Abstract
Two frequent single-nucleotide-polymorphisms (SNPs) are present in the indoleamine 2,3-dioxygenase 2 (IDO2) gene that influence its enzymatic activity. Thus, one SNP (R248W) is associated with a reduction in IDO2 catalytic activity, whereas the other SNP (Y359stop) generates a premature stop codon abolishing activity completely. In the present study, we describe the presence of a specific cellular immune response in the periphery which correlated with the functional status of the IDO2 protein. Hence, the induction of IDO2-specific T cells in peripheral blood requires the presence of a functional IDO2 protein and, consequently, is restricted to individuals that are not homozygous for the stop codon. Furthermore, we detected stronger T-cell responses in donors with the homozygous Y wild type at position 359 when compared with the heterozygous genotype. Interestingly, we found a higher number of immune responses against IDO2 in patients homozygous for the 248W giving reduction in IDO2 activity compared with the 248R. Hence, spontaneous immune responses against IDO2 seem to be correlated with reduced enzymatic activity of IDO2. The patient IDO2 genotype may well influence the outcome of IDO2-based anti-cancer vaccination.
Keywords: IDO2, SNP, T-cell antigen, genotype, spontanous immune responses
Introduction
Indoleamine 2,3-dioxygenase (IDO) is an immunoregulatory enzyme that can foster an immunotolerant environment. Hence, IDO has been implicated in suppressing T-cell immunity in normal and pathological settings, including cancer.1 Several studies have shown that indoleamine 2,3-dioxygenase (IDO) functions in facilitating conversion of naïve T lymphocytes into T regulatory cells.2 Consistent with a role for IDO in mediating tolerance to tumor, preclinical studies have shown the promise of IDO inhibitors in targeting several cancers.3-7 Especially, 1-methyl-tryptophan (1MT) has been widely studied as an inhibitor of IDO activity. Interestingly, recent studies have shown that the racemer D-1MT has superior antitumor activity compared with L-1MT.8 The D-1MT preferentially target a paralog of IDO named indoleamine 2,3-dioxygenase-2 (IDO2).9 Several lines of data have suggested a possible link between the IDO-like protein IDO2 and cancer. First, IDO2 expression has been described in human tumors, including renal and pancreatic tumors. In pancreatic tumors IDO2 expression was described both at the level of the tumor cell as well as immune cells in tumor-draining lymph nodes.10 Second, the apparent selective inhibition of IDO2 by the D stereoisomer of the IDO blocker 1MT, which tends to be more active than the L-isomer in a variety of biological assays for IDO function, suggests that IDO2 may be important to sustain immune escape and growth of tumors.8 Especially, D-1MT heighten chemotherapeutic efficacy in mouse models of cancer in a nontoxic fashion.
Two frequent single-nucleotide-polymorphisms (SNPs) have been revealed in the IDO2 gene. Thus, one SNP (R248W) was associated with a reduction in IDO2 catalytic activity, whereas the other SNP (Y359stop) generates a premature stop codon abolishing activity completely.9 It was recently described that IDO2-reactive T cells were present in peripheral blood of cancer patients and that these T cells were able to recognize and kill tumor cells.11 In the current study, we examined the association between the presence of spontaneous IDO2-specific T cells and the functional status of the IDO2 protein based on genotyping of the two IDO2 SNPs. Our data demonstrate that spontaneous cytotoxic T-cell reactivity against IDO2 requires a full-length IDO2 protein.
Results
Genotyping
To investigate the association between spontaneous IDO2-specific T-cell responses and the functional status of the IDO2 protein, we genotyped DNA from HLA-A2+ individuals for the two SNPs: Y359stop and R248W using RT-PCR and sequencing. We genotyped 21 healthy donors, 12 malignant melanoma patients, 5 renal cell carcinoma patients and 6 breast cancer patients for the SNP Y359stop. For the SNP R248W we genotyped 19 healthy donors, 12 malignant melanoma patients, 5 renal cell carcinoma patients and 6 breast cancer patients. The combined genotypes are shown in Table 1. The number of individuals found in the combined groups did not seem to be independently distributed. Thus, some groups had a limited number of individuals. In particular, no individuals were homozygous on both SNPs for the stop/decreased activity alleles (combined genotype of 359 stop/stop and 248 low/low) and just 2 out of 42 individuals had a genotype encoding a fully active IDO2 protein (359 wt/wt and 248 wt/wt).
