Abstract
Loss of OVCA1/DPH2L1 correlates with ovarian and breast cancer. To study its in vivo role, we generated Ovca1 mutant alleles in mice. Ovca1 heterozygotes spontaneously develop cancer. Ovca1 mutant mice die during embryonic development and at birth with developmental delay and defects in multiple organ systems. Cell proliferation defects were observed in Ovca1 mutant mouse embryonic fibroblasts (MEFs). p53 deficiency can rescue these Ovca1 mutant MEF proliferation defects and partially rescue Ovca1 mutant embryonic phenotypes. Furthermore, Ovca1; p53 double heterozygotes developed tumors quicker than p53 heterozygotes and with an increased carcinoma incidence. Multiple tumor burden in Ovca1 heterozygotes that were also p53 deficient was significantly higher than in p53 homozygous mutants. These in vivo findings demonstrate that Ovca1 is a tumor suppressor that can modify p53-induced tumorigenesis and suggest that it acts as a positive regulator for cell cycle progression. The close linkage of OVCA1 and p53 on human Chromosome 17 suggests that coordinated loss may be an important mechanism for the evolution of ovarian, breast, and other tumor phenotypes.
Keywords: Tumor suppressor, ovarian cancer, DPH2L1, Ovca2, HIC1, Miller-Dieker syndrome
Ovarian cancer is a leading cause of cancer-related death in gynecological malignancy. Chromosome 17 exhibits the most common abnormalities of this disease. A subregion defined by the D17S28 and D17S5 loci on Chromosome 17p13.3 is deleted in 80% of ovarian tumors, indicating that a putative tumor suppressor(s) is located in this region that when lost results in ovarian cancer (Phillips et al. 1993, 1996a). In several studies, this deletion region was not accompanied by loss of heterozygosity (LOH) at the linked p53 locus located at 17p13.1, suggesting the presence of an additional tumor suppressor gene(s) located in 17p13.3 (Saxena et al. 1992; Phillips et al. 1993, 1996a; Cornelis et al. 1994; Merlo et al. 1994; Schultz et al. 1996; Chattopadhyay et al. 1997; Liscia et al. 1999; Hoff et al. 2001). Interestingly, allelic loss at D17S28 and D17S5 also occurs in many other types of cancer, including breast, lung, and brain malignancies (Saxena et al. 1992; Merlo et al. 1994; Konishi et al. 1998; Phelan et al. 1998; Liscia et al. 1999; Hoff et al. 2001).
Within this deletion region, two candidate genes, OVCA1/DPH2L1 and HIC1, have been studied. The focus of this report is OVCA1/DPH2L1. OVCA1/DPH2L1 is a candidate tumor suppressor gene for ovarian cancer identified on the basis of mapping allelic loss, positional cloning, and exon trapping methods (Phillips et al. 1996b; Schultz et al. 1996). Forced overexpression of OVCA1 caused suppression of colony formation of ovarian cancer cell lines (Bruening et al. 1999). An observed reduction in cell proliferation rate was due to an increase in the cell population at G1 of the cell cycle (Bruening et al. 1999), suggesting that OVCA1 is involved in cell cycle progression.
We previously cloned and characterized the mouse ortholog of human OVCA1 (Chen and Behringer 2001). The mouse Ovca1 gene is expressed throughout embryogenesis and in most adult mouse tissues. We also found that another protein, OVCA2, is encoded within the Ovca1 locus as it is in humans (Schultz et al. 1996; Chen and Behringer 2001). Ovca2 is encoded by a unique exon and a second exon that encodes the 3′-untranslated region of Ovca1. Thus, Ovca1 and Ovca2 do not share any protein-coding information (Fig. 1A). The deduced protein sequences of both mouse and human OVCA1 share a low level of identity (∼25%) with the yeast DPH2 (diphthamide biosynthesis protein-2) gene product (Mattheakis et al. 1993). It is unlikely that OVCA1 is the mammalian ortholog of yeast DPH2 because human DPH2L2 has higher similarity to yeast DPH2 than does OVCA1 (Schultz et al. 1998). Although OVCA1 contains the conserved DPH2 domain (NCBI, conserved domain database, pfam01866) and has similarity among different species (human, mouse, fly, worm, yeast), the in vivo role of OVCA1 is still unknown.
Figure 1.
Generation of Ovca1-2- and Ovca1-specific knockout mice. (A) Gene targeting strategy to generate Ovca1 mutant alleles. The Ovca1-Ovca2 locus is shown at the top. (Boxes) Exons; (light shaded boxes) Ovca1 coding region; (dark shaded boxes) Ovca2 coding region; (open boxes) untranslated sequences. (P1 to P5) Primers used for RT-PCR analysis and genotyping. The targeting vectors and resulting Ovca1-specific and Ovca1-2 mutant alleles with the neo expression cassette removed are shown below. (neo-flox) loxP-flanked neomycin resistant cassette; (tk) HSV thymidine kinase expression cassette; (3X stop) XbaI linker containing multiple stop codons; (H) HindIII; (N) NotI; (S) StuI; (X) XbaI; (Xh) XhoI. (B) RT-PCR analysis of Ovca1 expression in Ovca1-2+/+, Ovca1-2+/-, and Ovca1-2-/- MEFs. The P3 and P4 primers were used to amplify Ovca1 transcripts spanning exons 1 to 13. P4 and P5 were used to amplify Ovca1 transcripts spanning exons 1 to 6. (RT +) With reverse transcriptase; (RT -) without reverse transcriptase. (C) RT-PCR analysis of Ovca1 and Ovca2 expression in Ovca1+/+, Ovca1+/-, and Ovca1-/- MEFs. The P3 and P4 primers were used to detect Ovca1 expression. The primers P2 and P3 were used to amplify Ovca2 transcripts. (D) Western blot showing OVCA1 protein in multiple organs (brain, lung, liver, and kidney) of E18.5 Ovca1+/+ but not Ovca1-/- mice. Arrow, 50-kD OVCA1 protein.
To directly examine the role of OVCA1 in regulating cell proliferation and tumor suppression, we generated Ovca1 mutant mice. Our results support the idea that Ovca1 regulates cell cycle progression and tumor formation. In addition, we found that Ovca1 can modify p53-induced tumor formation. Ovca1 and p53 are linked on the same chromosome in both mouse (Chromosome 11) and human, suggesting that alterations of this chromosome during tumor formation may be a mechanism for the evolution of tumor phenotypes. This may be particularly significant because there is a high association of p53 mutations with human ovarian cancer (Bast and Mills 2000; Schuijer and Berns 2003).
Results
Generation of Ovca1-2- and Ovca1-specific knockout mice
The initial targeting vector simultaneously disrupted both Ovca1 and Ovca2 (Fig. 1A). Upon homologous recombination, Ovca1 exons 7 to 12 and a portion of exon 13 will be deleted. This targeted mutation also results in the deletion of the first exon of Ovca2, resulting in an allele designated Ovca1-2. Targeted ES cell clones were identified by Southern analysis and mice carrying the Ovca1-2 allele were produced from chimeras using standard procedures (Supplemental Fig. 1).
Ovca1 expression was analyzed in Ovca1-2 mutant mice by RT-PCR using RNA from mouse embryonic fibroblasts (MEFs) derived from embryonic day 13.5 (E13.5) Ovca1-2+/+, Ovca1-2+/-, and Ovca1-2-/- embryos. Primers P4 and P3 located in Ovca1 exons 1 and 13, respectively, amplified a 1.4-kb product from Ovca1-2+/+ and Ovca1-2+/- MEFs, but not from Ovca1-2-/- MEFs (Fig. 1B). We also examined potential transcripts that could be generated between the remaining Ovca1 exons 1 and 6. Primers P4 (exon 1) and P5 located in exon 6 amplified a 617-bp product from Ovca1-2-/- MEFs, indicating that the Ovca1-2 allele can generate an Ovca1 transcript containing exon 1 to 6 sequences (Fig. 1B).
In parallel to these studies, we also generated an Ovca1-specific knockout allele. Because the Ovca2 gene is embedded within the Ovca1 locus, we generated an Ovca1-specific targeted allele by insertion of an XbaI oligonucleotide linker containing multiple stop codons in all three reading frames into the third exon of Ovca1 (Fig. 1A). This should disrupt the translation of Ovca1, but not interfere with Ovca1 transcription or Ovca2 transcription and translation. Mice carrying the Ovca1-specific allele, designated Ovca1, were generated as above (Supplemental Fig. 1). RT-PCR analysis of Ovca1+/+, Ovca1+/-, and Ovca1-/- MEFs demonstrated that Ovca1 and Ovca2 transcripts were expressed (Fig. 1C). However, Western blot analysis using a polyclonal antibody to OVCA1 demonstrated an absence of OVCA1 protein in several organs (brain, lung, liver, and kidney) from E18.5 Ovca1-/- embryos in comparison to wild-type controls (Fig. 1D). This indicates that the Ovca1 allele is a null allele for Ovca1 but likely still expresses OVCA2.
