Abstract
Our understanding of eukaryotic transcriptional activation mechanisms has been hampered by an inability to identify the direct in vivo targets of activator proteins, primarily because of lack of appropriate experimental methods. To circumvent this problem, we have developed a fluorescence resonance energy transfer (FRET) assay to monitor interactions with transcriptional activation domains in living cells. We use this method to show that the Tra1 subunit of the SAGA (Spt/Ada/Gcn5/acetyltransferase) complex is the direct in vivo target of the yeast activator Gal4. Chromatin-immunoprecipitation experiments demonstrate that the Gal4-Tra1 interaction is required for recruitment of SAGA to the upstream activating sequence (UAS), and SAGA, in turn, recruits the Mediator complex to the UAS. The UAS-bound Mediator is required for recruitment of the general transcription factors to the core promoter. Thus, our results identify the in vivo target of an activator and show how the activator-target interaction leads to transcriptional stimulation. The FRET assay we describe is a general method that can be used to identify the in vivo targets of other activators.
Keywords: FRET, SAGA, Tra1, Gal4, Mediator, activator target
Transcription initiation by RNA polymerase II involves the assembly of general transcription factors (GTFs) on the core promoter to form a preinitiation complex (PIC). A variety of studies indicate that promoter-specific activator proteins (activators) work, at least in part, by increasing PIC formation (Orphanides et al. 1996; Roeder 1996; Ptashne and Gann 1997; Lee and Young 2000). Activator-mediated stimulation of PIC assembly is believed to result from a direct interaction between the activation domain (AD) and one or more components of the transcription machinery, termed the “target.” The unambiguous identification of the direct in vivo targets of activators has been a major challenge in the field.
Transcriptional induction of genes involved in galactose utilization (GAL genes) has been a model experimental system for studying transcriptional activation mechanisms. The well-characterized acidic activator Gal4 is responsible for the transcriptional stimulation of GAL genes, such as GAL1, which contain Gal4-binding sites in their promoters (Johnston 1987; Johnston and Carlson 1992; Dudley et al. 1999). A variety of transcriptional components have been proposed to be the target of Gal4 including TBP (Melcher and Johnston 1995; Wu et al. 1996), TFIIB (Wu et al. 1996), Srb4 (Koh et al. 1998; Park et al. 2000), Gal11 (Jeong et al. 2001), the Swi/Snf complex (Neely et al. 2002), and the SAGA (Spt/Ada/Gcn5/acetyltransferase) complex (Bhaumik and Green 2001; Brown et al. 2001; Larschan and Winston 2001). These proposals are based on either in vitro protein interaction studies or inferences from various indirect in vivo experiments, and to date there is no definitive evidence that Gal4 directly interacts with any of these putative targets in vivo. The major obstacle has been the lack of an experimental strategy to measure direct interactions with transcriptional ADs in vivo.
The development of spectral variants of green fluorescent protein (GFP) has made it possible to perform fluorescence resonance energy transfer (FRET) analysis in living cells. FRET occurs when two compatible GFP-derived fluorophores are positioned within a critical distance of one another (Patterson et al. 2000). For one well-characterized pair of compatible GFP-derived fluorophores, enhanced cyan fluorescent protein (ECFP) and enhanced yellow fluorescent protein (EYFP), this distance is ∼50 Å (Heim and Tsien 1996). When the structure of GFP is considered (Ormo et al. 1996; Yang et al. 1996), an interfluorophore distance of 50 Å corresponds to a separation between ECFP and EYFP of 25-35 Å (Damelin and Silver 2000). Therefore, if a pair of ECFP-EYFP fusion proteins gives rise to a FRET signal, the probability of direct binding is extremely high. Accordingly, FRET can be used as an assay to detect direct protein-protein interactions in living cells (Berney and Danuser 2003).
We have been studying the yeast SAGA complex and its potential role as an activator target (Bhaumik and Green 2001, 2002). Yeast SAGA is a 1.8-MD complex, which contains at least 14 subunits: Ada1, Ada2, Ada3, Ada5/Spt20, Gcn5, Spt3, Spt7, Spt8, TAF5, TAF6, TAF9, TAF10, TAF12, and Tra1 (Hampsey 1997; Grant et al. 1998; Brown et al. 2000). To determine whether SAGA is in fact a direct target of Gal4 and, if so, to identify the SAGA subunit directly contacted, we developed a FRET assay to monitor interactions with the Gal4 AD in vivo. The results of these experiments reveal that a single SAGA subunit, Tra1, is the direct in vivo target of Gal4. Using chromatin-immunoprecipitation (ChIP) assays in conjunction with appropriate yeast mutants, we show that the Gal4-Tra1 interaction initiates an ordered protein interaction network culminating in PIC assembly and transcription activation.
Results
Development of a FRET assay for detecting interactions with Gal4 in vivo
Figure 1A illustrates the experimental strategy for detecting proteins that interact with Gal4 by FRET. We derived yeast strains in which enhanced cyan fluorescent protein (ECFP) was fused to Gal4, and enhanced yellow fluorescent protein (EYFP) was fused to a potential interacting protein. Following excitation of the donor fluorophore (ECFP) with the 405-nm laser line, fluorescence emission intensity is measured over a range of wavelengths. Interaction between two fluorophore-tagged proteins permits energy transfer between the fluorophores, which leads to quenching of ECFP (donor) emission and sensitized emission of EYFP (acceptor), resulting in a peak of fluorescence emission at 525 nm.
Figure 1.