Table 1. Distribution of Y359stop and R248W genotypes. The table shows distribution of the Y359stop allele and R248W allele and the association of the two. p value calculated by Spearman rank correlation.
| Y359stop | R248W | ||
|---|---|---|---|
| 359 wt/wt |
21 (48%) |
248 wt/wt |
7 (17%) |
| 359 wt/stop |
17 (39%) |
248 wt/low |
23 (55%) |
| 359 stop/stop | 6 (14%) | 248 low/low | 12 (29%) |
| Combined genotypes | 248 wt/wt | 248 wt/low | 248 low/low |
|---|---|---|---|
| 359 wt/wt |
2 |
13 |
6 |
| 359 wt/stop |
2 |
8 |
6 |
| 359 stop/stop |
3 |
2 |
0 |
| p = 0.13 |
Association between IDO2 genotype and IDO2 specific T-cell responses
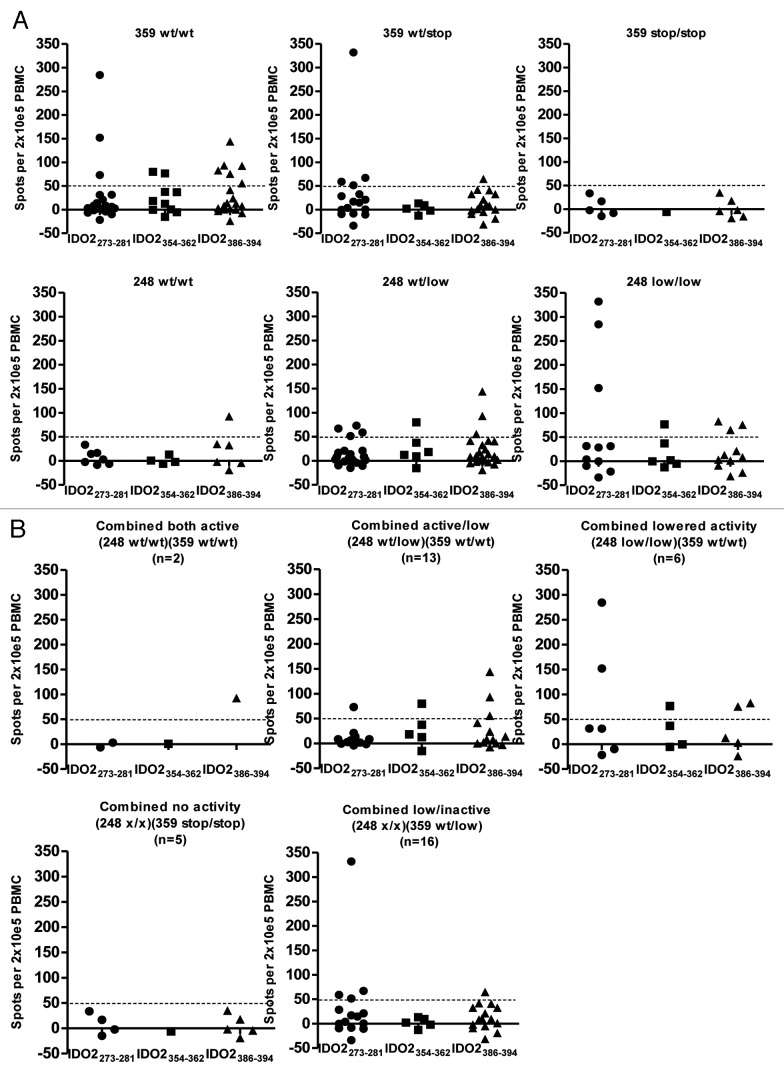
ELISPOT assays were performed to detect specific T cells against three IDO2-derived peptide epitopes: IDO2273–281, IDO2354–362 and IDO2386–394. Positive T-cell responses against the peptides were defined as > 50 spots/2 × 105 PBMC. We found that T-cell responses against the 3 IDO2 epitopes were restricted to individuals being SNP Y359stop homozygous or heterozygous wildtype (359 wt/wt or 359 wt/stop) (Fig. 1A). Thus, we did not detect any responses in the group with the 359 stop/stop genotype for any of the examined epitopes. Individuals being homozygous Y359 wild type (359 wt/wt) had a significantly higher number of responses against peptide IDO2386–394 (located down-stream of the Y359stop SNP) than individuals having a heterozygous wild type (359 wt/stop) or homozygous stop (359 stop/stop) genotype (p = 0.04). Furthermore we found a significant association between the numbers of T-cell responses in each of the three genotypes by comparing them in an ordered manner to the number of responses (p = 0.04). Hence, more responses were detected in the donors with the homozygous wildtype (359 wt/wt) genotype than the heterozygous genotype (359 wt/stop). Likewise, more responses were detected in the donors with the heterozygous (359 wt/stop) genotype than the homozygous stop (359 stop/stop) (Fig. 1A).
Figure 1.
