Trumpp, pers. comm.).
Our results confirm results reported last year that p53 status affects mammalian aging (Tyner et al. 2002). Because the mice described in that report carry a large genomic deletion that encompasses substantial sequence upstream of the p53 locus in addition to the p53 gene itself, however, the link between p53 and premature aging could not be made with certainty. Our results utilizing a very precisely mapped 3-kb deletion that removed exon 2 and the flanking intron sequences on either side firmly identify p53 as the cause of the early-aging phenotype. Translation of this mutant p53 gene initiates at the ATG codon in exon 4 (Met41; Rovinski et al. 1987) and produces a 44-kD protein. This is the same internal initiation codon that produces the short form of p53 from the endogenous human p53 locus, as reported by Courtois et al. (2002) and Yin et al. (2002). Where our data disagree with those reports, however, is that we find evidence that, at least in the mouse, the protein is encoded by its own RNA transcript, leaving open the possibility of a mechanism in addition to alternative translation that can generate the shorter protein product (Yin et al. 2002). In mice with one mutant p53 allele coding for a truncated gene product and one intact allele, cells lose the ability to regenerate aging or damaged tissue (Tyner et al. 2002). Mice carrying additional copies of the entire p53 locus exhibit normal growth and aging (Garcia-Cao et al. 2002), which together suggest that the ratio of short to long forms of p53 must be preserved in order for p53 to function normally. As even a slight increase in the level of p44 boosts the level of p53 (Fig. 5C), one consequence of upsetting the normal p44:p53 ratio is an increase in the overall activity of p53. We have limited ourselves to defining the consequences of this imbalance on IGF signaling; presumably there are also consequences on other pathways on which p53 impinges.
The need for both p44 and p53 for full expression of the phenotype described in this paper suggests that the short and long forms of p53 act in concert. As reported by Courtois et al. (2002), the short form of p53 co-immuno-precipitated with the full-length form of p53 in human cell lines, clearly demonstrating protein-protein interaction between them. As the oligomerization domain is present in both isoforms, the most likely explanation at the molecular level is that they form heterotetramers. The participation of p44 in heterotetramers with p53 would tend to stabilize the overall structure, because the lack of an Mdm2-binding domain would mean that Mdm2 cannot shuttle p44 to the proteasome for degradation, as is clear from the much longer half-life of p44 compared to p53 (>9.5 h vs. 2 h; Rovinski et al. 1987). The relative contribution of p44 to these tetramers could potentially alter functional p53 status in more than one way, and even in opposite directions. This could explain, for example, why some p53 targets are hyperactivated by p44 (p21Cip1, Mdm2, IGFBP-3) whereas at other targets p44 interferes with p53 activity (Gadd45 and IGF-1R). It could also resolve the apparent discrepancy between suppression of trans-activation by DeltaNp53 in transfected cells, as reported by Courtois et al. (2001), and enhancement of trans-activation by p44 reported in this paper. Experiments with DeltaNp53 were performed either in the absence of full-length p53 or in a vast excess of the short form of p53, and thus are equivalent to our results in animals when the p44 transgene was crossed onto a p53 null background.
Our work identifies one major molecular pathway that is affected by alteration in p53 status, the pathway activated by IGF-1. The significance of this finding is that it provides a mechanism by which the same pathway that governs life span in lower organisms can govern mammalian life span. It also provides a molecular link between the daf (INS/IGF) pathway that modulates life span in C. elegans, and the activity of Sir2, a gene shown to mediate life-span extension by caloric restriction in yeast (Lin et al. 2000). Sir2 is a protein deacetylase whose activity is dependent on the energy status of a cell. In addition to its histone deacetylase activity, the mammalian homolog of the yeast Sir2 gene [Sir2α in mouse (Imai et al. 2000) and SIRT1 in human (Frye 1999)] can regulate the activity of p53 (Luo et al. 2001; Vaziri et al. 2001). This should render p53 activity in mammalian cells—including transcriptional control of genes in the IGF signaling pathway—responsive to the same cues that modulate longevity in yeast. By positioning p53 function between these two longevity determinants (IGF signaling and Sir2), mammalian cells can remain mitotically active and contribute to long life span without growing out of control and killing the organism.