Development of a FRET assay for detecting interactions with Gal4 in vivo. (A) Experimental strategy. Direct interaction between Gal4-ECFP and an EYFP-tagged protein (black) results in a peak of fluorescence emission at 525 nm and a concomitant reduction in donor (ECFP) emission. (B) Fluorescence emission spectra in yeast strains coexpressing Gal4-ECFP and Gal80-EYFP, or expressing Gal4-ECFP alone, Gal80-EYFP alone, or Gal4-ECFP and unfused EYFP expressed from the GAL80 promoter. (C) Fluorescence emission spectra in Gal4-ECFP/Gal80-EYFP cells in which the EYFP acceptor fluorophore was photobleached at 514 nm prior to excitation of the ECFP donor fluorophore with the 405-nm laser line. (PB) Photobleaching.
To test the feasibility of this approach, we first performed a series of control experiments that monitored the interaction between Gal4 and its negative regulator Gal80. In the absence of galactose, Gal80 binds to the Gal4 AD and inhibits its activity (Johnston 1987; Johnston and Carlson 1992). We derived a yeast strain in which endogenous Gal4 and Gal80 were fused to ECFP and EYFP, respectively, and expressed under their native promoters from their natural chromosomal loci. The results of Figure 1B show that in dextrose medium, Gal4-ECFP/Gal80-EYFP cells displayed a strong peak of fluorescence emission at 525 nm, as expected for a FRET signal. Significantly, a FRET signal was not detected in yeast strains expressing Gal4-ECFP alone, Gal80-EYFP alone, or Gal4-ECFP and unfused EYFP.
A true FRET signal should be reduced by photobleaching the acceptor fluorophore with an accompanying increase in donor emission (Karpova et al. 2003). Figure 1C shows that after photobleaching the acceptor, EYFP, the emission intensity of ECFP (donor) observed in Gal4-ECFP/Gal80-EYFP cells substantially increased, whereas EYFP emission was reduced. Thus, the FRET signal observed in Gal4-ECFP/Gal80-EYFP cells is due to EYFP sensitized emission and not ECFP leak-through emission. Collectively, the results of Figure 1 demonstrate that FRET can be used to identify proteins that directly interact with the Gal4 AD in vivo.
Identification of the SAGA subunit that is contacted by Gal4
Having established the validity of the experimental approach, we constructed a panel of yeast strains coexpressing Gal4-ECFP and one of the 14 SAGA subunits fused to EYFP. Each strain was grown in galactose medium, in which Gal4 is active, and analyzed for FRET as described above. The results of Figure 2A reveal that a FRET signal was observed only when the SAGA subunit Tra1 was fused to EYFP. Quantitation of these FRET results are presented in Figure 2B. Significantly, the FRET signal in Gal4-ECFP/Tra1-EYFP cells (FRET efficiency; 0.78) was higher than that in Gal4-ECFP/Gal80-EYFP cells (FRET efficiency; 0.55 [galactose] or 0.66 [dextrose]).
Figure 2.
Identification of the SAGA subunit that is contacted by Gal4. (A) Fluorescence emission spectra in yeast strains coexpressing Gal4-ECFP and one of the 14 SAGA subunits fused to EYFP. (B) FRET efficiency in yeast strains coexpressing Gal4-ECFP and Gal80-EYFP or one of the 14 SAGA subunits fused to EYFP. FRET efficiency (EFRET) was calculated using the formula EFRET = (Ipost - Ipre)/Ipost, where Ipre and Ipost are the fluorescence intensities of ECFP before and after photobleaching, respectively (see Materials and Methods for details). Spectra were recorded from three cells; the average FRET efficiency and standard deviation are shown. (C) Fluorescence emission spectra for all 14 Gal4-ECFP/SAGA subunit-EYFP strains following excitation of the EYFP acceptor fluorophore with the 488-nm laser line. (D) Growth of strains expressing SAGA subunit-EYFP fusions in dextrose (YPD) and galactose (YPG) media. (E) Analysis of PHO84 transcription in strains expressing SAGA-EYFP fusion proteins. Transcription was monitored by primer-extension analysis, quantitated, and presented as the percent PHO84 transcription relative to the untagged strain FY23.
Several experiments confirmed that the inability to detect a FRET signal between Gal4-ECFP and the remaining 13 SAGA subunit-EYFP fusions was not due to a failure to express sufficient levels of functional fusion proteins. First, the EYFP emission spectra of Figure 2C show that all 14 SAGA subunit-EYFP fusion proteins were expressed and at roughly comparable levels. Second, six of the SAGA subunits are essential for viability (TAF5, TAF6, TAF9, TAF10, TAF12, and Tra1; Saleh et al. 1998; Green 2000) and five are required for GAL1 transcription (Ada1, Spt3, Spt7, Spt20, and Tra1; Roberts and Winston 1997; Dudley et al. 1999; Sterner et al. 1999; and see below). Figure 2D shows that strains expressing all 14 EYFP fusions grew on dextrose and galactose media, indicating that the fused subunits were functional. Third, all SAGA subunits are required for PHO84 transcription (Bhaumik and Green 2002; our unpublished data), which occurred normally in all 14 SAGA subunit-EYFP fusion strains (Fig. 2E).
Figure 3A presents several control spectra. First, as expected, no FRET signal was detected in cells expressing Tra1-EYFP alone. Second, following photobleaching of the acceptor in Gal4-ECFP/Tra1-EYFP cells, the 525-nm fluorescence emission peak was substantially reduced, and there was a concomitant increase in donor emission, as expected for a true FRET signal. As described above, only five of the 14 SAGA subunits (Ada1, Spt3, Spt7, Spt20, and Tra1) are required for GAL1 transcription. To verify the occurrence and specificity of the Gal4-Tra1 interaction, for these five critical SAGA subunits and two additional SAGA subunits, Ada2 and Gcn5, we switched the fluorophore tag from the C to the N terminus and retested interaction with Gal4 in the FRET assay. The emission spectra of Figure 3B (quantitated in Fig. 3C) indicate, once again, that a FRET signal was detected only with Tra1.
