Abstract
Background
Histology and/or culture are generally considered the gold standard for the detection of H. pylori infection. Especially in children, these tests may result in a false negative outcome because of patchy distribution of the organism in the stomach mucosa. We have developed a PCR assay utilizing nested primer pairs directed against a subunit of the H. pylori urease gene (ureA). As part of a prospective evaluation of diagnostic tests to aid in detecting H. pylori infection in children, the aim of this study was to compare our PCR and Western blot assays with results obtained from histologic examination of biopsy specimens, rapid urease tests, and an FDA approved serologic assay and published PCR results to determine if we could validate the assays for diagnostic use on our patient population.
Results
Gastric biopsy specimens obtained from 101 pediatric patients were evaluated for the presence of H. pylori using histologic techniques, rapid urease (CLOtest) test and the PCR assay. Serum samples from each patient were assayed using both ELISA and Western Blot for antibodies to H. pylori. A total of 32 patients tested were positive by at least one of the methods evaluated. Thirteen patients had positive histology, 13 had a positive CLOtest, and 17 patients had positive H. pylori PCR. Out of the 13 CLO positive patients, 12 were positive by histologic analysis and all 13 were positive by PCR. Results of serologic tests on the same population did not correlate well with other assays. Twenty-eight patients showed serologic evidence of H. pylori infection, of which 9 were both CLO and histology positive and 12 were positive by PCR. Of the seropositive patients, 26 were ELISA positive, 13 were positive by Western blot, and 11 by both serologic methods.
Conclusions
The results obtained suggest that our nested PCR assay has the specificity and sensitivity necessary for clinical application when compared to standard histologic examination and rapid urease test. In addition, we found the current commercially available approved ELISA method appears unable to accurately detect H. pylori in this population. The Western blot assay yielded better concordance with CLOtest and histology, but not as good as the nested PCR assay.
Background
Helicobacter pylori is a gram-negative curved or spiral bacterium which colonizes the human gastric mucosa. Work by Warren and Marshall in the early 1980s demonstrated the organism's association with gastritis and peptic ulceration. Chronic H. pylori infection has also been linked to the development of gastric cancers [1-3].
Diagnosis of H. pylori infection generally includes a combination of both invasive and non-invasive methods. Invasive tests include endoscopy, with biopsy of the affected region followed by histologic examination of stained specimens to demonstrate the presence of the bacterium and culture of the bacterium from the tissue. Culture of the organism, although not routinely performed, is considered the gold standard for the diagnosis of H. pylori infection. Biopsy specimens are also used to detect the presence of the H. pylori urease. This biopsy urease test (Campylobacter-like organism or CLOtest) is considered rapid and economical, and is in widespread use in clinical practice. Non-invasive tests include detection of the bacterial urease by the 13C- or 14C-urea breath tests, which can be expensive and may involve exposure to radioactivity, and serologic testing of the patient's serum to detect IgG and IgA antibody response to the organism [4-6]. Recently, a rapid EIA to detect H. pylori antigen in stool has also become available [7].
Polymerase chain reaction (PCR) has been demonstrated to be a reliable and highly sensitive tool for detection of bacterial gene sequences in a variety of clinical specimens. Samples used in PCR assays to detect H. pylori include gastric biopsies, saliva, dental plaque, and stool, although the latter has met with mixed success [8-10]. As part of a prospective evaluation of tests used to diagnose H. pylori infection in the pediatric population, we developed a PCR assay to detect H. pylori sequences in gastric biopsy specimens from children undergoing upper endoscopy. Patients included in this study were all assessed for H. pylori infection as part of their physician's suspicion of H. pylori infection or as part of a rule out for another diagnosis. The PCR assay uses a nested set of primer pairs directed against a region of the urease A gene. Results obtained using this assay were compared to those obtained by histologic examination of hematoxylin-eosin stained gastric biopsy specimens, CLOtest, serologic analyses including ELISA and Western Blot, and results of published studies using similar methods.
Gastric mucosal biopsy specimens from pediatric patients were evaluated for the presence of H. pylori by standard histologic techniques, CLOtest, and our nested PCR assay. Serum samples from each patient were tested for antibodies to H. pylori by ELISA and by an in-house Western Blot assay. Our results indicate that while the nested PCR assay correlates well with results obtained using other invasive tests and has the sensitivity and specificity necessary for routine clinical use, results of current serologic methods may not accurately reflect the H. pylori status of pediatric patients.