Materials and methods
Production of transgenic mice
The transgene was microinjected into pronuclear-stage B6SJL embryos by standard procedures. To generate p44 mice on an ICR background, the original B6SJL transgenics were bred for at least four generations with ICR mice, then intercrossed to produce homozygotes. To generate p44 mice on a p53 null background, p44 (ICR) homozygotes were crossed to C57Bl/6 p53(-/-) mice (Taconic).
Bone mineral density measurement
Bone mineral density (BMD) measurements were performed on the dissected bones using the small-animal densitometer PIXI-mus II (Faxitron X-Ray). Measurements were conducted on the total body bones, as well as on separate femora, tibiae, and lumbar spines using the image analysis software provided by the system. A region of interest (ROI) was generated in the secondary spongiosa of the above-mentioned bones. The BMD (g/cm2) is defined as the integral mass of bone mineral per unit of projected area.
Bone histology and histomorphometry
After fixation in 10% neutral buffered formalin, all four limbs and spine were decalcified, processed, and embedded in paraffin. Longitudinal midsagittal sections 7 μm in thickness from the tibia, femur, and lumbar spines were cut, stained with hematoxylin and eosin (H&E), and prepared for histomorphometric analysis. Images for all the tissues were captured and analyzed using Metamorph software (Universal Imaging) and viewed on a Leica DM LB compound microscope outfitted with a Q-Imaging Micropublisher Cooled CCD color digital camera. Trabecular bone volume (TBV), defined as bone volume/tissue volume % (BV/TV %), was assessed on the secondary spongiosa of the proximal tibia, distal femur, and lumbar spine at 50× magnification on H&E-stained sections. Bothosteoblast and osteoclast numbers were assessed on the same section at 200× magnification and expressed as number of cells/mm bone surface (N.Ob/BS; /mm) and (N.Oc/BS;/mm), respectively. Nomenclature and symbols of histomorphometric parameters used are as described by Parfitt et al. (1987).
Histology
Tissues were fixed in Bouin's solution, embedded in paraffin, and sectioned. Following routine H&E staining, sections were viewed on a Zeiss Axiophot light microscope outfitted with a SONY DSC-S75 digital camera with an 8.93-mm color CCD image device and 3× zoom lens (Zeiss). Images were prepared using Adobe Photoshop v.7 (Adobe Systems).
Survival curves
Survival data on p44 transgenic mice were obtained from our own colony. The NT data were provided by Harlan Sprague Dawley. The background strain of both the transgenic mice and the NT mice is Hsd:ICR(CD-1).
RNase protection assay
The probe is a 251-bp StyI fragment spanning exons 2-4 from pp53-176 (a gift from Dr. Greg Matlashewski, McGill University, Montreal, QC, Canada) that was cloned into pGEM-5Zf(+) (Promega). To prepare the probe, the p44/p53 riboprobe plasmid was linearized with SpeI and transcribed using SP6 RNA polymerase in the presence of 32P-UTP (800 Ci/mmole; ICN). The probe was gel-purified and eluted overnight in 0.5 M NH4OAc/1 mM EDTA/0.2% SDS. To perform the assay, 2.5-10 μg total RNA was hybridized overnight with 2 × 105 cpm p44/p53 riboprobe and 1 × 105 cpm cyclophilin riboprobe (Ambion) at 50°C in 80% formamide/40 mM PIPES buffer/0.4 M NaCl/1 mM EDTA. The following day, the hybridization reactions were digested with a solution of RNase A/RNase T1 for 1 h at 30°C, precipitated, and resuspended in gel loading buffer (95% formamide/0.025% each xylene cyanol and bromophenol blue/0.5 mM EDTA/and 0.025% SDS). The samples were electrophoresed through a 6% polyacrylamide/8 M urea gel, which was then dried and subjected to autoradiography. The probe spans the deleted exon 2 in p44 and protects fragments of 140 and 220 bases of p44 and p53, respectively.