Figure 3.
Confirmation of the Gal4-Tra1 interaction and analysis of other putative Gal4 targets. (A) Fluorescence emission spectra in Gal4-ECFP/Tra1-EYFP cells that have been photobleached as described in Fig. 1C. (PB) Photobleaching. (B) Fluorescence emission spectra of yeast strains expressing Gal4-EYFP and an N-terminal ECFP-tagged SAGA subunit. (C) FRET efficiency in yeast strains coexpressing Gal4-EYFP and ECFP-SAGA fusion proteins. FRET efficiency was calculated as described in Fig. 2B. (D) Fluorescence emission spectra in cells expressing Gal4-ECFP and one of four EYFP-tagged possible targets, TBP, TFIIB, Srb4, or Gal11.
Finally, as discussed above, several other factors have been proposed to be direct targets of Gal4. We analyzed four such factors (TBP, TFIIB, Srb4, and Gal11) in the FRET assay. The results of Figure 3D show that a FRET signal was not detected in Gal4-ECFP/TBP-EYFP, Gal4-ECFP/TFIIB-EYFP, Gal4-ECFP/Srb4-EYFP, and Gal4-ECFP/Gal11-EYFP cells. These FRET data are consistent with the results of ChIP experiments presented here and in our previous study (Bhaumik and Green 2001), which also suggest that TBP, TFIIB, and Mediator subunits are not direct in vivo targets of Gal4.
The Gal4-Tra1 interaction occurs only in galactose medium
The interaction between Gal4 and its relevant target should occur only in galactose medium, in which the Gal4 AD is relieved from Gal80 inhibition (Johnston 1987; Johnston and Carlson 1992). To test this prediction, Gal4-ECFP/Tra1-EYFP cells were shifted from raffinose to galactose medium, and a series of emission spectra were recorded at various time points. Figure 4A,B shows that the FRET signal was evident in galactose but not raffinose medium and increased linearly for ∼50 min after the shift and then leveled off. Significantly, the kinetics of the Gal4-Tra1 interaction, as measured by FRET, and transcriptional induction of GAL1, as measured by primer extension, were very similar (Fig. 4B).
Figure 4.
The Gal4-Tra1 interaction occurs only in galactose medium. (A) Fluorescence emission spectra in Gal4-ECFP/Tra1-EYFP cells at various time points following a shift from raffinose to galactose medium. (B) Kinetics of the Gal4-Tra1 interaction, monitored by FRET (data from A), and transcriptional induction of GAL1, monitored by primer-extension (Bhaumik and Green 2002; inset), following a shift from raffinose to galactose medium. (C) Fluorescence emission spectra in Gal4-ECFP/Gal80-EYFP cells in dextrose, raffinose, and galactose media.
Notably, in Gal4-ECFP/Gal80-EYFP cells, a FRET signal was evident in dextrose, raffinose, and galactose media (Fig. 4C). These results are in excellent agreement with previous biochemical experiments showing that in galactose media Gal80 remains physically associated with Gal4 at a second site (Sil et al. 1999), again validating the FRET assay for detecting Gal4 interactions in vivo.
The Gal4-Tra1 interaction is required for SAGA recruitment and transcriptional activation
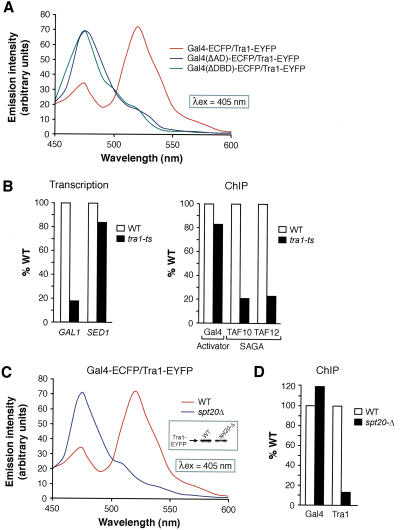
Collectively, the results described above demonstrate that Gal4 and Tra1 interact in vivo. To test whether this interaction was dependent on the Gal4 AD, as expected for an interaction between Gal4 and its relevant target, we derived a yeast strain coexpressing Tra1-EYFP and an ECFP fusion to a Gal4 derivative lacking its AD, Gal4(ΔAD)-ECFP. Figure 5A shows that an emission spectrum recorded from Gal4(ΔAD)-ECFP/Tra1-EYFP cells lacked the characteristic 525-nm emission peak, indicating that the Gal4 AD is required for the Gal4-Tra1 interaction. A FRET signal was also not observed using a Gal4 mutant lacking its DNA-binding domain [Gal4(ΔDBD)-ECFP/Tra1-EYFP cells]. Significantly, both of these Gal4 mutants are unable to support transcription activation.
Figure 5.
The Gal4-Tra1 interaction is required for SAGA recruitment and transcriptional activation. (A) Fluorescence emission spectra of yeast strains expressing Tra1-EYFP and ECFP fused to either Gal4 or derivatives lacking its AD, Gal4(ΔAD)-ECFP, or its DNA-binding domain, Gal4(ΔDBD)-ECFP. (B) Transcriptional analysis of GAL1 and SED1 by primer-extension (left) and ChIP analysis of Gal4 and SAGA recruitment to the GAL1 UAS (right) following inactivation of Tra1. Formaldehyde-based in vivo cross-linking and ChIP was performed as previously described (Bhaumik and Green 2001). Immunoprecipitated (IP) DNA was amplified by PCR using primer pairs corresponding to the GAL1 UAS, and IP DNA was quantitated and presented as the ratio of IP to input relative to wild type. (C) Fluorescence emission spectra in Gal4-ECFP/Tra1-EYFP cells in the presence and absence of the SAGA subunit Spt20. (Inset) Immunoblot showing Tra1-EYFP levels in wild-type and spt20Δ strains. (D) ChIP analysis of Tra1 recruitment to the GAL1 UAS in wild-type cells and in an spt20Δ deletion mutant.