Phenotype of Ovca1 mutant mice
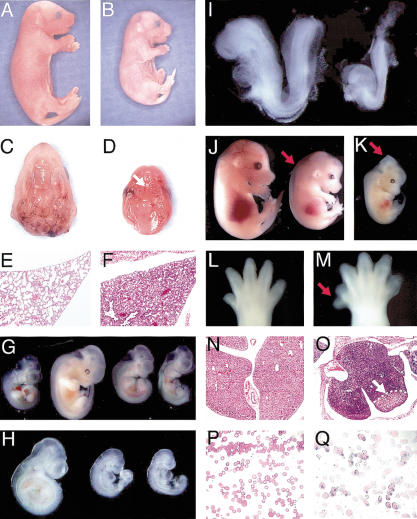
Ovca1-2 or Ovca1 homozygous mutants, on a B6/129 mixed genetic background, died soon after birth with a smaller body size (Fig. 2A,B; Supplemental Table 1). The average body weight of Ovca1-2 -/- or Ovca1-/- mice at E18.5 was 0.66 ± 0.08 g (n = 7), that was ∼50% the body weight of wild-type (1.28 ± 0.10 g; n = 9) or heterozygous (1.27 ± 0.12 g; n = 9) littermates. The Ovca1-2-/- or Ovca1-/- mice had no overt gross structural defects. However, all of the newborn mutants had cleft palates (Fig. 2C,D). Caesarian section revealed that the mutants had respiratory distress. The mutant lungs did not float in water, indicating that the mutants did not breathe prior to death. Histological analysis of the lungs of E18.5 Ovca1-2-/- mutants showed an immature lung phenotype (Fig. 2E,F). There was hypercellularity and thickened mesenchyme in the mutants in comparison to wild-type controls.
Figure 2.
Inactivation of Ovca1 or Ovca1-2 causes developmental abnormalities. Gross morphology of E18.5 (A) wild-type and (B) Ovca1-2-/- pups. Ventral view of palate of E18.5 (C) wild-type and (D) Ovca1-2-/- mice, showing cleft palate in the mutant (arrow). (E,F) H&E-stained histological sections of lung from E18.5 wild-type (E) and Ovca1-2-/- (F) pups. (G) E10.5-E11.5 embryos. E10.5 wild-type embryo (left); E11.5 wild-type embryo (second from left); two E11.5 Ovca1-2-/- embryos (right). (H) E9.5 embryos. Wild-type embryo (left); two Ovca1-2-/- embryos (right). (I) E8.5 embryos. Wild-type embryo (left); Ovca1-2-/- embryo (right). (J) E14.5 embryos. Wild-type embryo (left); Ovca1-/- embryo (right) with edema (arrow). (K) E13.5 Ovca1-2-/- embryo with midbrain exencephaly (arrow). (L) Hindlimb of an E14.5 wild-type embryo. (M) Preaxial polydactyly (arrow) in right hindlimb of E14.5 Ovca1-2-/- embryo. (N,O) H&E-stained histological sections of E16.5 fetal liver from wild-type (N) and Ovca1-2-/- (O) embryos. The mutant liver is highly basophilic and has a focal area of necrosis (arrow). (P,Q) Representative Wright-Giemsa-stained blood smears of E16.5 wild-type (P) and Ovca1-2-/- or Ovca1-/- (Q) embryos. Note that many of the mutant erythrocytes are nucleated.
In addition to the lethality at birth, we found that ∼30%-50% of Ovca1-2-/- and Ovca1-/- embryos died after E11.5 to E13.5 on a B6/129 mixed genetic background (Supplemental Table 1). However, on a 129/SvEv inbred genetic background, the Ovca1-2-/- and Ovca1-/- mutants died before E13.5. These findings suggest that there are genetic modifiers that differ between C57Bl/6 (B6) and 129 that influence mutant viability during the second half of gestation.
The Ovca1-2-/- or Ovca1-/- embryos appeared to be delayed in development by 12 to 24 h based on their body size and developmental characteristics. As shown in Figure 2G, the Ovca1-2-/- embryos at E11.5 had similar body size to E10.5 wild-type embryos. Eye pigmentation at this stage was also found to be delayed in the Ovca1-2-/- embryos. Occasionally, an absence of the eye or abnormal eye pigmentation was observed (data not shown). At E9.5, most of the Ovca1-2-/- embryos had turned but were smaller (Fig. 2H). At E8.5, Ovca1-2-/- embryos had zero to two somites compared with wild-type littermates, which had six to seven somites (Fig. 2I).
In addition to the global developmental delay, abnormalities in various organs were observed. Edema was observed in both the Ovca1-/- (Fig. 2J) and Ovca1-2-/- (data not shown) embryos at E13.5 and E14.5. Dilated vessels and myxoid stroma were observed histologically in the mutants (data not shown). Neural tube formation in the midbrain region was also occasionally found to be abnormal at this stage (Fig. 2K). In addition, preaxial polydactyly of the right hindlimb was found in the majority (60%∼70%) of Ovca1-/- and Ovca1-2-/- embryos (Fig. 2L,M). Focal liver degeneration was observed in ∼30%-50% of the Ovca1-2-/- embryos at E16.5 (Fig. 2N,O). Numerous hepatoblasts containing basophilic/elongated nuclei and a high nuclear/cytoplasmic ratio were present in Ovca1-2-/- livers. Interestingly, nucleated erythrocytes were also observed at E16.5 in both Ovca1-/- and Ovca1-2-/- fetuses, whereas wild-type controls had enucleated erythrocytes (Fig. 2P,Q). The nucleated erythrocytes in the Ovca1-/- and Ovca1-2-/- fetuses may be due to abnormal hepatic development because the liver is the major hematopoietic organ at this stage of mouse embryogenesis.
Behavior of Ovca1 mutant MEFs
The primary defect of Ovca1-2 or Ovca1 mutant embryos is a generalized reduction in size compared with their control littermates. The smaller embryos could reflect a decrease in cell proliferation and/or increase in apoptosis. Therefore, we established primary mouse embryonic fibroblast (MEF) cultures to investigate the cellular growth behavior of wild-type and heterozygous and homozygous mutants for the Ovca1-2 and Ovca1 alleles at E13.5. In addition, we bred the Ovca1-2+/- mice with Ovca1+/- mice to generate Ovca1/Ovca1-2 embryos that would be equivalent to an Ovca1-/- Ovca2+/- genotype. Interestingly, the Ovca1/Ovca1-2 embryos exhibited the same phenotypes (edema, smaller body size, developmental delay, and lethality) that we observed in the Ovca1-2-/- or Ovca1-/- embryos (data not shown).
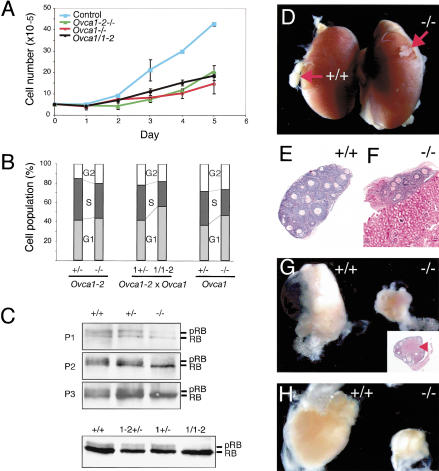
Ovca1-2-/-, Ovca1-/-, or Ovca1/Ovca1-2 MEFs grew poorly in comparison to controls (Fig. 3A). Cell cycle analysis showed a reduction in the S-phase population (20%-30%) in Ovca1-2-/-, Ovca1-/-, or Ovca1/Ovca1-2 MEFs in comparison to heterozygous controls (Fig. 3B). No obvious sub-G1 cell population was observed in all three genotypes, indicating that cell death was not a major contributing factor to the mutant MEF growth defects (data not shown). We also examined cell size using forward side scatter to examine gated G1 or G2 cells by flow cytometry but found no differences (data not shown). In addition, p21, cyclin D, cyclin E, and Cdk4 protein levels did not differ significantly in Ovca1 +/+, +/-, and -/- MEFs, indicating that Ovca1 loss does not activate p53 (Supplemental Fig. 2). We further examined the phosphorylation status of retinoblastoma (RB) in MEFs because of its known role in regulating cell cycle progression (Classon and Harlow 2002). Phosphorylation of RB in late G1 promotes entrance into the Sphase by releasing the E2F family transcription factors for activation of downstream cell cycle regulators. In contrast, dephosphorylated RB present in G0/G1 binds E2F family members, repressing their transcriptional activation. As shown in Figure 3C, phosphorylated RB (pRB) was reduced in Ovca1-/- MEFs in comparison to Ovca1+/+ or Ovca1+/- controls at early passages (P1, P2, and P3). The reduced phosphorylated RB status was also observed in Ovca1/Ovca1-2 MEFs (Fig. 3C). The phosphorylation status of RB is consistent with the proliferation defect and reduced S-phase cell population in Ovca1-2-/- and Ovca1-/- MEFs.