GH rescue
Male and female p44 and NT mice were weighed daily and treated with100 μL saline or 100 μL of 100 ng/g body weight recombinant rat growth hormone (kindly provided by Genetech). Volume was kept constant at 100 μL with saline. Mice were injected subcutaneously over the shoulder area once daily beginning at 10:00 am for 35 d. They were killed at the end of the 35-d treatment period. Blood was collected for serum, and tissues were collected and placed immediately in liquid nitrogen, then stored at -80°C.
Serum IGF-1 assay
Mouse serum samples were assayed for IGF-1 levels using a Rat IGF-1 kit from Diagnostic Systems Laboratories according to insert instructions and reagents provided. Mouse IGF-1 and rat IGF-1 differ by only one amino acid and are highly cross-reactive in this assay. This enzyme immunoassay uses the competitive enzyme immunoassay format. Standard, controls, and unknowns containing IGF-1 are incubated with biotin-labeled rat IGF-1 and goat anti-rat IGF-1 antiserum in microtiter wells coated with rabbit anti-goat gamma-globulins. Biotin-labeled antigens and unlabeled antigens compete for a limited number of anti-rat IGF-1 binding sites. After incubation and washing, the wells are incubated with streptavidin-horseradish peroxidase (HRPO), which binds to the biotinylated rat IGF-1. The unbound HRPO is washed and followed by incubation with substrate tetramethylbenzidine (TMB). An acidic stopping solution is added to stop the reaction, and the degree of enzymatic turnover was measured using a Phenix GENios at wavelength 450 nm. Data were analyzed using nonlinear least-squared fitting to a binding competition model using Graph Pad Prism.
Western blots
Protein extracts were prepared with RIPA lysis buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) or with NP-40 lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris, pH 8.0) from cells routinely grown in DMEM/10% fetal calf serum (FCS). On the day before harvest, cells were switched to 1% FCS in DMEM and grown for an additional 18 h, then switched to 0% FCS for 3 h. Following serum starvation, the cells were treated with rhIGF-1 (10 nM in DMEM; rhIGF-1 Lot # 01 was obtained through NHPP, NIDDK from Dr. Parlow) for 15 min or 3 h and harvested immediately afterwards. To inhibit MEK activity, UO126 (Promega) was added 30 min before addition of IGF-1 and maintained in cultures at a final concentration of 10 μM. Western blots were generated using standard procedures. Antibodies used were: CM5 (Novocastra Laboratories/Vector Labs) to detect total p53 and #9284 (Cell Signalling Technology) against phospho-p53(Ser15); SC713 (Santa Cruz Biotechnology) against the IGF-R; #9551 against phospho-PTEN(Ser380) and #9556 (both Cell Signalling Technology) to detect total PTEN; #9271 against phospho-Akt-(Ser473) and # 9272 (both Cell Signalling Technology) to detect total Akt/PKB; #9461 (Ser256) and #9464 (Thr24) against phospho-FKHR, and #9462 (all Cell Signalling Technology) to detect total FKHR; 691001 (JCN) against cytoplasmic actin, and Sc-6246 (Santa Cruz Biotechnology) against p21/Cip1. Antigen-antibody complexes were further reacted with HRP-conjugated secondary antibodies (Jackson Immuno-Research Laboratories) and detected by chemiluminescence (SuperSignal Pico reagent, Pierce). Signals were quantified by laser densitometry on a Molecular Dynamics instrument running ImageQuant software. The blots were stripped and re-reacted with an anti-actin or anti-GAPDH antibody to normalize the signals.
Transfections
Lipofectamine reagent (Invitrogen) was used to transfect embryonic fibroblasts according to the manufacturer's recommendations.