To determine the role of Tra1 in Gal4-mediated transcriptional activation, we analyzed a tra1 temperature-sensitive mutant (Kulesza et al. 2002). Figure 5B shows that inactivation of Tra1 substantially decreased transcription of GAL1 but not SED1, a SAGA-independent gene (Bhaumik and Green 2002). A ChIP assay shows that following inactivation of Tra1, Gal4 was still bound to the GAL1 upstream activating sequence (UAS), whereas SAGA recruitment, as monitored by association of SAGA TAFs with the UAS (Bhaumik and Green 2001), decreased significantly, explaining the transcriptional defect.
The SAGA subunit Spt20 is required for complex integrity (Grant et al. 1997; Sterner et al. 1999) and thus transcription of SAGA-dependent genes such as GAL1 (Roberts and Winston 1997; Dudley et al. 1999; Bhaumik and Green 2001). Figure 5C shows that in an spt20Δ deletion mutant, the interaction between Gal4-ECFP and Tra1-EYFP, as measured by FRET, failed to occur even though Tra1-EYFP was expressed at normal levels (inset). These results suggest that Tra1 must be incorporated within SAGA for interaction with Gal4 in vivo. To confirm this result, we used a ChIP assay to analyze Tra1 recruitment to the GAL1 UAS in the presence or absence of an intact SAGA complex. Figure 5D shows that in the spt20Δ deletion mutant, recruitment of Tra1 to the GAL1 UAS was substantially reduced.
The Gal4-Tra1 interaction initiates an ordered protein interaction network at the GAL1 UAS
The results of the FRET experiments indicate that Tra1, and thus SAGA, is directly targeted by the Gal4 AD and recruited to the UAS. We next attempted to determine how recruitment of SAGA to the UAS by Gal4 increases PIC assembly. The SRB/Mediator complex was of particular interest because, like SAGA, Mediator has been implicated as a target of some transcriptional activators (Myers and Kornberg 2000; Naar et al. 2001). We performed a ChIP assay using two sets of promoter-specific primer pairs that we have previously shown can distinguish binding to the GAL1 UAS or core promoter (Bhaumik and Green 2001). To monitor SAGA recruitment to the GAL1 UAS, we analyzed the association of subunits present exclusively in SAGA (e.g., Spt3) or TAFs that are specifically recruited to the GAL1 UAS as part of the SAGA complex (e.g., TAF10 and TAF12; Bhaumik and Green 2001). Mediator recruitment was monitored by analyzing either Srb2 or Srb4. Figure 6A shows that, like Gal4 and SAGA (see also Bhaumik and Green 2001), Mediator was bound to the GAL1 UAS and not the core promoter.
Figure 6.
An ordered protein interaction network at the GAL1 UAS facilitates PIC assembly. (A) ChIP analysis of Gal4, SAGA, and Mediator binding at GAL1. IP DNA was amplified by PCR using primer pairs corresponding to the UAS or core promoter of GAL1 (Bhaumik and Green 2001). (B) SAGA is required for recruitment of Mediator to the GAL1 UAS. A ChIP assay was performed using primer pairs corresponding to the GAL1 UAS, and IP DNA was quantitated and presented as described in Fig. 5B. (C) ChIP analysis of Gal4, SAGA, and Mediator recruitment at the GAL1 UAS following inactivation of Srb4.
We next analyzed recruitment of Gal4 and Mediator to the GAL1 UAS in four SAGA subunit deletion mutants (ada1Δ, spt3Δ, spt7Δ, and spt20Δ) known to compromise GAL1 transcription (Roberts and Winston 1997; Dudley et al. 1999; Bhaumik and Green 2001). Figure 6B shows, as expected, that in all four mutants, Gal4 was bound but recruitment of Mediator to the UAS was dramatically reduced compared with the wild-type strain. Thus, SAGA is required for efficient recruitment of Mediator to the GAL1 UAS.
To determine whether Mediator has a role in recruitment of SAGA or Gal4, we performed ChIP assays in a strain harboring a temperature-sensitive mutation in the Mediator subunit Srb4, which is required for complex integrity (Koh et al. 1998; Kang et al. 2001). Figure 6C shows, as expected, that inactivation of Srb4 substantially decreased recruitment of Mediator (Srb2) to the UAS. However, inactivation of Srb4 did not significantly affect recruitment of SAGA or Gal4. These data, in conjunction with our previous study (Bhaumik and Green 2001) and the results presented above, reveal the following interaction network at the GAL1 UAS: Gal4-SAGA-Mediator.
Recruitment of SAGA and Mediator to the GAL1 UAS is required for PIC assembly at the core promoter
Finally, we analyzed the roles of SAGA and Mediator in PIC assembly at the core promoter. Figure 7A shows that in cells lacking functional SAGA (an spt20Δ deletion strain) or Mediator (Srb4 inactivation), the GTFs TBP, TFIIB, and RNA polymerase II (Rbp1) were not recruited to the core promoter. In contrast, inactivation of TBP or TFIIB had only a negligible (less than two fold) effect on recruitment of Mediator, SAGA, or Gal4 to the UAS (Fig. 7B; Bhaumik and Green 2001) but, as expected, virtually eliminated PIC assembly, as measured by RNA polymerase II recruitment.
Figure 7.