Figure 3.
Ovca1 mutant MEF cell cycle analysis and ovary grafts. (A) Growth dynamics of Ovca1 mutant MEFs. Genotypes are indicated in upper left corner: (Control) wild-type or Ovca1+/-. (B) Histogram showing the percentage of the MEF population in G1 (G0/G1), S, and G2 (G2/M). (C) Western blot showing RB and its phosphorylation (pRB) status in Ovca1 mutant MEFs. Analysis of passages 1 to 3 (P1, P2, P3) was derived from Ovca1+/+ (+/+), Ovca1+/- (+/-), and Ovca1-/- (-/-) embryos. Analysis of MEFs at passage 4 (P4) was derived from embryos obtained from Ovca1+/- × Ovca1-2+/- matings. (D) Gross morphology of Ovca1-2+/+ (+/+) and Ovca1-2-/- (-/-) ovaries (arrows) grown under the kidney capsule for 1 mo. (E,F) Histology of wild-type and Ovca1-2-/- ovary grafts shown in D. (G) Gross morphology of wild-type and Ovca1-2-/- ovaries grown in the ovarian bursa for 3 mo. (Inset) Histology of the 3-mo-old Ovca1-2-/- ovary graft. Corpus luteum (arrowhead) indicates ovulation. (H) Gross morphology of wild-type and Ovca1-2-/- ovaries grown in the ovarian bursa for 9 mo.
Transplantation of Ovca1 mutant ovaries
To investigate the role of Ovca1 in ovarian development and disease, we surgically transferred the ovaries from Ovca1-2-/- or Ovca1-/- knockout fetuses at E18.5 to either the kidney capsule or ovarian bursa of histocompatible female mice. The ovary grafts were examined grossly and histologically after different periods of time. As shown in Figure 3D, grafts of Ovca1-2-/- ovaries grown for 1 mo under the kidney capsule exhibited smaller size in comparison to wild-type ovary grafts. Normal follicle formation was observed histologically in both wild-type and Ovca1-2-/- ovary grafts (Fig. 3E,F). Ovary grafts grown for 3 and 9 mo in the ovarian bursa were isolated (Fig. 3G,H). Consistently, the Ovca1-2-/- ovaries were smaller than the control grafts at both time points examined. Corpus lutea were observed in wild-type (data not shown) and Ovca1-2-/- ovary grafts (Fig. 3G, inset), indicating evidence of ovulation. No ovarian carcinomas were found in the transplanted Ovca1 mutant ovaries during the period of analysis.
Genetic study of Ovca1 and p53
The cell proliferation defect (poor growth, reduced S-phase cell population and reduced RB phosphorylation status) in the Ovca1 mutant MEFs indicated that Ovca1 regulates cell cycle progression. Therefore, we examined whether elimination of a cell cycle checkpoint gene, such as p53, in an Ovca1-deficient background could override the Ovca1-2-/- and Ovca1-/- MEF proliferation defect phenotype. Ovca1 and p53 are closely linked, residing within 8 cM of each other on mouse Chromosome 11. Therefore, to obtain mice homozygous for both Ovca1-2 and p53, we first generated mice heterozygous for both mutations on the same chromosome referred to as cis Ovca1-2+/- p53+/- by a chromosomal recombination event. In addition, double-heterozygous mutant mice for Ovca1-2+/- and p53+/- in trans were generated, referred to as trans Ovca1-2+/- p53+/-.
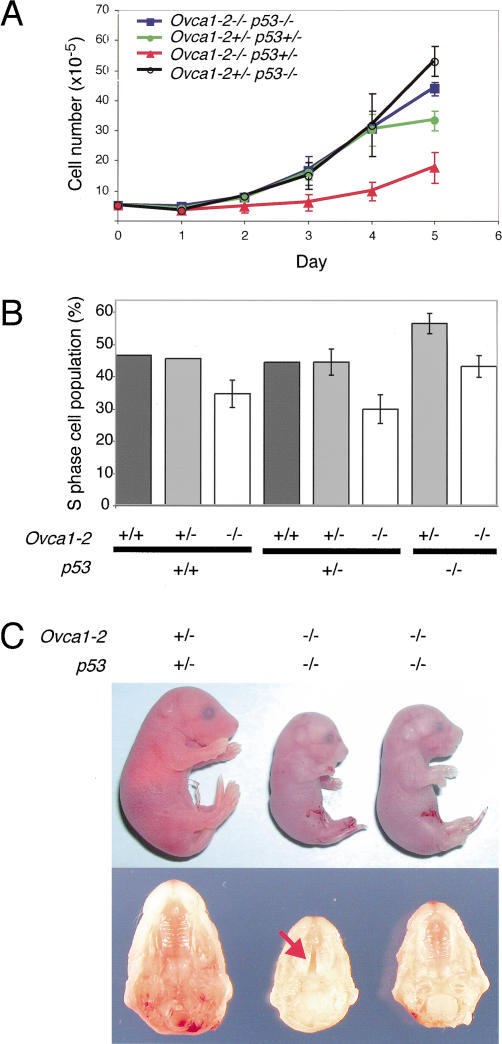
We established MEFs derived from various genotypes (Ovca1-2-/- p53-/-, Ovca1-2+/- p53+/-, Ovca1-2-/- p53+/-, Ovca1-2+/- p53-/-). As shown in Figure 4A, the Ovca1-2-/- p53+/- MEFs grew poorly like Ovca1-2-/- MEFs (Figs. 3A and 4A). Interestingly, the Ovca1-2-/- p53-/- MEFs had growth curves similar to the heterozygous control (cis Ovca1-2+/- p53+/-) MEFs, indicating that loss of p53 rescued the Ovca1-2-/- MEF proliferation defect. Cell cycle analysis revealed that the S-phase population (20%-30%) of Ovca1-2-/- MEFs was reduced in either p53+/+ or p53+/-, but was not reduced by p53-/- (Fig. 4B). Our findings suggest that the Ovca1-2-/- MEF proliferation defect can be rescued in a p53-deficient background.
Figure 4.
Effect of p53-deficiency on Ovca1 mutant MEF growth and embryo development. (A) Growth dynamics of Ovca1 mutant MEFs. Genotypes are indicated in the upper left corner. (B) Percentage of S-phase cell population derived from MEFs with various Ovca1 and p53 genotypes. Two to three independent MEF lines were analyzed (Ovca1-2+/+ p53+/+, n = 2; Ovca1-2+/- p53+/+, n = 2; Ovca1-2-/- p53+/+, n = 3; Ovca1-2+/+ p53+/-, n = 2; Ovca1-2+/- p53+/-, n = 3; Ovca1-2-/- p53+/-, n = 3; Ovca1-2+/- p53-/-, n = 3; Ovca1-2-/- p53-/-, n = 3). (C) Gross morphology of E18.5 cis Ovca1-2+/- p53+/- pup (left) and Ovca1-2-/- p53-/- pups (two pups on the right; top); normal palate fusion was found in 50% of Ovca1-2-/- p53-/- pups (n = 4; representative shown at right) in comparison to cleft palate (arrow) found in 50% of the same genotype (n = 4, representative shown in the middle). Normal palate morphology of representative cis Ovca1-2+/- p53+/- littermate (n > 4) is shown at the left as a control.
We also asked whether loss of p53 could rescue the lethal phenotype of Ovca1-2-/- mice. At E18.5, we found that Ovca1-2 p53 double-homozygous mutant mice still died soon after birth, but some of them showed different gross phenotypes compared with Ovca1-2-/- mutants. Approximately 50% of the Ovca1-2-/- p53-/- fetuses (n = 8) were ∼10%-20% larger than Ovca1-/- or Ovca1-2-/- fetuses (Figs. 2A and 4C). The Ovca1-2-/- p53-/- fetuses were in two weight categories, either 0.7 g (n = 4) or 0.9 g (n = 4) in comparison to their cis Ovca1-2+/- p53+/- littermates at 1.3 g (n > 4). Notably, palate shelf fusion was found in the Ovca1-2-/- p53-/- fetuses that were of larger body size (n = 4; Fig. 4C). Based on the phenotypes of Ovca1-2-/- p53-/- at E18.5, we conclude that the perinatal lethality of Ovca1-2-/- mice was not rescued by loss of p53. However, some Ovca1-2-/- developmental abnormalities (smaller body size and cleft palate) were partially rescued in a p53-deficient background. This partial rescue was never observed in Ovca1-2-/- mice (n = 16) on the same B6/129 mixed genetic background. Taken together, these data suggest a genetic interaction between Ovca1 and p53.
Spontaneous tumorigenesis in Ovca1-2+/- and Ovca1+/- mice in p53+/+, p53+/-, and p53-/- backgrounds
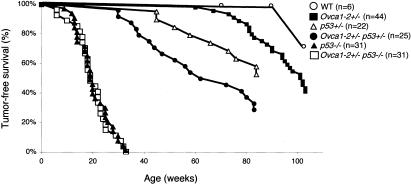
We monitored the occurrence of spontaneous tumor formation in male and female Ovca1-2+/- mice on p53+/+, p53+/-, and p53-/- genetic backgrounds for ∼2 yr (104 wk). The tumor-free survival curves are shown in Figure 5. In a cohort of Ovca1-2+/- mice (n = 44), we found 25 (56.8%) animals, both males and females, which developed various tumors by 2 yr of age.