Luciferase assays
Cells were plated, transfected, and assayed in triplicates for each different condition. After overnight transfection, cells were starved for 24 h in medium containing 1% FCS. Luciferase activity was detected using the Dual Luciferase Reporter Assay System (Promega).
Northern blots
RNA extraction using TRI-Reagent (Molecular Research Products), Northern blotting, and hybridizations were performed as described (Cronin et al. 2001).
Cell size determination
Testis and liver Testes from prepubertal mice of each genotype were fixed in Bouin's fixative, embedded in paraffin, and sectioned. For counting, the sections were stained with H&E and imaged on a Zeiss Axiophot microscope equipped with a Sony DSC-S75 3.3 megapixel CCD camera. The images were printed onto paper, and individual tubule cross-sections were cut out of the paper and weighed. This controlled for the varying dimensions of the tubules and thus was a more precise way of comparing cell size between animals of different genotypes than would have been allowed by a simple cells-per-tubule basis. To simplify the method even further, we used testes from 3-wk-old mice. At this time, the seminiferous epithelium is undergoing the first wave of spermatogenesis, and all tubules are at approximately the same stage of differentiation. We counted only the large, round primary spermatocytes, and determined their mass relative to the mass of their paper image. The only other stage present at this time consists of small cells at the periphery of the seminiferous epithelium, which represent the beginning of the next wave of spermatogenesis and were not counted. We used a similar technique to determine the relative size of cells in the liver. Equivalent sections of liver parenchyma were stained with DAPI to label cell nuclei, and images were prepared in which the entire field was filled with uniformly stained cells. The number of nuclei in the entire field was taken as the number of cells per fixed area of tissue for each of the genotypes of p44 transgenic and NT adult animals.
Spleen Single-cell suspensions were obtained by disrupting the spleen in a glass homogenizer in Dulbecco minimal essential medium (DMEM, Gibco) containing 10% FCS. Red blood cells were lysed by hypertonic shock using a 150 mM ammonium chloride lysis buffer (8.29 g ammonium chloride, 1 g potassium bicarbonate, and 37 mg EDTA in 1000 mL H2O) for 5 min. Cells were washed three times in Dulbecco phosphate-buffered saline (DMEM, Gibco) and analyzed using a FACSCalibur flow cytometer (Becton Dickinson). The acquired data of 20,000 cells per sample were analyzed using Cell Quest v3.1 analysis software (Becton Dickinson).
Proliferation assays of embryonic cells in vitro
Embryos from timed pregnant females were collected at E12.5-E14, minced, and incubated in a collagenase solution (2 mg/mL collagenase B and 2 U/mL DNase in RPMI 1640) for 1 h at 37°C. The heads were saved for genotyping. The embryos were triturated and incubated for an additional 1 h at 37°C. The cells were collected into a centrifuge tube, spun at 1000 rpm for 5 min, and the medium was poured off. The cells were washed and spun again, and then plated in DMEM/10% FCS. The cells were grown overnight, and the next day they were frozen in 90% serum/10% DMSO as P0, and plated for the P0 assay. Ten duplicate wells containing 5×104 cells each of homozygous, hemizygous, and NT embryonic cells were plated on day 0. On each of 10 successive days, the cells were harvested and counted in a hemacytometer, and the duplicates averaged. For later passages, cells were carefully monitored and passaged on the first day the plate was seen to be confluent. In this way, the cells were never allowed to overgrow or undergrow the plate, so that the growth characteristics of cells in later passages reflect the growth characteristics they would have had if allowed to grow continuously in unlimited space. Each assay was carried out as described for P0.
To determine the rate of cell death associated with each genotype, we harvested cells in duplicate wells every 2 d and stained them with the Live/Dead kit from Molecular Probes. The live (green) and dead (red) cells were counted in a hemocytometer on a fluorescence microscope, and the ratio of dead:live was calculated for each genotype.