Association between IDO2 genotype and T-cell responses. (A) T-cell responses against 3 peptides from IDO2 (IDO2273–281, IDO2354–362, IDO2386–394) as measured by indirect ELISPOT are shown for individuals grouped according to their SNP Y359stop (upper row) and SNP 248W (lower row) genotype. Responses were only found in individuals having homozygous or heterozygous wildtype genotype (359 wt/wt or 359 wt/stop). Homozygous 359 wild type Y/Y (wt/wt; upper left), heterozygous Y/STOP (wt/stop; upper middle) and homozygous STOP/STOP (stop/stop; upper right). Homozygous R248 wild type R/R (wt/wt, lower left), heterozygous R/W (wt/low, lower middle) and homozygous W/W (low/low, lower right). (B) T-cell responses against three peptides from IDO2 (IDO2273–281, IDO2354–362, IDO2386–394) are shown for individuals grouped according to their combined genotype of the two polymorphic SNP variants. Two out of forty-two individuals were Y359 wt/wt – R248 wt/wt (upper left). None was Y359 stop/stop–R248 low/low (data not shown).
T-cell responses against peptide IDO2273–281 (located up-stream of the Y359stop SNP) were equally only found in individuals being homozygous Y359 wild type or heterozygous. Two T-cell responses out of 16 tested were detected against the third epitope; IDO2354–362 (this peptide spans the position of Y359stop SNP) and both were detected in individuals being homozygous Y359 wildtype (359 wt/wt). However, more individuals need to be analyzed to firmly conclude on the association between the IDO2 genotype and the T-cell responses against this peptide.
For the SNP R248W the C genotype gives rise to the amino acid Arginine (R) (248wt) whereas the T genotype gives rise to the amino acid Tryptophan (W) (248low). We found that 7 T-cell responses out of 28 tested (~25%) against the 3 IDO2 peptides were detected in the homozygous low group; 248 low/low, 8 out of 49 tested (~16%) were found in the 248 wt/low group, and 1 out of 17 tested (~5%) was detected in the 248 wt/wt group (Fig. 1A).
Next, we investigated the association between IDO2 T-cell responses and the combined genotype of the two IDO2 SNP variants. As previously mentioned the number of individuals found in the combined groups did not seem to be independently distributed. Thus, only 2 out of 42 individuals were homozygous for both functional alleles (248 wt/wt/359 wt/wt) (Fig. 1B), and no individuals were homozygous for the non-functional alleles (248 low/low and 359 stop/stop) (data not shown).