Figure 5.
Tumor incidence in Ovca1 mutant mice.
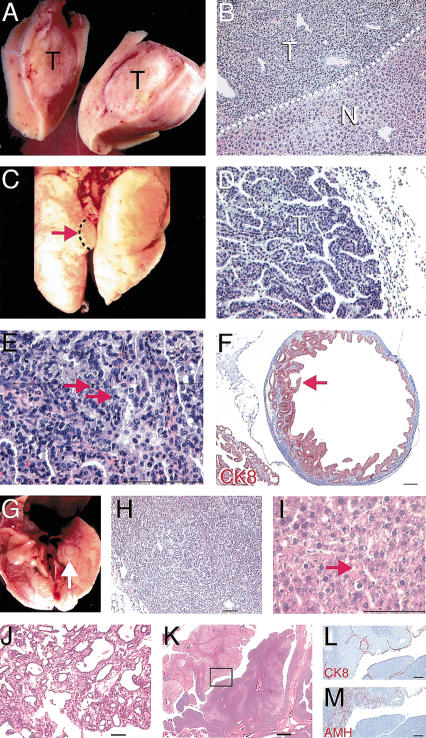
Adenomas/adenocarcinomas (15 of 25 animals; 60%) were the predominant neoplasm observed in Ovca1-2+/- mice followed by lymphomas (12 of 25; 48%) and soft tissue sarcomas (3 of 25; 12%). The mean latency of Ovca1-2+/- tumor occurrence was ∼92 wk. Five mice were found to have multiple tumor burden (Table 1). Two mice developed hepatocellular adenoma/adenocarcinomas (Fig. 6A,B) and lung adenocarcinomas (Fig. 6C-E) at the age of 97 wk. Another mouse developed a lung adenoma and a malignant stromal tumor showing liver, mesenteric, spleen, and lymph node involvement at the age of 101 wk (data not shown). One female mouse developed ovarian cystadenoma (Fig. 6F) and lung adenoma (data not shown) at 80 wk of age.
Table 1.
Spontaneous tumorigenesis of Ovca 1-2+/− mice in p53+/+, p53+/−, and p53−/− backgrounds in comparison to p53+/− and p53−/− mice
| Ovca 1-2+/− | Ovca 1-2+/−p53+/− | Ovca 1-2+/−p53−/− | p53+/− | p53−/− | |
|---|---|---|---|---|---|
| Total number | 44 | 25 | 31 | 22 | 31 |
| Male | 29 | 6 | 15 | 6 | 21 |
| Female | 15 | 19 | 16 | 16 | 10 |
| Tumor incidence (%) | 25 (56.8) | 18 (72) | 31 (100) | 11 (50.0) | 31 (100) |
| Tumor spectrum (%) | |||||
| Lymphoma | 12 (40) | 9 (50.0) | 23 (62.2) | 4 (36.4) | 25 (80.6) |
| Osteosarcoma | 0 | 2 (11.1) | 0 | 6 (54.5) | 0 |
| Soft tissue sarcomaa | 3 (10) | 2 (11.1) | 11 (33.3) | 1 (9.1) | 4 (12.9) |
| Adenoma/carcinomab | 15 (50) | 5 (27.8) | 1 (3.03) | 0 | 2 (6.5) |
| Teratoma | 0 | 0 | 1 (3.03) | 0 | 1 (3.2) |
| Brain tumor | 0 | 0 | 1 (3.03) | 0 | 0 |
| Total tumor number | 30c | 18 | 37d | 11 | 32e |
| Mean latency (weeks) | 92 | 52 | 21 | 70 | 20 |
The tumor-free survival of wild-type mice included two females and four males that were age-matched B6129 mixed genetic background sampled from our colony (n > 500).
Soft tissue sarcomas include fibrosarcoma, malignant histiocytoma, and hemangioma/hemangiosarcoma.
All types of epithelium-derived tumors are included. One pulmonary adenoma with increased mitotic figures was found in an Ovca 1-2+/− p53−/− mouse. Two p53−/− mice were found with adenomatous polyp (adenoma) in the intestine and adenoma in the lung, respectively. Two mammary gland adenosquamous carcinomas, two lung adenocarcinomas, and one keratinized squamous tumor (keratoacanthoma) were identified in Ovca 1-2+/− p53+/− animals (5/18). Adenoma or carcinoma in liver (n = 4) and lung (n = 10) were the major epithelial tumors of Ovca 1-2+/− mice. One ovarian cystadenoma was found in an Ovca 1-2+/− female.
Five mice were found with multiple tumor burden. Two mice were found with hepatoma and lung adenoma. One mouse had a lung adenoma nd a malignant stromal tumor. One mouse developed lung adenoma and lymphoma. One mouse had a lung adenoma and ovarian cystadenoma.
Six mice were found with multiple tumor burden. Five mice were found with lymphoma and soft tissue sarcomas. One mouse had three types of neoplasms, including lymphoma, hemangioma, and adenoma.
One p53−/− mouse had lymphoma and lung adenoma.
Figure 6.
Spontaneous tumor formation in Ovca1-2+/- and Ovca1+/- mice. (A) Tumor mass (T) found in the liver of an Ovca1-2+/- mouse. (B) H&E-stained section of tumor shown in A. Hepatocellular adenoma (T) showing basophilic, focal eosinophilic, and clear cell appearance with compression of the normal hepatic parenchyma (N). Dashed line indicates the boundary between tumor and normal hepatic parenchyma. Scale bar, 100 μm. (C) Tumor mass (arrow) found in the lung of an Ovca1-2+/- mouse. (D) H&E-stained section of bronchiolo-alveolar carcinoma (T) with mixed papillary and solid patterns shown in C. Scale bar, 100 μm. (E) Higher magnification of D, showing mitotic figures (arrowheads). (F) Cystadenoma found in the ovary of an Ovca1-2+/- mouse at 80 wk. Papillary-like epithelium immunostained for cytokeratin 8 (CK8, red) projects into the cyst, which is lined with tall columnar cells (arrow). Scale bar, 400 μm. (G) Tumor mass (arrow) found in a ventral lung lobe of an Ovca1+/- mouse at 78 wk of age. (H) H&E-stained section of tumor shown in G, showing a predominantly glandular arrangement of the tumor cells. (I) H&E-stained section of hepatocellular ademoma of an Ovca1+/- mouse at 66 wk of age. There is an abnormal presence of many hyaline bodies (arrow). Scale bar, 100 μm. (J) Mammary gland tumor found in an Ovca1+/- female at 70 wk of age. Adenocarcinoma type B was indicated because of collagenousstroma and glandular components consisting of focal cystic, hemorrhagic, and papillary growth patterns. Scale bar, 100 μm. (K) Tubular adenoma found in the ovary of an Ovca1+/- female at 82 wk of age. (Box) Region shown in L and M. Scale bar, 400 μm. (L) The neoplastic cells were lined by surface mesothelial cells which expressed CK8 (red). Scale bar, 100 μm. (M) AMH (red) was detected focally in the tumor. Scale bar, 100 μm.
At this time, we have also found that 17 of 40 (42.5%) mice, heterozygous for the Ovca1-specific mutation, had tumors from 53 to 85 wk of age. Four of these 17 Ovca1+/- mice had multiple tumor burden. Consistently, we identified lymphoma (data not shown), lung adenoma (Fig. 6G,H), and hepatocellular adenoma (Fig. 6I), similar to the Ovca1-2+/- tumor spectrum. Notably, one Ovca1+/- breeding female mouse developed a mammary gland tumor (Fig. 6J) at 70 wk of age. In addition, one ovarian tubular adenoma (∼1 cm in size) was found in one Ovca1+/- mouse at 82 wk of age. The ovarian tumor cells were lined by surface mesothelial cells, which were identified by cytokeratin 8 (CK8) immunostaining (Fig. 6K). Focal neoplastic cells expressed anti-Müllerian hormone (AMH; Fig. 6K), indicating a granulosa cell origin. Thus, both Ovca1-2+/- and the Ovca1+/- mice spontaneously form tumors, supporting the idea that Ovca1 is a tumor suppressor gene.
Ovca1-2+/- p53+/- (n = 25) mice developed tumors quicker than p53+/- (n = 22) or Ovca1-2+/- mice (n = 44). The mean latency for tumor development in Ovca1-2+/- p53+/- mice was 52 wk of age in comparison to p53+/- mice at the age of 70 wk (Table 1). In addition, the tumor incidence of Ovca1-2+/- p53+/- mice was 72% (18/25), which is higher than the tumor incidence of p53+/- mice (50%; 11/22) by 90 wk of age (Fig. 5; Table 1). Interestingly, mammary gland tumors (n = 2; Fig. 7A), pulmonary tumors (n = 2; Fig. 7B), and a skin keratoacanthoma were found in Ovca1-2+/- p53+/- mice but not in p53+/- mice. These results suggest that loss of one copy of Ovca1-2 in a p53+/- background significantly increased carcinoma incidence (p = 0.047) in comparison to p53+/- mice.