Senescence-associated-β-galactosidase assay and BrdU incorporation
Senescent mouse embryo fibroblasts (MEFs) were identified by their ability to stain positively for acidic β-galactosidase (pH 6), as described by Dimri et al. (1995). After a second passage (P2), explanted cells were plated at 3×105 in 60-mm dishes, grown to ∼70% confluency, and stained in SA-associated-β-galactosidase staining solution overnight at 37°C. For prevention or reversion of replicative senescence, MEFs were passaged twice and plated each time at 105 cells/100-mm petri dishand grown to 70% confluency. Presenescent cells (<10% of the culture showing DNA synthesis monitored by BrdU incorporation) were cultured for 48 h in 10 μM U0126 (with daily medium exchange). Cells were pulsed for 1 h with BrdU at 5 μg/mL medium before fixation in 2% formaldehyde + 0.2% glutaraldehyde (for simultaneous assays for replicative senescence and BrdU incorporation). Cells were fixed with 4% paraformaldehyde when assayed only for BrdU incorporation. BrdU was detected by the monoclonal antibody G3G4 (developed by Stephen J. Kaufman and obtained from the Developmental Studies Hybridoma Bank under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences).
Acknowledgments
We thank Drs. Alan Bernstein and Alain Lavigueur for the gift of DNA encoding p44 (plasmids pMR44 and pE44); Harlan Sprague Dawley for sharing survival data on Hsd:ICR(CD-1) mice; Drs. Moshe Oren and Bert Vogelstein for the Mdm2-luc and p21Cip1-luc reporter constructs, respectively; and Drs. Greg Matlashewski for p53, Al Fornace for Gadd45, Michael Weber for IGF-1R, Frank Furnari for PTEN, and Donna George for Mdm2 cDNAs. The antibody against the phosphorylated form of ERK (pTEpY) was a gift from Dr. Thomas W. Sturgill. We thank Amy Ryan, Pattie Hellmann, Dr. Jianhua Liu, Marc Caligtan, Elaine Chen, and Patrick Martin for technical assistance on critical experiments; Dr. Carmen Sapienza (Fels Institute) for critical reading of an early version of this manuscript; and Dr. Scott Eblen for informative discussions about MAPK pathways. This work was supported by grants RR11102 and AG20915 to H.S. and DK07766 to B.M. from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1162404.
References
- Beausejour C.M., Krtolica, A., Galimi, F., Narita, M., Lowe, S.W., Yaswen, P., and Campisi, J. 2003. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J. 22: 4212-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellido T., Jilka, R.L., Boyce, B.F., Girasole, G., Broxmeyer, H., Dalrymple, S.A., Murray, R., and Manolagas, S.C. 1995. Regulation of interleukin-6, osteoclastogenesis, and bone mass by androgens. The role of the androgen receptor. J. Clin. Invest. 95: 2886-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M., Kahn, B.B., and Kahn, C.R. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572-574. [DOI] [PubMed] [Google Scholar]
- Bulavin D.V., Saito, S., Hollander, M.C., Sakaguchi, K., Anderson, C.W., Appella, E., and Fornace Jr, A.J. 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18: 6845-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D.J., Gems, D., Harshman, L.G., Oldham, S., Stocker, H., Hafen, E., Leevers, S.J., and Partridge, L. 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104-106. [DOI] [PubMed] [Google Scholar]
- Courtois S., Verhaegh, G., North, S., Luciani, M.G., Lassus, P., Hibner, U., Oren, M., and Hainaut, P. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21: 6722-6728. [DOI] [PubMed] [Google Scholar]