Recruitment of SAGA and Mediator to the GAL1 UAS is required for PIC assembly at the core promoter. (A) PIC assembly at the GAL1 core promoter is dependent on Gal4, SAGA, and Mediator. (B) Analysis of Gal4, SAGA, and Mediator recruitment and PIC assembly following inactivation of TBP and TFIIB. (C) Schematic summary. (Top) At the GAL1 UAS, SAGA serves as an “adaptor” that recruits Mediator to the Gal4 AD, and ultimately results in PIC assembly at the core promoter. The interaction between Gal4 and SAGA occurs directly via Tra1. Other interactions, indicated by arrows, have not been demonstrated to be direct. (Bottom) At some promoters, the activator (Act) functions by direct contact with Mediator.
Discussion
Tra1 is the in vivo target of the Gal4 AD
In this study, we have shown that FRET can be used to identify proteins that interact with a transcriptional AD in vivo. The power of this experimental approach is that protein-protein interactions are analyzed in vivo on intact complexes and not isolated subunits. Using this method, we found that the SAGA subunit Tra1 is the direct target of Gal4. Several findings presented here and in previous studies are consistent with this conclusion. First, transcriptional activation by Gal4 requires SAGA (Bhaumik and Green 2001; Larschan and Winston 2001) and, specifically, the SAGA subunit Tra1 (Fig. 5B). Second, ChIP experiments have shown that SAGA is recruited by Gal4 to the UAS of several GAL genes (Bhaumik and Green 2001; Larschan and Winston 2001) and to minimal Gal4-binding sites in vivo (Bhaumik and Green 2001). Finally, in vitro protein interaction experiments have shown that Tra1 can interact with the Gal4 AD (Brown et al. 2001).
Interestingly, we found that the Gal4-Tra1 interaction did not occur in an spt20Δ deletion mutant in which the SAGA complex is disrupted (Fig. 4C). Thus, Tra1 does not have an intrinsic ability to interact with the Gal4 AD but rather requires proper presentation within the SAGA complex. It seems likely that this finding is relevant to the selective interaction of Gal4 with SAGA and not the NuA4 complex (Reid et al. 2000), which also contains Tra1. In this regard, our in vivo results differ from those of Brown et al. (2001), who reported, using an in vitro protein-protein interaction assay, that Gal4 interacted with isolated Tra1.
The yeast Tra1 protein is homologous to the human transformation/transcription domain-associated protein TRRAP, a member of the ataxia telangiectasia mutated (ATM) family. TRRAP has been reported to interact with the ADs of several oncogenic transcription factors, such as c-Myc and E2F (McMahon et al. 1998). Thus, Tra1 and TRRAP may function as direct targets of multiple transcriptional ADs in yeast and humans, respectively.
An ordered protein-protein interaction pathway for transcription complex assembly on the GAL1 promoter
Using a combination of biophysical, molecular biological, and genetic experiments, we have delineated an ordered protein interaction pathway by which the activator Gal4 stimulates transcription (summarized in Fig. 7C, top). The functional consequence of the Gal4-Tra1 interaction is recruitment of SAGA to the UAS, which initiates a series of protein-protein interactions culminating in PIC assembly at the core promoter. One of the earliest events is the SAGA-directed recruitment of Mediator to the UAS. Consistent with our model, a recent publication has shown, using ChIP assays, that Mediator is associated with the UAS of GAL genes (Kuras et al. 2003). Also in support of our model, Bryant and Ptashne (2003) found that the transcriptional machinery assembles at the promoters of GAL genes in three stages: SAGA is recruited first, followed by Mediator, and finally TBP, RNA polymerase II, and other GTFs. Recruitment of SAGA also precedes Mediator on the HO promoter (Cosma et al. 1999), although in this case, SAGA provides an essential histone acetyltransferase activity, whereas on the GAL1 promoter Gcn5 is dispensable and SAGA acts solely as an adaptor (Bhaumik and Green 2001).
Mediator is a conserved multisubunit complex that has been implicated as a target of several transcriptional activators in yeast and mammalian cells (for review, see Myers and Kornberg 2000; Naar et al. 2001). Our finding that SAGA serves as an “adaptor” that recruits Mediator to the Gal4 AD fits in well with these previous studies and demonstrates how different activator-target interactions can trigger similar activation pathways (Fig. 7C, bottom).
In addition to the protein interaction network we describe here, several other transcription complex assembly pathways have been proposed. For example, in some assays direct interactions between Gal4 and Mediator have been detected (Koh et al. 1998; Park et al. 2000; Jeong et al. 2001). Consistent with this possibility, Bryant and Ptashne (2003) reported that Mediator was recruited to the GAL1 promoter in an spt20Δ mutant, prompting the suggestion that in the initial stages of activation, SAGA and Mediator are independently contacted and recruited by Gal4. In contrast, using four independent SAGA deletion mutants, we found that recruitment of Mediator to the UAS required SAGA (Fig. 6B). We note that in the Bryant and Ptashne (2003) study, Mediator recruitment was not specifically measured at the UAS, and the amount of bound Mediator in wild-type and spt20Δ strains was not directly compared. Perhaps in the absence of SAGA, Mediator is recruited at a low level or to a region of the GAL1 promoter other than the UAS. However, because SAGA is absent, such a putative complex would be nonproductive and fail to support PIC assembly and transcription.
Previous studies have suggested that SAGA interacts with TBP via the Spt3 subunit (Larschan and Winston 2001). This proposal is based largely on genetic experiments showing partial suppression of a tbp mutant by particular spt3 mutations, which has been interpreted as evidence for an Spt3-TBP interaction (Eisenmann et al. 1992; Larschan and Winston 2001). However, this type of extragenic suppression is not always the result of a direct protein-protein interaction (see, e.g., Guarente 1993). Nonetheless, our results do not rule out the possibility of parallel pathways to the protein interaction network we have described.