Figure 7.
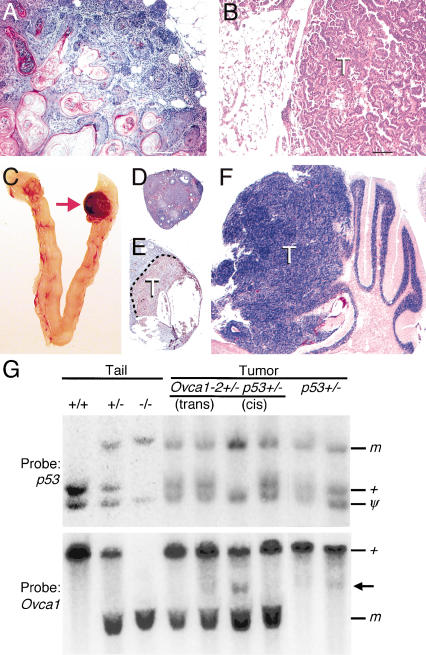
Interaction between Ovca1 and p53 mutations for tumor formation. (A) H&E-stained section of a representative mammary gland adenosquamous carcinoma was found in two Ovca1-2+/- p53+/- mice at 38 and 78 wk of age. There are keratinized well-differentiated squamous cells surrounded by the glandular elements of the mammary gland. Scale bar, 100 μm. (B) H&E-stained section shows a bronchiolo-alveolar carcinoma with a papillary growing pattern in an Ovca1-2+/- p53+/- male mouse at 80 wk of age. Scale bar, 100 μm. (C) Reproductive tract from an Ovca1-2+/- p53-/- female mouse at 33 wk of age. One ovary (arrow) shown at the right is enlarged and hemorrhagic. H&E-stained sections of ovaries shown in C with normal histology (D) and a hemangiosarcoma immunostained with CD34 (brown; E). (F) H&E-stained section of a brain tumor (T) showing a transition from the cerebellar granular layer to a neoplastic appearance reminiscent of human medulloblastoma, replacing the cerebellar folia. (G) Southern blot analysis of p53 and Ovca1 in tumors from Ovca1-2+/- p53+/- and p53+/- mice. Tail DNA from wild-type (+/+), cis Ovca1-2+/- p53+/- (+/-), and Ovca1-2-/- p53-/- (-/-) mice. Genomic DNA from individual tumors, including trans Ovca1-2+/- p53+/-, cis Ovca1-2+/- p53+/-, and p53+/-. (m) Mutant allele; (+) wild-type allele; (ψ) p53 pseudogene. One cis Ovca1-2+/- p53+/- tumor shows LOH of the p53 locus and an additional minor band (arrow), suggesting a rearrangement of the Ovca1 locus in a subset of cells in the tumor.
The mean latency of tumor incidence was about the same (20 wk) in Ovca1-2+/- p53-/- (n = 31) and p53-/- mice (n = 31; Fig. 5; Table 1). However, multiple tumor types in single animals were found to be significantly higher in Ovca1-2+/- p53-/- mice than in p53-/- mice (6 of 31 in Ovca1-2+/- p53-/- mice vs. 1 of 31 mice in p53-/- mice; p = 0.045). For example, we found a female Ovca1-2+/- p53-/- mouse with thymic lymphoma and ovarian hemangiosarcoma (Fig. 7C-E) at 33 wk of age. Another female Ovca1-2+/- p53-/- mouse developed lymphoma, hemangioma, and pulmonary carcinoma at 32 wk of age. We only found one p53-/- male mouse that developed lymphoma and pulmonary adenoma at 30 wk of age. The tumor spectrum of Ovca1-2+/- p53-/- animals remained similar to p53-/- mice. However, there were increasing soft tissue sarcoma (33.3% vs. 12.9%) and decreasing lymphoma incidences (62.2% vs. 80.6%) in Ovca1-2+/- p53-/- tumors in comparison to p53-/- tumors. In addition, one medulloblastoma was found (Fig. 7F) in an Ovca1-2+/- p53-/- mutant. These findings indicate that loss of one copy of Ovca1 can modify tumor spectrum in a p53-deficient background.
To determine whether there is Ovca1 or p53 allelic loss in Ovca1-2+/- p53+/- or p53+/- tumors, we performed Southern blot analysis. The p53 wild-type allele was lost in one cis Ovca1-2+/- p53+/- tumor but not in two trans Ovca1-2+/- p53+/- or two p53+/- tumors that were examined (Fig. 7G). In the same tumor with p53 LOH, we found that the Ovca1 wild-type allele was present, but an additional distinct minor was detected, indicating a potential rearrangement of the Ovca1 locus in a subset of tumor cells (Fig. 7G). Interestingly, a similar pattern was also detected in one of the p53+/- tumors.
Discussion
Correlative human cancer genetic studies and forced overexpression in vitro assays suggested OVCA1 as a candidate tumor suppressor gene located on Chromosome 17 associated with ovarian and other cancers (Phillips et al. 1996b; Schultz et al. 1996). Here we show that loss of Ovca1 leads to an increased frequency of tumor formation in mice, demonstrating in vivo that Ovca1 is a tumor suppressor gene. We found that ∼20% of Ovca1-2+/- mice developed tumors, predominantly lymphomas, by 80 wk of age. By 2 yr, there was a >50% tumor incidence. It appears that the Ovca1+/- mice will have a similar frequency of tumor incidence. The data suggest that Ovca1 acts as a tumor suppressor. This is in contrast to the high correlation of OVCA1 deletion observed in human ovarian and other cancers (Phillips et al. 1996b; Schultz et al. 1996; Miller et al. 2003). Loss of one copy of Ovca1 can accelerate tumorigenesis and increase carcinoma incidence in a p53+/- background. In addition, Ovca1 heterozygosity can modify tumor spectrum and increase multiple tumor burden in a p53-/- background.
Although OVCA1 and p53 are strongly associated with human ovarian cancer (for review, see Bast and Mills 2000), ovarian epithelial tumors were not readily identified during the period of analysis in this study. However, we did identify one ovarian serous cystadenoma in an Ovca1-2+/- female at 80 wk of age and one tubular adenoma in an Ovca1+/- female at 82 wk of age. Interestingly, serous cystadenoma is a relatively common benign epithelial tumor of the ovary in humans (Zaloudek 2000). Perhaps Ovca1 plays a role in the development of benign ovarian tumors that in combination with other genetic alterations contributes to the transition from low- to high-grade ovarian tumors. We also identified one ovarian hemangiosarcoma in an Ovca1-2+/- p53-/- female at only 33 wk of age. In a separate study, we have found hemangiosarcoma in p53-/- ovaries transplanted into wild-type hosts but only after ∼1 yr (C.-M. Chen and R.R. Behringer, unpubl.). This suggests that Ovca1 heterozygosity may decrease the latency of p53-induced ovarian tumors in mice. Primary ovarian hemangiosarcoma in humans is rare (Zaloudek 2000). Thus, species-specific differences between human and mouse will likely dictate the utility of mouse models of human ovarian cancer.
BRCA1 and BRCA2 are associated with breast and ovarian cancer in humans (Gayther et al. 1995, 1997). However, Brca1 and Brca2 mutant mice die as early embryos, and heterozygotes do not develop breast or ovarian cancer (Gowen et al. 1996; Hakem et al. 1996; Liu et al. 1996; Sharan et al. 1997). It is interesting to note that mammary carcinoma is only observed in mice that conditionally express a hypomorphic Brca1 allele in mammary glands (Xu et al. 1999). In addition, an increase in mammary carcinoma incidence and decrease latency was observed in Brca1 p53 mutant mice, indicating that Brca1 and p53 mutations interact in mammary carcinoma formation (Cressman et al. 1999; Xu et al. 1999; Cao et al. 2003). Likewise, mammary epithelium-specific inactivation of Brca2 in mice did not result in mammary tumor formation. However, when combined with p53-deficiency, mammary gland tumors formed (Jonkers et al. 2001). Furthermore, p53+/- mice on a BALB/c genetic background develop mammary carcinoma, whereas this type is only 1% of tumors found when the same mutation is on a B6 or 129 genetic background (Kuperwasser et al. 2000; Blackburn et al. 2003). Thus, the development of ovarian cancer in Ovca1 mutant mice may require specific genetic manipulations.