- Cronin C.A., Gluba, W., and Scrable, H. 2001. The lac operator-repressor system is functional in the mouse. Genes & Dev. 15: 1506-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry W.B., Putzke, A.P., and Rothman, J.H. 2001. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science 294: 591-595. [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E.E., Linskens, M., Rubelj, I., Pereira-Smith, O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. 92: 9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben R.G., Eberle, J., Stahr, K., and Goldberg, M. 2000. Androgen deficiency induces high turnover osteopenia in aged male rats: A sequential histomorphometric study. J. Bone Miner. Res. 15: 1085-1098. [DOI] [PubMed] [Google Scholar]
- Flurkey K., Papaconstantinou, J., Miller, R.A., and Harrison, D.E. 2001. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl. Acad. Sci. 98: 6736-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R.A. 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260: 273-279. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao I., Garcia-Cao, M., Martin-Caballero, J., Criado, L.M., Klatt, P., Flores, J.M., Weill, J.C., Blasco, M.A., and Serrano, M. 2002. `Super p53' mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21: 6225-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Lynch, J., and Brecher, P. 2000. Nitric oxide increases p21(Waf1/Cip1) expression by a cGMP-dependent pathway that includes activation of extracellular signal-regulated kinase and p70(S6k). J. Biol. Chem. 275: 11389-11396. [DOI] [PubMed] [Google Scholar]
- Guarente L. and Kenyon, C. 2000. Genetic pathways that regulate ageing in model organisms. Nature 408: 255-262. [DOI] [PubMed] [Google Scholar]
- Guevara-Aguirre J., Rosenbloom, A.L., Vasconez, O., Martinez, V., Gargosky, S.E., Allen, L., and Rosenfeld, R.G. 1997. Two-year treatment of growth hormone (GH) receptor deficiency with recombinant insulin-like growth factor I in 22 children: Comparison of two dosage levels and to GH-treated GH deficiency. J. Clin. Endocrinol. Metab. 82: 629-633. [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Dupont, J., Ducos, B., Leneuve, P., Geloen, A., Even, P.C., Cervera, P., and Le Bouc, Y. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421: 182-187. [DOI] [PubMed] [Google Scholar]
- Imai S., Armstrong, C.M., Kaeberlein, M., and Guarente, L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795-800. [DOI] [PubMed] [Google Scholar]
- Jin S., Martinek, S., Joo, W.S., Wortman, J.R., Mirkovic, N., Sali, A., Yandell, M.D., Pavletich, N.P., Young, M.W., and Levine, A.J. 2000. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. 97: 7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A., Maltby, V., Mock, D., Rossant, J., Pawson, T., and Bernstein, A. 1989. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol. Cell. Biol. 9: 3982-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.J., Defossez, P.A., and Guarente, L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126-2128. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev, A.Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107: 137-148. [DOI] [PubMed] [Google Scholar]
- Lupu F., Terwilliger, J.D., Lee, K., Segre, G.V., and Efstratiadis, A. 2001. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 229: 141-162. [DOI] [PubMed] [Google Scholar]
- Meloche S., Seuwen, K., Pages, G., and Pouyssegur, J. 1992. Biphasic and synergistic activation of p44mapk (ERK1) by growth factors: Correlation between late phase activation and mitogenicity. Mol. Endocrinol. 6: 845-854. [DOI] [PubMed] [Google Scholar]
- Miller S.J., Lou, D.Y., Seldin, D.C., Lane, W.S., and Neel, B.G. 2002. Direct identification of PTEN phosphorylation sites. FEBS Lett. 528: 145-153. [DOI] [PubMed] [Google Scholar]
- Mowat M., Cheng, A., Kimura, N., Bernstein, A., and Benchimol, S. 1985. Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature 314: 633-636. [DOI] [PubMed] [Google Scholar]
- Ollmann M., Young, L.M., Di Como, C.J., Karim, F., Belvin, M., Robertson, S., Whittaker, K., Demsky, M., Fisher, W.W., Buchman, A., et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91-101. [DOI] [PubMed] [Google Scholar]