Several aspects of the SAGA-mediated protein interaction network remain to be clarified. For example, what is the basis by which SAGA recruits Mediator? Functional interactions between SAGA and two Mediator subunits, Sin4 and Gal11, have been reported (Roberts and Winston 1997), and may help explain how Mediator is recruited by SAGA. An equally important issue is how UAS-bound Mediator facilitates PIC assembly at the core promoter. Presumably, this results from interactions between specific Mediator subunits and one or more GTFs. An understanding of such Mediator-GTF interactions is likely also relevant to those instances in which Mediator is the direct activator target.
GAL1 is a TAF-independent (TAFind) promoter as defined by the dispensability of TBP-associated factors (TAFs) for transcription (Li et al. 2000). Activator-mediated recruitment of TBP to a TAFind promoter requires multiple GTFs, including Mediator (Li et al. 2000), and the interaction pathway we have delineated (Fig. 7C, top) explains this requirement. Like GAL1, several TAFind promoters require SAGA for transcription (Lee et al. 2000; Bhaumik and Green 2002). It will be important to determine whether the other SAGA-dependent promoters use Tra1 as the activator target and a similar interaction network.
In contrast to TAFind promoters, TAF-dependent (TAFdep) promoters require multiple TAFs for transcription (Kuras et al. 2000; Li et al. 2000), and TBP recruitment is dependent on TAFs but not Mediator or other GTFs (Li et al. 2000). It thus seems likely that activation of TAFdep promoters will involve a different activator target and protein interaction pathway. Moreover, unlike GAL1, the majority of yeast genes are not SAGA-dependent (Lee et al. 2000), and thus their expression must involve activator targets other than SAGA. The FRET assay we describe here is a general method that can be used to identify the in vivo targets of other activators.
Materials and methods
Plasmids
The plasmids TSGP1 (pFA6a-ECFP-kanMX6) and SGP27 (pFA6a-EYFP-TRP1) were derived from pFA6a-GFP(S65T)kanMX6 and pFA6a-GFP(S65T)-TRP1 (Longtine et al. 1998) by replacing GFP(S65T) with ECFP and EYFP, respectively. TSGP1 and SGP27 were then used to C-terminally tag each protein of interest expressed from its natural chromosomal locus as previously described (Longtine et al. 1998). For N-terminally tagging proteins with ECFP, we constructed plasmid TSGP2, in which the Kan resistance marker and ECFP fragment were cloned under the GAL1 promoter; therefore, a protein N-terminally tagged by this method is expressed from its natural chromosomal locus but under the GAL1 promoter. The plasmids pFA6a-13Myc-KanMX6 and pFA6a-3HA-His3MX6 (Longtine et al. 1998) were used for genomic myc- and HA-epitope tagging proteins of interest.
Yeast strains
All strains used in the FRET experiments were derived from the haploid strain FY23 (Roberts and Winston 1996). We first tagged Gal4 with ECFP at the C terminus to generate TSGY1 (Gal4-ECFP), and then genomically tagged each of the 14 SAGA subunits, other possible Gal4 targets, or Gal80 at the C terminus in TSGY1 to generate a panel of double-tagged strains: TSGY2 (Gal4-ECFP/Ada1-EYFP), TSGY3 (Gal4-ECFP/Ada2-EYFP), TSGY4 (Gal4-ECFP/Ada3-EYFP), TSGY5 (Gal4-ECFP/Gcn5-EYFP), TSGY6 (Gal4-ECFP/Spt20-EYFP), TSGY7 (Gal4-ECFP/Spt3-EYFP), TSGY8 (Gal4-ECFP/Spt7-EYFP), TSGY9 (Gal4-ECFP/Spt8-EYFP), TSGY10 (Gal4-ECFP/TAF5-EYFP), TSGY11 (Gal4-ECFP/TAF6-EYFP), TSGY12 (Gal4-ECFP/TAF9-EYFP), TSGY13 (Gal4-ECFP/TAF10-EYFP), TSGY14 (Gal4-ECFP/TAF12-EYFP), TSGY15 (Gal4-ECFP/Tra1-EYFP), TSGY16 (Gal4-ECFP/Gal80-EYFP), TSGY31 (Gal4-ECFP/TBP-EYFP), TSGY32 (Gal4-ECFP/TFIIB-EYFP), TSGY33 (Gal4-ECFP/Srb4-EYFP), and YDA038 (Gal4-ECFP/Gal11-EYFP). To generate a panel of N-terminally tagged ECFP-SAGA subunit strains, we first tagged Gal4 with EYFP at the C terminus to generate TSGY22 (Gal4-EYFP), and then genomically tagged SAGA subunits in TSGY22 to generate double-tagged strains: TSGY23 (Gal4-EYFP/ECFP-Ada1), TSGY24 (Gal4-EYFP/ECFP-Ada2), TSGY25 (Gal4-EYFP/ECFP-Gcn5), TSGY26 (Gal4-EYFP/ECFP-Spt3), TSGY27 (Gal4-EYFP/ECFP-Spt7), TSGY28 (Gal4-EYFP/ECFP-Spt20), and TSGY29 (Gal4-EYFP/ECFP-Tra1). The endogenous SPT20 gene in TSGY15 was disrupted using standard PCR-based methods to generate TSGY17 (Gal4-ECFP/Tra1-EYFP/spt20Δ). To generate TSGY18 [Gal4(ΔAD)-ECFP/Tra1-EYFP], the ECFP cassette, which contained the ADH terminator, was inserted into the GAL4 open reading frame (ORF) to produce a truncated protein lacking the AD. To generate TSGY30 [Gal4(ΔDBD)-ECFP/Tra1-EYFP], the cassette for ECFP N-terminal tagging was inserted into the GAL4 ORF, replacing amino acids 1-147 corresponding to the DNA-binding domain of Gal4. To generate TSGY21 (Gal4-ECFP/EYFP expressed from the GAL80 promoter), the EYFP cassette was integrated into the 5′-end of the GAL80 ORF. Single-tagged strains containing Tra1-EYFP (TSGY19) and Gal80-EYFP (TSGY20) were also generated. All strains were verified for expression of ECFP and/or EYFP fusion proteins by microscopy and by immunoblotting with a GFP antibody (SC-9996; Santa Cruz Biotechnology, Inc.).