Lung and mammary gland tumors were found in Ovca1-2+/- p53+/- animals, implying a tumor suppressor role for human OVCA1 and p53 loci in lung and breast cancer. Consistent with this observation is the fact that OVCA1 LOH is also one of the most prevalent chromosomal alterations in breast carcinoma (Miller et al. 2003). Multiple tumor burden in Ovca1-2+/- p53-/- animals was found to be significantly higher than in p53-/- animals. In addition, an increasing incidence of soft tissue sarcoma and one medulloblastoma was found in Ovca1-2+/- p53-/- mice, suggesting that tumor spectrum in a p53-deficient background can be modified by loss of one copy of Ovca1. A recent report correlated human papilloma virus infection and expression of the p53 inhibitor HPV E6 in ovarian cancer (Wu et al. 2003). Perhaps HPV infection and E6 expression causing p53 inhibition coupled with the loss of OVCA1 may be a contributing factor to ovarian carcinogenesis.
The mechanism that causes accelerated tumorigenesis in Ovca1-2+/- p53+/- mice versus p53+/- is unclear. However, it seems reasonable to suggest that the role of Ovca1 in regulating cell cycle progression may be a key factor for the accelerated tumorigenesis in Ovca1+/- p53+/- mice. Recently, OVCA1 has been identified as a BRCA1-inducible gene, using a forced BRCA1 overexpression assay in the MCF breast cancer cell line (Atalay et al. 2002). BRCA1 mutations are associated with the formation of both breast and ovarian cancer (Gayther et al. 1995). Thus, this raises the possibility of a link between BRCA1 and OVCA1 in breast and ovarian carcinogenesis. In addition, other genetic lesions may be required for the acceleration of tumorigenesis in Ovca1-2+/- p53+/- mice because we observed p53 LOH and perhaps Ovca1 rearrangement in only one of the four Ovca1-2+/- p53+/- tumors examined.
The Ovca1-2 mutant phenotypes were identical to those of the Ovca1-specific mutants, suggesting that these phenotypes are predominantly contributed by Ovca1 inactivation not by Ovca2. Forced overexpression of OVCA2 did not alter cell cycle progression or suppress ovarian cancer cell line focus formation (Bruening et al. 1999). Thus, the role of Ovca2 remains unclear because the Ovca2-null phenotype may be subtle and therefore difficult to distinguish from the Ovca1-specific mutant phenotypes of Ovca1-2-/- mice.
Our findings demonstrate that Ovca1 is required for embryonic and postnatal viability. The timing of death varied from midgestation to just after birth. Developmental delay, smaller body size, and abnormalities in multiple organ systems most likely contributed to the lethal phenotype. The developmental delay is consistent with the autonomous cell proliferation defect revealed in the MEF and ovary transplantation assays. The alterations in the proportion of Ovca1 mutant cells in the S phase of the cell cycle and reduction of phosphorylated RB protein indicate a role for Ovca1 in regulating cell cycle progression at the G1- to S-phase transition by down-regulating the activity of cyclin-dependent kinases (CDKs). p53 deficiency would reduce CDK inhibitor activity, shifting the RB to phosphorylated RB balance, thereby rescuing the Ovca1 proliferation defect.
Loss of p53 did not rescue the perinatal lethality, although in ∼50% of the cases there was rescue of the smaller body size and cleft palate of Ovca1-2-/- p53-/- mice. These findings suggest that there are genetic differences between B6 and 129 that can modify the Ovca1 mutant embryo phenotype. Homozygosity for loss of function mutations in other tumor suppressors associated with ovarian cancer, including Brca1 and Brca2, also leads to embryo lethality (Gowen et al. 1996; Hakem et al. 1996; Liu et al. 1996; Sharan et al. 1997). Thus, the cellular abnormalities caused by loss of these tumor suppressors compromises the complex developmental pathways that must be coordinated to produce a viable embryo.
Heterozygous deletion of 17p13.3 is also associated with Miller-Dieker syndrome (MDS; OMIM 247200) in humans (Chong et al. 1997; Hirotsune et al. 1997, 1998; Cardoso et al. 2003; Dobyns and Ledbetter 2003). Carter et al. (2000) reported that the phenotype of Hic1 (hypermethylated in cancer 1) knockout mice resembled most but not all of the clinical features of MDS, including craniofacial abnormalities, limb abnormalities, exencephaly, and omphalocele. We found that there are several similarities between Ovca1-/- and Hic1-/- embryos. Both types of mutant mice showed global developmental delay, reduction of body size, cleft palate phenotypes, and limb defects. This suggests that MDS may be a contiguous gene-deletion syndrome that involves both HIC1 and OVCA1.
In summary, our findings suggest that Ovca1 acts as a positive regulator for cell cycle progression, is a tumor suppressor, and is a tumor modifier in conjunction with p53. Human Chromosome 17 and mouse Chromosome 11 are laden with numerous tumor suppressor genes, including p53, OVCA1, BRCA1, and NF1. In addition to Ovca1+/- p53+/- and Brca1+/- p53+/- mice, double-heterozygous mice for Nf1 and p53 also showed an acceleration of tumorigenesis and alteration of tumor spectrum (Vogel et al. 1999). Our studies suggest that the linkage of these tumor suppressor genes and their potential coordinated loss by chromosomal rearrangements may be an important mechanism for the phenotypic changes observed in tumors as they evolve.
Materials and methods
Gene targeting and generation of Ovca1 and Ovca1-2 mice
The isolation of Ovca1 DNA from a 129/SvEv mouse genomic library was described previously (Chen and Behringer 2001). A loxP-flanked Pgk-Neo-bpA (Neo) expression cassette and a herpes simplex virus thymidine kinase (TK) expression cassette were used to generate the gene targeting vectors (Fig. 1A). For the Ovca1-specific targeting vector, an XbaI linker with stop codons (underlined) in all three reading frames (CTAGTCTAGACTAG; Stratagene) was ligated 5′ to the Neo cassette and introduced into the StuI site of the third exon of Ovca1 to block Ovca1 translation. Then 25 μg of linearized targeting vectors was electroporated into 1 × 107 PC3 ES cells (O'Gorman et al. 1997; Nagy et al. 2003). After 10 d of selection in G418 and FIAU, ES cell colonies were picked and genotyped by Southern analysis. The Ovca1-specific (1D4) and Ovca1-2 (3C7 and 3F10) targeted ES cell clones were subsequently injected into C57BL/6J (B6) blastocysts and transferred to pseudopregnant Swiss females. Male chimeric mice were bred with B6 females for germ-line transmission of the Ovca1 and Ovca1-2 targeted alleles. Upon germ-line transmission, the Neo cassettes were deleted by Cre recombinase expressed in the spermatids of the chimeras (O'Gorman et al. 1997). The phenotypic analysis of the Ovca1 and Ovca1-2 mutants, MEFs, and tumors was performed on a B6 × 129/SvEv mixed genetic background. The lethality of Ovca1 and Ovca1-2 mutants was also examined on a 129/SvEv inbred genetic background.
MEF isolation, culture, and cell cycle analysis
MEFs were isolated from E13.5 embryos as described (Nagy et al. 2003). Passage 0 (P0) was defined as the primary isolate. MEFs at passage 1 (P1) were plated at 5 × 105 cells in 10-cm2 gelatinized plates in complete medium (Dulbecco's modified Eagle's medium [DMEM; GIBCO; Invitrogen], 10% fetal bovine serum [FBS; Hyclone], 2 mM L-glutamine, 100 units/mL penicillin G, and 100 μg/mL streptomycin sulfate). Cells were counted daily for 5 d using a trypan blue dye exclusion method. MEF growth dynamics were measured from two to three independent isolates for each genotype.
For cell cycle analysis, MEFs (P1) were plated at 106 cells in 10-cm2 gelatinized plates and harvested the next day. The MEFs were washed twice with PBS containing 0.1% Tween-20 (PBST) and resuspended in ice-cold 70% ethanol. After ethanol fixation, the MEFs were washed twice with PBST, resuspended in a PI solution (50 μg/mL propidium iodide, 10 μg/mL RNase A, PBST) and incubated for 30 min at 37°C before analysis by flow cytometry. The percentages of cells in G0/G1, S, and G2/M phases were analyzed using the MultiCycle software program.
Ovca1 and Ovca2 expression analysis
For RT-PCR, 106 MEFs were plated at P1 and harvested the next day. Total RNA was extracted using TRIzol (Life Technology), according to the manufacturer's instructions. First-strand cDNA was generated by RT from total RNA using the Superscript II first-strand synthesis system (Life Technology). The Ovca1 cDNA spanning exons 1 to 13 was amplified by the forward primer P4 (5′-CCCCAAGCTTGTGGGCTCTTGGGT GATGGC-3′) and reverse primer P3 (5′-CACTGTGGACTCTT CCAGAGC-3′). The Ovca1 cDNA spanning from exon 1 to exon 6 was amplified by forward primer P4 and reverse primer P5 (5′-CGGGATCCGGGACACTGATGTGATAATCAGCTT TCAGC-3′). The Ovca2 partial cDNA was amplified by forward primer P2 (5′-CTGCGAGTGCTGTGTCTAGCCGG-3′) and reverse primer P3. The PCR conditions were 4 min at 94°C, 35 cycles for 1 min at 94°C, 1 min at 58°C, 1 min at 72°C, and a final 10-min extension at 72°C.