Results
Complete data for this study was obtained on 101 pediatric patients (49 males, 52 females). Three patients had samples taken from two separate locations at the time of biopsy while one patient underwent endoscopic examination twice in a four month time period, yielding 105 biopsies and 102 serum samples for testing. Of these 105 samples, a total of 34 (32.4%) gave positive results for at least one of the methods examined (Figure 1). Fourteen samples were both CLOtest positive (13.3%) and positive by our nested PCR assay. Fourteen biopsies were positive by histologic staining (13.3%), of which 2 gave a negative result by PCR. A total of 18 samples were positive by the nested PCR assay (17.1%). Patient samples were considered to be positive for H. pylori by PCR amplification if the 279 bp band was seen in the initial reaction and/or the 120 bp was seen in the internal nested reaction. Of the 18 PCR positive samples, 12 showed both the 279 and the 120 bp bands, while the remaining 6 gave a positive reaction only after the internal nested amplification. When invasive methods were considered together, 20 samples were positive by one or more of these tests (19.0%). Twenty-eight (27.4%) of the matching serum specimens on this patient population showed reactivity to H. pylori. Of these, 26 were positive by ELISA, 13 were positive by Western blot, and 11 were positive by both serologic methods. For 14 (13.3%) of the serum samples, serologic testing yielded the only H. pylori positive results. Interestingly, one patient had clearly negative CLOtest and PCR results, while the histologic examination of both of this child's biopsies (obtained from endoscopies performed on two separate dates) showed the presence of organisms consistent with H. pylori. At both examination dates, both ELISA and Western blot on this patient were also positive. Conversely, our PCR assay yielded the only positive result for another patient shown included in the study. Using our nested assay, we were able to detect H. pylori-specific sequences at an estimated concentration of 1 picogram (pg) after just the initial round of amplification. This corresponds to approximately 540 H. pylori bacteria, based on a size of 1.67 Mbp for the complete H. pylori genome [11]. Amplification of the internal gene segment using the nested primer pair further increased the reliable detection limit sensitivity to approximately 30 femtograms (fg), corresponding to about 16 bacterial genomes.
Figure 1.
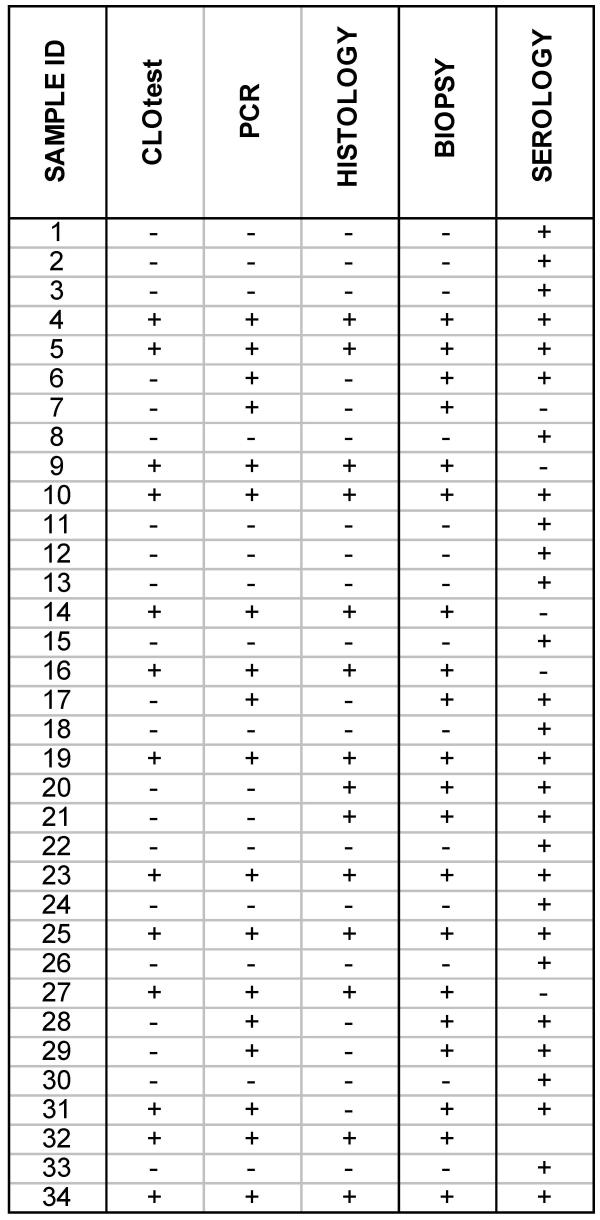
Results of patient testing for H. pylori. Results of all testing (including biopsy-based methods and serology) are shown for the thirty-four samples included in the study which gave at least one positive result. Samples 20 and 21 were biopsies collected during endoscopic examinations performed 4 months apart on the same patient. Matched serum samples for this patient were obtained at each visit. Sample 31 was a biopsy collected from the antrum of a patient undergoing endoscopy, while sample 32 was from the body of the stomach of the same patient. Only one serum sample from this patient was received for analysis. The remaining sixty-seven patients (71 samples) were negative for all assays performed (data not shown). The BIOPSY column includes all specimens for which at least one test result was positive (CLOtest, PCR, or histology). The SEROLOGY column includes all serum samples considered reactive by either H. pylori ELISA (IgG and/or IgA), Western Blot (IgG and/or IgA), or both.