- Parfitt A.M., Drezner, M.K., Glorieux, F.H., Kanis, J.A., Malluche, H., Meunier, P.J., Ott, S.M., and Recker, R.R. 1987. Bone histomorphometry: Standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2: 595-610. [DOI] [PubMed] [Google Scholar]
- Roovers K. and Assoian, R.K. 2000. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays 22: 818-826. [DOI] [PubMed] [Google Scholar]
- Rosenfeld R.G., Rosenbloom, A.L., and Guevara-Aguirre, J. 1994. Growth hormone (GH) insensitivity due to primary GH receptor deficiency. Endocr. Rev. 15: 369-390. [DOI] [PubMed] [Google Scholar]
- Rovinski B. and Benchimol, S. 1988. Immortalization of rat embryo fibroblasts by the cellular p53 oncogene. Oncogene 2: 445-452. [PubMed] [Google Scholar]
- Rovinski B., Munroe, D., Peacock, J., Mowat, M., Bernstein, A., and Benchimol, S. 1987. Deletion of 5′-coding sequences of the cellular p53 gene in mouse erythroleukemia: A novel mechanism of oncogene regulation. Mol. Cell. Biol. 7: 847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster N., Gotz, C., Faust, M., Schneider, E., Prowald, A., Jungbluth, A., and Montenarh, M. 2001. Wild-type p53 inhibits protein kinase CK2 activity. J. Cell. Biochem. 81: 172-183. [DOI] [PubMed] [Google Scholar]
- Stambolic V., MacPherson, D., Sas, D., Lin, Y., Snow, B., Jang, Y., Benchimol, S., and Mak, T.W. 2001. Regulation of PTEN transcription by p53. Mol. Cell 8: 317-325. [DOI] [PubMed] [Google Scholar]
- Tang E.D., Nunez, G., Barr, F.G., and Guan, K.L. 1999. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274: 16741-16746. [DOI] [PubMed] [Google Scholar]
- Tatar M., Kopelman, A., Epstein, D., Tu, M.P., Yin, C.M., and Garofalo, R.S. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107-110. [DOI] [PubMed] [Google Scholar]
- Torres J. and Pulido, R. 2001. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 276: 993-998. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G.R., and Bishop, J.M. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414: 768-773. [DOI] [PubMed] [Google Scholar]
- Tyner S.D., Venkatachalam, S., Choi, J., Jones, S., Ghebranious, N., Igelmann, H., Lu, X., Soron, G., Cooper, B., Brayton, C., et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415: 45-53. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain, S.K., Ng Eaton, E., Imai, S.I., Frye, R.A., Pandita, T.K., Guarente, L., and Weinberg, R.A. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149-159. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Grossman, S.R., Takahashi, Y., Rokas, M.V., Nakamura, N., and Sellers, W.R. 2001. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 276: 48627-48630. [DOI] [PubMed] [Google Scholar]
- Werner H., Karnieli, E., Rauscher, F.J., and LeRoith, D. 1996. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc. Natl. Acad. Sci. 93: 8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D., Parry, D., Cherwinski, H., Bosch, E., Lees, E., and McMahon, M. 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 17: 5598-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.Q., Malcolm, K., Worthen, G.S., Gardai, S., Schiemann, W.P., Fadok, V.A., Bratton, D.L., and Henson, P.M. 2002. Cross-talk between ERK and p38 MAPK mediates selective suppression of pro-inflammatory cytokines by transforming growth factor-β. J. Biol. Chem. 277: 14884-14893. [DOI] [PubMed] [Google Scholar]
- Yin Y., Stephen, C.W., Luciani, M.G., and Fahraeus, R. 2002. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 4: 462-467. [DOI] [PubMed] [Google Scholar]
- Zhao R., Gish, K., Murphy, M., Yin, Y., Notterman, D., Hoffman, W.H., Tom, E., Mack, D.H., and Levine, A.J. 2000. The transcriptional program following p53 activation. Cold Spring Harb. Symp. Quant. Biol. 65: 475-482. [DOI] [PubMed] [Google Scholar]