A recombinant GST/OVCA1 (81-435) fusion protein was synthesized using methods previously described (Chen et al. 1997). Antiserum against the GST/OVCA1 (81-435) fusion protein was raised in rabbits (Covance Research Products). IgG was purified from this antiserum using protein A Sepharose and absorbed with GST protein. OVCA1 protein was examined in embryonic tissues collected from E18.5 fetuses. Tissues were homogenized in lysis buffer (PBS containing 1% Triton X-100). Then 40-μg protein extracts were boiled in 2× Laemmli buffer, separated by SDS-PAGE (12% gel) and detected by Western blot using the ECL detection system (Amersham). Anti-OVCA1 IgG and peroxidase-conjugated anti-rabbit IgG (Zymed Laboratories, Inc.) were used at 1:500 and 1:2000 dilutions, respectively. For the detection of RB phosphorylation status, 106 MEFs (P1) were plated in gelatinized 10-cm2 plates and harvested the next day, counted, and lysed in 2× Laemmli buffer. Equal amounts of cell lysates were separated by SDS-PAGE (7% gel) and detected by Western blot. The anti-RB antibody (M153, Santa Cruz Biotechnology) was used at 1:200 dilution.
Ovary transplantation
Newborn ovaries were transferred under the kidney capsules or in the ovarian bursae of 3- to 5-wk-old B6/129 F1 females (Nagy et al. 2003).
Histology
Embryos or tissue samples were fixed in 4% paraformaldehyde or 10% neutral-buffered formalin overnight at 4°C or for 2-3 h at room temperature, respectively. The samples were then dehydrated through a graded series of ethanols, cleared in xylene, and infiltrated with paraffin; embedded. 4-μm sections were cut and stained with hematoxylin and eosin (H&E). Blood from 16.5-dpc embryos was smeared onto slides and stained with Wright-Giemsa (Fisher Scientific).
Generation of Ovca1-2 and p53 compound mice
Ovca1 and p53 are located ∼8 cM apart on mouse Chromosome 11. Thus, the generation of Ovca1/p53 double-homozygous mutants requires the generation of a recombinant chromosome containing mutations in both genes in cis. Therefore, Ovca1-2 heterozygotes were bred to C57BL/6-p53 heterozygotes (Jacks et al. 1994) to generate double-heterozygous progeny, with both mutations in trans (designated trans Ovca1-2+/- p53+/-). Subsequently, trans Ovca1-2+/- p53+/- mice were bred with B6 mice or intercrossed with trans Ovca1-2+/- p53+/- mice to generate progeny with the Ovca1-2 and p53 mutations on the same chromosome (designated cis Ovca1-2+/- p53+/-). Progeny with both mutations on the same chromosome were obtained at an ∼1 % frequency. The recombinant chromosome was maintained using an engineered balancer chromosome (Zheng et al. 1999).
Southern blot analysis
For Southern blot analysis, 10 μg of genomic DNA isolated from tail or frozen tumors was digested with EcoRI/StuI or EcoRI, separated on a 0.8% agarose gel, and transferred to Hybond-N+ nylon transfer membranes (Amersham Biosciences). The EcoRI/StuI blot was hybridized with a p53 cDNA probe spanning exons 7 to 10 derived from plasmid LTR-XA. The EcoRI blot was hybridized with a 5′ Ovca1 probe, a 2-kb XbaI fragment located at the 5′-end of bacterial phage clone 15-3-1 (Chen and Behringer 2001).
Immunohistochemistry
Tissue sections were placed in boiling antigen retrieval buffer (DAKO) for 10 min after deparaffinization and rehydration. Endogenous peroxidase activity was quenched by 3% hydrogen peroxide for 10 min followed by incubation with primary antibodies to CD34 (1:50; Cedarlane), cytokeratin 8 (CK8; TROMA-1, 1:100; NICHD supported Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), and AMH (MISC-20; 1:100; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Appropriate biotinylated antibodies (anti-rat IgG against antibodies to CD34 and TROMA-1; anti-goat IgG against antibody to AMH) were subsequently incubated with slides followed by ABC reaction (ABC kit; Vector Laboratories, Inc.). Staining was performed using substrate-chromagen solution containing 3% 3-amino-9-ethylcarbazole (AEC) and hydrogen peroxide. The slides were lightly counterstained with hematoxylin and then mounted.
Analysis of tumor formation
The mice used in this study were maintained in a colony that was serologically positive for mouse hepatitis virus. Up to five mice were housed in microisolator cages using specific pathogen free husbandry. The animals were provided with autoclaved water and food (Rodent NIH-31 #7027, Harlan Teklad) ad libidum. Mice were inspected for tumors twice per week. Tumor samples were isolated and processed for histopathology. If tumors were not grossly apparent, then a full necropsy was performed. In most cases, lung, liver, and kidney were also collected to histologically screen for metastatic lesions. Tumor diagnosis was based on the international classification of mouse tumors (Mohr 2001) and in consultation with pathologists.
Acknowledgments
We thank Steve O'Gorman for PC3 ES cells, Allan Bradley for SNL 76/7 STO cells, and Jinsong Liu (M.D. Anderson Cancer Center) and Junn-Liang Chang (Armed Forces Taoyuan General Hospital, Taiwan) for consultation on histopathology. We are grateful to Gigi Lozano for providing plasmid LTR-XA and helpful comments on the manuscript. This work was supported by a Development Grant from the National Cancer Institute Specialized Programs of Research Excellence in Ovarian Cancer CA83639 and grants from the National Institutes of Health (NIH) HD30284 and Department of Defense Ovarian Cancer Research Program OC020187 to R.R.B. DNA sequencing, flow cytometry, and veterinary resources were supported by the NIH Cancer Center Support (Core) Grant, CA16672.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1162204.
Supplemental material is available at http://www.genesdev.org.
References
- Atalay A., Crook, T., Ozturk, M., and Yulug, I.G. 2002. Identification of genes induced by BRCA1 in breast cancer cells. Biochem. Biophys. Res. Commun. 299: 839-846. [DOI] [PubMed] [Google Scholar]
- Bast R.C. and Mills, G.B. 2000. Alterations in oncogenes, tumor suppressor genes, and growth factors associated with epithelial ovarian cancers. In Ovarian cancer: Methods and protocols (ed. J.M.S. Bartlett), pp. 37-48. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Blackburn A.C., Brown, J.S., Naber, S.P., Otis, C.N., Wood, J.T., and Jerry, D.J. 2003. BALB/c alleles for Prkdc and Cdkn2a interact to modify tumor susceptibility in Trp53+/- mice. Cancer Res. 63: 2364-2368. [PubMed] [Google Scholar]
- Bruening W., Prowse, A.H., Schultz, D.C., Holgado-Madruga, M., Wong, A., and Godwin, A.K. 1999. Expression of OVCA1, a candidate tumor suppressor, is reduced in tumors and inhibits growth of ovarian cancer cells. Cancer Res. 59: 4973-4983. [PubMed] [Google Scholar]
- Cao L., Li, W., Kim, S., Brodie, S.G., and Deng, C.X. 2003. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes & Dev. 17: 201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C., Leventer, R.J., Ward, H.L., Toyo-Oka, K., Chung, J., Gross, A., Martin, C.L., Allanson, J., Pilz, D.T., Olney, A.H., et al. 2003. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am. J. Hum. Genet. 72: 918-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.G., Johns, M.A., Zeng, X., Zhou, L., Zink, M.C., Mankowski, J.L., Donovan, D.M., and Baylin, S.B. 2000. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum. Mol. Genet. 9: 413-419. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay P., Rathore, A., Mathur, M., Sarkar, C., Mahapatra, A.K., and Sinha, S. 1997. Loss of heterozygosity of a locus on 17p13.3, independent of p53, is associated with higher grades of astrocytic tumours. Oncogene 15: 871-874. [DOI] [PubMed] [Google Scholar]
- Chen C.M. and Behringer, R.R. 2001. Cloning, structure, and expression of the mouse Ovca1 gene. Biochem. Biophys. Res. Commun. 286: 1019-1026. [DOI] [PubMed] [Google Scholar]
- Chen C.M., You, L.R., Hwang, L.H., and Wu Lee, Y.H. 1997. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-β receptor modulates the signal pathway of the lymphotoxin-β receptor. J. Virol. 71: 9417-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S.S., Pack, S.D., Roschke, A.V., Tanigami, A., Carrozzo, R., Smith, A.C., Dobyns, W.B., and Ledbetter, D.H. 1997. A revision of the lissencephaly and Miller-Dieker syndrome critical regions in Chromosome 17p13.3. Hum. Mol. Genet. 6: 147-155. [DOI] [PubMed] [Google Scholar]