As culture of H. pylori from biopsy specimens was not performed in our study, other methods based on endoscopic examination (specifically CLOtest and histologic staining of the biopsy specimens) were considered the gold standard for determination of the sensitivity and specificity of the PCR and serologic assays. Based on the stated criteria, 12 of the patients examined as part of this study (11.9%) were diagnosed as H. pylori infected. Overall, the invasive methods were shown to be more sensitive and specific than the serologic methods used for this population. The data indicates identical sensitivity and comparable specificity of the nested PCR assay compared to the widely used gold standard methods (Figure 2).
Figure 2.
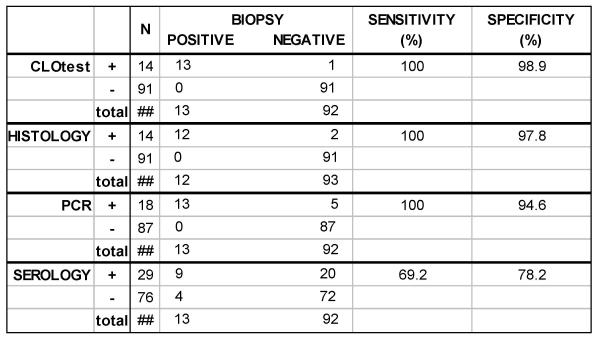
Sensitivity and specificity of the test methods evaluated for detection of H. pylori infection. The sensitivity and specificity of each test method was compared to our accepted biopsy gold standard – patients were considered to be infected with H. pylori if both rapid urease and histology tests were positive.
Discussion
It is generally accepted that most H. pylori infections are acquired in childhood or adolescence, and it has been postulated that infection with this common pathogen at a young age increases the risk of H. pylori-related complications later in life. In light of these issues, accurate diagnosis of infection with H. pylori is important in the symptomatic pediatric patient. In this study, we examined the clinical usefulness of a both a nested PCR assay developed in-house to detect H. pylori sequences in pediatric gastric biopsy specimens, and an in-house Western blot assay for H. pylori. We compared these assays to commonly used FDA approved tests to diagnose infection with this organism.
One hundred and one children undergoing endoscopy were tested using invasive (biopsy-based) methods and non-invasive (serologic) tests. Each of the assays examined has some limitations associated with its use, as evidenced by the results observed for a number of patients included in the study. Of the 101 pediatric patients tested, 12 were considered to be positive based on established criteria, but only 7 of those were positive by all of the methods used. Results observed using these invasive methods could be influenced by a variety of factors, including load and distribution of the organism, virulence and urease production in the infecting strain, and pathologist experience.
In general, CLOtest and histology showed the best correlation and proved to be the most sensitive and specific methods. None of the biopsy samples tested were positive by CLOtest only; all samples positive in the rapid urease test were also PCR positive, and all but one were positive by histological examination. One patient included in the study (each of two samples obtained 4 months apart) was positive only by histology. Matching serum samples were positive at both time points. Nested PCR directed against ureA also showed good correlation with the established biopsy methods. For five of the patients, PCR was the only biopsy-based test yielding positive results, while four of these children were also positive by serologic methods. Our data indicate that our nested assay has comparable sensitivity (100%) and specificity (94.6%) to the biopsy-based methods currently available, results in line with previously published reports for molecular diagnostic methods [9,10,12].
Review of the current literature reveals that in general, commercially available serologic assays for diagnosis of H. pylori are limited in their ability to reliably detect the infection in children [3,12]. Our data appears to support this, as the commercial serologic assay in use in our laboratory was shown to be less sensitive (46% of CLOtest and histology positives were identified as such) in our patient population compared to the invasive methods studied. In our patient population, immunologic reactivity to H. pylori was seen in 28 patients, however only 9 of those met the established criteria selected for documented infection. In four cases, all testing on biopsy specimens gave positive results, but the corresponding serologic assays were shown to be negative. Fourteen of the serums that showed reactivity matched to biopsy samples proven to be negative, further supporting the use of established invasive methods for accurate detection. The Western blot assay appeared to yield improved results, detecting 69.2% of the defined positives as such with 4 (31%) probable false positives, versus 14 (54%) for the commercial ELISA.