For ChIP experiments, we generated a series of strains in which Srb4 and Srb2 were epitope-tagged at the C terminus and expressed from their natural chromosomal loci. Srb4 was myc-tagged to generate: SGY216 (Srb4-myc; ada1Δ) and its wild-type equivalent, SGY211; SGY209 (Srb4-myc; spt7Δ) and its wild-type equivalent, SGY210; SGY215 (Srb4-myc; spt3Δ) and its wild-type equivalent, SGY212; and SGY191 (Srb4-myc; tfIIb-ts) and SGY192 (Srb4-myc; tbp-ts) and their wild-type equivalent, SGY190. Srb4 was also HA-tagged to generate SGY179 (Srb4-HA; spt20Δ) and its wild-type equivalent, SGY178. The parental strains for the Srb4-tagged derivatives were as follows: FY1559 (ada1Δ) and its wild-type equivalent, FY602 (Sterner et al. 1999); FY963 (spt7Δ) and its wild-type equivalent, FY61 (Roberts and Winston 1996); FY294 (spt3Δ) and its wild-type equivalent, FY631 (Sterner et al. 1999); JZY102 (tfIIb-ts; Li et al. 1999) and tbp-ts (Cormack and Struhl 1992) and their wild-type equivalent JZY101; and FY1097 (spt20Δ) and its wild-type equivalent, FY67 (Roberts and Winston 1996). Srb2 was C-terminally tagged with the myc epitope in the srb4-ts strain Z628 and its wild-type equivalent, Z579 (Thompson and Young 1995) to generate SGY203 and SGY204, respectively. Tra1 was C-terminally tagged with the myc epitope in the spt20Δ strain FY1272 and its wild-type equivalent, FY251 (kindly provided by Fred Winston, Harvard Medical School) to generate TSGY30 and TSGY31, respectively. The tra1-ts (Kulesza et al. 2002) and gal4Δ (SGY14; Bhaumik and Green 2001) strains have been described.
FRET analysis
Cells were grown at 30°C to log phase in dextrose- (YPD), galactose- (YPG), or raffinose-containing rich medium, transferred to poly-lysine-coated glass slides, and examined immediately on a Leica confocal laser scanning microscope system (TCS SP2 AOBS; Leica Microsystems), which contained 405-, 458-, 476-, 488-, 514-, 543-, and 633-nm laser lines. The 458-nm laser line was used to excite the focused cells, and the images were captured with a liquid-cooled CCD camera. The analysis of digitized microscope images allowed selection of a certain region of interest (ROI) of a single cell and thus optimized the signal-to-noise ratio. For all FRET experiments, spectra were recorded from three cells and the three spectra were averaged and plotted. To record ECFP emission spectra, the ECFP donor in the selected ROI of a single cell was excited with the 405-nm laser line to avoid leak-through of ECFP emission over EYFP emission, and emission spectra were generated by scanning the ROI eight times with 10-nm spectral resolution and recorded using Leica Confocal Software (LCS). To record EYFP emission spectra, the EYFP-fluorophore was excited by the 488-nm laser line. For EYFP acceptor photobleaching experiments, cells were photobleached by scanning an ROI eight times using the 514-nm argon laser line. Fluorescence emission spectra were recorded before and after photobleaching following ECFP excitation at 405 nm to assess changes in donor ECFP fluorescence. FRET acceptor photobleaching software was used to document that FRET occurred by showing that the intensity of donor ECFP fluorescence increased after the EYFP acceptor was photobleached. FRET efficiency (EFRET) was calculated using the formula EFRET = (Ipost - Ipre)/Ipost, where Ipre and Ipost are the fluorescence intensities of ECFP before and after photobleaching, respectively.
Acknowledgments
We thank M. Damelin and P. Silver for advice and discussions; F. Winston, M. Cole, K. Struhl, and R. Young for yeast strains; I. The, E. Latz, and T. Geil for advice on confocal microscopy; and S. Evans for editorial assistance. We are especially grateful to Florin Catrina for derivation of a mathematical solution for a molecular model relevant to the interpretation of the FRET results. This work was supported in part by a grant from the NIH to M.R.G. M.R.G. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1148404.