- Classon M. and Harlow, E. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2: 910-917. [DOI] [PubMed] [Google Scholar]
- Cornelis R.S., van Vliet, M., Vos, C.B., Cleton-Jansen, A.M., van de Vijver, M.J., Peterse, J.L., Khan, P.M., Borresen, A.L., Cornelisse, C.J., and Devilee, P. 1994. Evidence for a gene on 17p13.3, distal to TP53, as a target for allele loss in breast tumors without p53 mutations. Cancer Res. 54: 4200-4206. [PubMed] [Google Scholar]
- Cressman V.L., Backlund, D.C., Hicks, E.M., Gowen, L.C., Godfrey, V., and Koller, B.H. 1999. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ. 10: 1-10. [PubMed] [Google Scholar]
- Dobyns W.B. and Ledbetter, D.H. 2003. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am. J. Hum. Genet. 72: 918-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther S.A., Warren, W., Mazoyer, S., Russell, P.A., Harrington, P.A., Chiano, M., Seal, S., Hamoudi, R., van Rensburg, E.J., Dunning, A.M., et al. 1995. Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat. Genet. 11: 428-433. [DOI] [PubMed] [Google Scholar]
- Gayther S.A., Mangion, J., Russell, P.A., Seal, S., Barfoot, R., Ponder, B.A., Stratton, M.R., and Easton, D. 1997. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat. Genet. 15: 103-105. [DOI] [PubMed] [Google Scholar]
- Gowen L.C., Johnson, B.L., Latour, A.M., Sulik, K.K., and Koller, B.H. 1996. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 12: 191-194. [DOI] [PubMed] [Google Scholar]
- Hakem R., de la Pompa, J.L., Sirard, C., Mo, R., Woo, M., Hakem, A., Wakeham, A., Potter, J., Reitmair, A., Billia, F., et al. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85: 1009-1023. [DOI] [PubMed] [Google Scholar]
- Hirotsune S., Pack, S.D., Chong, S.S., Robbins, C.M., Pavan, W.J., Ledbetter, D.H., and Wynshaw-Boris, A. 1997. Genomic organization of the murine Miller-Dieker/lissencephaly region: Conservation of linkage with the human region. Genome Res. 7: 625-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S., Fleck, M.W., Gambello, M.J., Bix, G.J., Chen, A., Clark, G.D., Ledbetter, D.H., McBain, C.J., and Wynshaw-Boris, A. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19: 333-339. [DOI] [PubMed] [Google Scholar]
- Hoff C., Mollenhauer, J., Waldau, B., Hamann, U., and Poustka, A. 2001. Allelic imbalance and fine mapping of the 17p13.3 subregion in sporadic breast carcinomas. Cancer Genet. Cytogenet. 129: 145-149. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington, L., Williams, B.O., Schmitt, E.M., Halachmi, S., Bronson, R.T., and Weinberg, R.A. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4: 1-7. [DOI] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen, R., van der Gulden, H., Peterse, H., van der Valk, M., and Berns, A. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29: 418-425. [DOI] [PubMed] [Google Scholar]
- Konishi H., Takahashi, T., Kozaki, K., Yatabe, Y., Mitsudomi, T., Fujii, Y., Sugiura, T., Matsuda, H., Takahashi, T., and Takahashi, T. 1998. Detailed deletion mapping suggests the involvement of a tumor suppressor gene at 17p13.3, distal to p53, in the pathogenesis of lung cancers. Oncogene 17: 2095-2100. [DOI] [PubMed] [Google Scholar]
- Kuperwasser C., Hurlbut, G.D., Kittrell, F.S., Dickinson, E.S., Laucirica, R., Medina, D., Naber, S.P., and Jerry, D.J. 2000. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am. J. Pathol. 157: 2151-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscia D.S., Morizio, R., Venesio, T., Palenzona, C., Donadio, M., and Callahan, R. 1999. Prognostic significance of loss of heterozygosity at loci on Chromosome 17p13.3-ter in sporadic breast cancer is evidence for a putative tumour suppressor gene. Br. J. Cancer 80: 821-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.Y., Flesken-Nikitin, A., Li, S., Zeng, Y., and Lee, W.H. 1996. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes & Dev. 10: 1835-1843. [DOI] [PubMed] [Google Scholar]
- Mattheakis L.C., Sor, F., and Collier, R.J. 1993. Diphthamide synthesis in Saccharomyces cerevisiae: Structure of the DPH2 gene. Gene 132: 149-154. [DOI] [PubMed] [Google Scholar]
- Merlo G.R., Venesio, T., Bernardi, A., Cropp, C.S., Diella, F., Cappa, A.P., Callahan, R., and Liscia, D.S. 1994. Evidence for a second tumor suppressor gene on 17p linked to high S-phase index in primary human breast carcinomas. Cancer Genet. Cytogenet. 76: 106-111. [DOI] [PubMed] [Google Scholar]
- Miller B.J., Wang, D., Krahe, R., and Wright, F.A. 2003. Pooled analysis of loss of heterozygosity in breast cancer. Amer. J. Hum. Genet. 73: 748-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr U. 2001. International classification of rodent tumors: The mouse. Springer-Verlag, Berlin.
- Nagy A., Gertsenstein, M., Vintersten, K., and Behringer, R.R. 2003. Manipulating the mouse embryo: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- O'Gorman S., Dagenais, N.A., Qian, M., and Marchuk, Y. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. 94: 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan C.M., Borg, A., Cuny, M., Crichton, D.N., Baldersson, T., Andersen, T.I., Caligo, M.A., Lidereau, R., Lindblom, A., Seitz, S., et al. 1998. Consortium study on 1280 breast carcinomas: Allelic loss on Chromosome 17 targets subregions associated with family history and clinical parameters. Cancer Res. 58: 1004-1012. [PubMed] [Google Scholar]
- Phillips N.J., Ziegler, M.R., Saha, B., and Xynos, F. 1993. Allelic loss on chromosome 17 in human ovarian cancer. Int. J. Cancer 54: 85-91. [DOI] [PubMed] [Google Scholar]
- Phillips N.J., Ziegler, M.R., Radford, D.M., Fair, K.L., Steinbrueck, T., Xynos, F.P., and Donis-Keller, H. 1996a. Allelic deletion on Chromosome 17p13.3 in early ovarian cancer. Cancer Res. 56: 606-611. [PubMed] [Google Scholar]
- Phillips N.J., Ziegler, M.R., and Deaven, L.L. 1996b. A cDNA from the ovarian cancer critical region of deletion on Chromosome 17p13.3. Cancer Lett. 102: 85-90. [DOI] [PubMed] [Google Scholar]
- Saxena A., Clark, W.C., Robertson, J.T., Ikejiri, B., Oldfield, E.H., and Ali, I.U. 1992. Evidence for the involvement of a potential second tumor suppressor gene on Chromosome 17 distinct from p53 in malignant astrocytomas. Cancer Res. 52: 6716-6721. [PubMed] [Google Scholar]
- Schuijer M. and Berns, E.M. 2003. TP53 and ovarian cancer. Hum. Mutat. 21: 285-291. [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Vanderveer, L., Berman, D.B., Hamilton, T.C., Wong, A.J., and Godwin, A.K. 1996. Identification of two candidate tumor suppressor genes on Chromosome 17p13.3. Cancer Res. 56: 1997-2002. [PubMed] [Google Scholar]
- Schultz D.C., Balasara, B.R., Testa, J.R., and Godwin, A.K. 1998. Cloning and localization of a human diphthamide biosynthesis-like protein-2 gene, DPH2L2. Genomics 52: 186-191. [DOI] [PubMed] [Google Scholar]
- Sharan S.K., Morimatsu, M., Albrecht, U., Lim, D.S., Regel, E., Dinh, C., Sands A., Eichele, G., Hasty, P., and Bradley, A. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804-810. [DOI] [PubMed] [Google Scholar]
- Vogel K.S., Klesse, L.J., Velasco-Miguel, S., Meyers, K., Rushing, E.J., and Parada, L.F. 1999. Mouse tumor model for neurofibromatosis type 1. Science 286: 2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q.J., Guo, M., Lu, Z.M., Li, T., Qiao, H.Z., and Ke, Y. 2003. Detection of human papillomavirus-16 in ovarian malignancy. Br. J. Cancer 89: 672-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wagner, K.U., Larson, D., Weaver, Z., Li, C., Reid, T., Hennighausen, L., Wynshaw-Boris, A., and Deng, C.X. 1999. Conditional mutation of Brca1 in mammary epithelia cells results in blunted ductal morphogenesis and tumor formation. Nat. Genet. 22: 37-43. [DOI] [PubMed] [Google Scholar]
- Zaloudek C. 2000. Tumors of the female genital tract. Part A. Tumors of the Ovary. In Diagnostic histopathology of tumors (ed. C.D. Fletcher), pp. 567-642. Churchill, Livingstone, Harcourt Publishers Limited, Edinburgh.
- Zheng B., Sage, M., Cai, W.W., Thompson, D.M., Tavsanli, B.C., Cheah, Y.C., and Bradley, A. 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22: 375-378. [DOI] [PubMed] [Google Scholar]