PCR is currently considered a research tool in the H. pylori arena, although a number of studies have been performed using primers directed against a variety of bacteria-specific sequences and several different sample types from saliva to stool. Our data suggests that our in-house nested assay would be an accurate and reliable tool for the detection of H. pylori infection in the pediatric population. In addition, our simplified boiling method alleviates the need for expensive and time-consuming DNA isolation procedures while still allowing detection of specific bacterial sequences. Although at this time this assay is based on an invasive diagnostic procedure, further studies need to be performed to validate its use in a non-invasive manner, while maintaining levels of sensitivity and specificity.
Conclusions
◆ Results of the nested PCR assay for detection of the Helicobacter pylori urease A gene in pediatric gastric biopsies show good correlation with results obtained from CLOtest slides and histologic examination.
◆ The data show a statistically significant difference between number of positive results obtained through invasive procedures (biopsy followed by CLOtest, histology, and PCR) and those obtained using serology (p = 0.038).
◆ Compared to CLOtest and histology as the standard for proven Helicobacter pylori infection, the nested PCR assay was shown to have a sensitivity of 100% and a specificity of 94.6%. These results suggest that the nested assay would be a reliable tool for clinical use in the detection of Helicobacter pylori infections.
Methods
Clinical Specimens
Gastric Biopsy Tissue
A total of 105 biopsy specimens from 101 pediatric patients (49 males, 52 females) undergoing upper endoscopy at the A. I. DuPont Hospital for Children were obtained from the Gastroenterology Laboratory. Specimens were received in CLOtest slides (Ballard Medical Products, Draper, UT), the CLOtest result was recorded, and samples were processed for PCR analysis. Briefly, biopsy tissue was removed from the gel matrix of the slide and washed three times with sterile distilled water. A final aliquot of distilled water was added and samples were boiled for 10 minutes. Samples were centrifuged to collect the tissue and the supernatant was removed to a sterile microcentrifuge tube for use in PCR analysis. All samples were stored at -70°C until tested.
Serum
Serum samples for H. pylori testing were received in the Immunology Laboratory at the time of endoscopy or within one month of the procedure. Serologic detection of H. pylori-specific IgG and IgA antibodies was performed using both a commercially available ELISA kit and an in-house Western Blot method.
ELISA
ELISA tests were performed using the GAP-IgG and GAP-IgA ELISA test kits (Biomerica, Newport Beach, CA) according to the manufacturer's instructions.
Western Blot
For in-house production of the Western Blot test strips, H. pylori (ATCC strain 43504) grown on Skirrows medium (BD Biosciences, Sparks, MD) was harvested, washed, and resuspended in PBS with 5 mM MgCl2. Total protein concentration was determined using a Bradford microassay (BioRad, Richmond, CA). Harvested bacteria were diluted to a concentration of 4.0 mg/ml in sample buffer containing 0.1 M dithiothreitol and electrophoresed in 11% denaturing polyacrylamide gels using a modification of the procedure of Laemmli [13]. Separated antigens were transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH) that were subsequently blocked in PBS containing 0.5% BSA, dried, cut into strips, and stored dessicated until testing was performed. Sera were tested for IgG and IgA antibodies by incubating the strips with a 1:200 dilution of test serum in PBS containing 1% nonfat dry milk for 1 hour at room temperature. Strips were washed and then incubated with a 1:1000 dilution of biotinylated antisera (goat anti-human IgG or goat anti-human IgA; Kirkegaard and Perry Laboratories, Gaithersburg, MD) for 1 hour at room temperature. The strips were washed again and incubated with a 1:1000 dilution of peroxidase-conjugated streptavidin (Kirkegaard and Perry Laboratories) for 1 hour. Strips were washed and incubated with 4-chloro-1-napthol substrate (7.8 mM 4-chloro-1-napthol diluted 1:2 with a 1:1500 dilution of 30% H2O2 in citrate phosphate buffer). The reaction was stopped with distilled water, the strips were air-dried, and banding patterns were evaluated.