References
- Berney C. and Danuser, G. 2003. FRET or No FRET: A quantitative comparison. Biophys. J. 84: 3992-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green, M.R. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 15: 1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22: 7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Lechner, T., Howe, L., and Workman, J.L. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25: 15-19. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Howe, L., Sousa, K., Alley, S.C., Carrozza, M.J., Tan, S., and Workman, J.L. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333-2337. [DOI] [PubMed] [Google Scholar]
- Bryant G.O. and Ptashne, M. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell. 11: 1301-1309. [DOI] [PubMed] [Google Scholar]
- Cormack B.P. and Struhl, K. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69: 685-696. [DOI] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka, T., and Nasmyth, K. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299-311. [DOI] [PubMed] [Google Scholar]
- Damelin M. and Silver, P.A. 2000. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell 5: 133-140. [DOI] [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle, C., and Winston, F. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 13: 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D.M., Arndt, K.M., Ricupero, S.L., Rooney, J.W., and Winston, F. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 6: 1319-1331. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan, L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 11: 1640-1650. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Sterner, D.E., Duggan, L.J., Workman, J.L., and Berger, S.L. 1998. The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8: 193-197. [DOI] [PubMed] [Google Scholar]
- Green M.R. 2000. TBP-associated factors (TAFIIs): Multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25: 59-63. [DOI] [PubMed] [Google Scholar]
- Guarente L. 1993. Synthetic enhancement in gene interaction: A genetic tool come of age. Trends Genet. 9: 362-366. [DOI] [PubMed] [Google Scholar]
- Hampsey M. 1997. A SAGA of histone acetylation and gene expression. Trends Genet. 13: 427-429. [DOI] [PubMed] [Google Scholar]
- Heim R. and Tsien, R.Y. 1996. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6: 178-182. [DOI] [PubMed] [Google Scholar]
- Jeong C.J., Yang, S.H., Xie, Y., Zhang, L., Johnston, S.A., and Kodadek, T. 2001. Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry 40: 9421-9427. [DOI] [PubMed] [Google Scholar]
- Johnston M. 1987. A model fungal gene regulatory mechanism: The GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51: 458-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. and Carlson, M. 1992. Regulation of carbon and phosphate utilization. In The yeast Saccharomyces: Gene expression (ed. E.W. Jones et al.), pp. 193-281. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kang J.S., Kim, S.H., Hwang, M.S., Han, S.J., Lee, Y.C., and Kim, Y.J. 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276: 42003-42010. [DOI] [PubMed] [Google Scholar]
- Karpova T.S., Baumann, C.T., He, L., Wu, X., Grammer, A., Lipsky, P., Hager, G.L., and McNally, J.G. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209: 56-70. [DOI] [PubMed] [Google Scholar]
- Koh S.S., Ansari, A.Z., Ptashne, M., and Young, R.A. 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1: 895-904. [DOI] [PubMed] [Google Scholar]
- Kulesza C.A., Van Buskirk, H.A., Cole, M.D., Reese, J.C., Smith, M.M., and Engel, D.A. 2002. Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1. Oncogene 21: 1411-1422. [DOI] [PubMed] [Google Scholar]
- Kuras L., Kosa, P., Mencia, M., and Struhl, K. 2000. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288: 1244-1248. [DOI] [PubMed] [Google Scholar]
- Kuras L., Borggrefe, T., and Kornberg, R.D. 2003. Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. 100: 13887-13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E. and Winston, F. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 15: 1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. and Young, R.A. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34: 77-137. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Causton, H.C., Holstege, F.C., Shen, W.C., Hannett, N., Jennings, E.G., Winston, F., Green, M.R., and Young, R.A. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701-704. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Virbasius, A., Zhu, X., and Green, M.R. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399: 605-609. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Bhaumik, S.R., and Green, M.R. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288: 1242-1244. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk, H.A., Dugan, K.A., Copeland, T.D., and Cole, M.D. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363-374. [DOI] [PubMed] [Google Scholar]
- Melcher K. and Johnston, S.A. 1995. GAL4 interacts with TATA-binding protein and coactivators. Mol. Cell. Biol. 15: 2839-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L.C. and Kornberg, R.D. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69: 729-749. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Lemon, B.D., and Tjian, R. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70: 475-501. [DOI] [PubMed] [Google Scholar]
- Neely K.E., Hassan, A.H., Brown, C.E., Howe, L., and Workman, J.L. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22: 1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormo M., Cubitt, A.B., Kallio, K., Gross, L.A., Tsien, R.Y., and Remington, S.J. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273: 1392-1395. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange, T., and Reinberg, D. 1996. The general transcription factors of RNA polymerase II. Genes & Dev. 10: 2657-2683. [DOI] [PubMed] [Google Scholar]
- Park J.M., Kim, H.S., Han, S.J., Hwang, M.S., Lee, Y.C., and Kim, Y.J. 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20: 8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.H., Piston, D.W., and Barisas, B.G. 2000. Forster distances between green fluorescent protein pairs. Anal. Biochem. 284: 438-440. [DOI] [PubMed] [Google Scholar]
- Ptashne M. and Gann, A. 1997. Transcriptional activation by recruitment. Nature 386: 569-577. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Iyer, V.R., Brown, P.O., and Struhl, K. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297-1307. [DOI] [PubMed] [Google Scholar]
- Roberts S.M. and Winston, F. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R.G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21: 327-335. [PubMed] [Google Scholar]
- Saleh A., Schieltz, D., Ting, N., McMahon, S.B., Litchfield, D.W., Yates III, J.R., Lees-Miller, S.P., Cole, M.D., and Brandl, C.J. 1998. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J. Biol. Chem. 273: 26559-26565. [DOI] [PubMed] [Google Scholar]
- Sil A.K., Alam, S., Xin, P., Ma, L., Morgan, M., Lebo, C.M., Woods, M.P., and Hopper, J.E. 1999. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol. Cell. Biol. 19: 7828-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant, P.A., Roberts, S.M., Duggan, L.J., Belotserkovskaya, R., Pacella, L.A., Winston, F., Workman, J.L., and Berger, S.L. 1999. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19: 86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.M. and Young, R.A. 1995. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. 92: 4587-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Reece, R.J., and Ptashne, M. 1996. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 15: 3951-3963. [PMC free article] [PubMed] [Google Scholar]
- Yang F., Moss, L.G., and Phillips Jr., G.N. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14: 1246-1251. [DOI] [PubMed] [Google Scholar]