Polymerase chain reaction (PCR)
PCR was carried out in a Perkin Elmer 9600 thermocycler (Foster City, CA) using 10 μl boiled supernatant (prepared as described above) as the sample in each 100 μl reaction. Control reactions contained 10 ng purified H. pylori DNA (ATCC strain 43504) in a 100 μl reaction. Oligonucleotide primers used were purchased from Integrated DNA Technologies (Coralville, IA). Oligonucleotides HpF1 (5' GATAAG TTGATGCTCCA CTACGCTG 3') and HpB25 (5' CTCAATAGG GGTATGCACG GTTAC 3') were used to amplify a 279 nucleotide (nt) product of the H. pylori urease A gene. Oligonucleotides HpF2 (5' AAGCAGTAGCTTTGATTAGTG CCC 3') and HpB5 (5'CGCCATCCATC ACATCATCTG 3') are internal to HpF1 and HpB25 in the urease A gene and were used in a secondary reaction to amplify a 120 nt product from the 279 nt primary amplification product. For both amplifications, cycling conditions were as follows: initial denaturation at 95°C for 5 minutes, followed by 34 cycles of denaturation at 94°C for 1 minute, annealing at 53°C for 1 minute, and extension at 94°C for 1 minute. A final extension cycle was added, increasing the 72°C incubation to 5 minutes. Following amplification, 20 μl samples of each reaction were mixed with 5 μl gel loading buffer (Sigma Biochemical, St. Louis, MO) and electrophoresed in 3.0% Nusieve 3:1 agarose (FMC Bioproducts, Rockland, ME) gels.
Statistical analysis
Because culture of H. pylori (the generally accepted gold standard) was not included in this study, statistical analysis was limited to comparison of results obtained through invasive methods (biopsy followed by CLOtest, histology, and PCR) and those obtained using serology.
Author's contributions
PTF conceived of the study and was primarily responsible for study design. KMBV participated in study design and data analysis, designed the nested PCR assay, carried out the molecular analyses, and drafted the manuscript. KMG participated in study design, data analysis, and serologic assays. RP participated in study design. All authors participated in review and preparation of the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank Drs. Uwe Blecker and Christopher Justinich of the Gastroenterology Department for performing the chart reviews on our patient population.
Contributor Information
Kathleen MB Vinette, Email: kvinette@nemours.org.
Kathleen M Gibney, Email: Kathleen_Gibney@Dadebehring.com.
Roy Proujansky, Email: rproujan@nemours.org.
Paul T Fawcett, Email: pfawcett@nemours.org.
References
- Warren JR. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273. doi: 10.1016/S0140-6736(83)92719-8. [DOI] [PubMed] [Google Scholar]
- Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- Czinn S. Serodiagnosis of Helicobacter pylori in pediatric patients. J Pediatr Gastroenterol Nutr. 1999;28:132–134. doi: 10.1097/00005176-199902000-00006. [DOI] [PubMed] [Google Scholar]
- NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- CLOtest package insert. Ballard Medical Products. 1997.
- Fennerty MB. Helicobacter pylori. Arch Intern Med. 1994;154:721–727. doi: 10.1001/archinte.154.7.721. [DOI] [PubMed] [Google Scholar]
- Vaira D, Malfertheiner P, Megraud F, Axon ATR, Deltenre M, Hirschl AM, Gasbarrini G, O'Morain C, Pajares Garcia JM, Quina M, Tytgat GNJ, and the HpSA European Study Group Diagnosis of Helicobacter pylori infection with a new non-invasive antigen-based assay. Lancet. 1999;354:30–33. doi: 10.1016/S0140-6736(98)08103-3. [DOI] [PubMed] [Google Scholar]
- Clayton CL, Kleanthous H, Coates PJ, Morgan DD, Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol. 1992;30:192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ha T, Ferguson DA, Chi DS, Zhao R, Patel NR, Krishnaswamy G, Thomas E. A newly developed PCR assay of Helicobacter pylori in gastric biopsy, saliva, and feces: Evidence of high prevalence in saliva supports oral transmission. Dig Dis Sci. 1996;41:2142–2149. doi: 10.1007/BF02071393. [DOI] [PubMed] [Google Scholar]
- Bickley J, Owen RJ, Fraser AG, Pounder RE. Evaluation of the polymerase chain reaction for detecting the urease C gene of Helicobacter pylori in gastric biopsy samples and dental plaque. J Med Microbiol. 1993;39:338–344. doi: 10.1099/00222615-39-5-338. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson , Khalak HG, Glodek A, McKenney K, Fitzgerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Whattey L, Wallin W, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;338:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Chong SKF, Lou Q, Asnicar M, Zimmerman SE, Croffie JM, Lee CH, Fitzgerald JF. Helicobacter pylori infection in recurrent abdominal pain in childhood: comparison of diagnostic tests and therapy. Pediatrics. 1995;96:211–215. [PubMed] [Google Scholar]
- Fawcett PT, Gibney KM, Vinette KMB. Helicobacter pylori can be induced to assume the morphology of Helicobacter heilmannii. J Clin Microbiol. 1999;37:1045–1048. doi: 10.1128/jcm.37.4.1045-1048.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