Immune responses in healthy donors and cancer patients
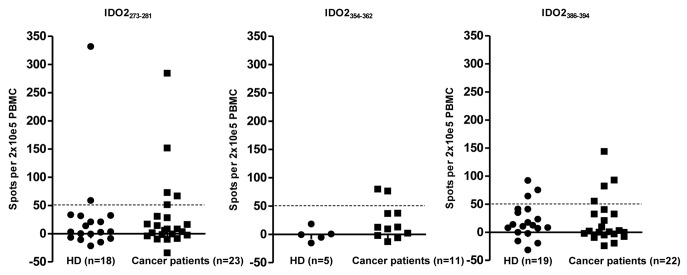
We analyzed the relevance of the measured T-cell responses in healthy donors (n = 21) and cancer patients (n = 23) separately. It seemed that the natural immunity against IDO2 was lower in healthy individuals compared with cancer patients (Fig. 2) although one healthy donor (248 low/low, 359 wt/stop) had a strong response against IDO2273–281.
Figure 2.
T-cell responses against IDO2 in healthy donors vs. cancer patients. T-cell responses against peptides IDO2273–281 (left), IDO2354–362 (middle) and IDO2386–394 (right) are shown for healthy donors (HD) and cancer patients (Cancer patients). T-cell immunity toward IDO2 was seemingly lower in healthy donors compared with cancer patients.
Association between IDO2 genotype and virus specific T-cell responses
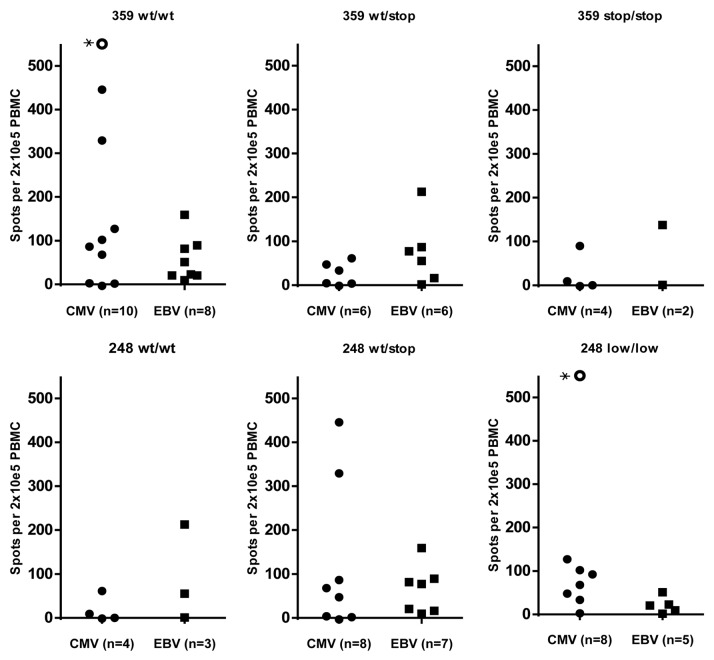
To investigate whether the association between IDO2 genotype and specific T-cell responses has any relevance to T-cell responses against alternative antigens, we performed direct ELISPOT to detect T cells against the well characterized HLA-A2 restricted peptide pp65 from CMV (NLVPMVATV) and the peptide from BMLF1 from EBV (GLCTLVAML). The presence of EBV specific T cells in the periphery did not seem to be associated with IDO2 SNP genotype as specific T cells were detected in all genotype groups (Fig. 3). However, notably the strongest CMV responses were detected in donors having a homozygous wildtype (359 wt/wt) IDO2.
Figure 3.
IDO2 genotype and virus specific T-cell responses. T-cell responses against HLA-A2 restricted peptides from CMV (NLVPMVATV) and EBV (GLCTLVAML) were detected by use of direct ELISPOT. The strongest CMV specific T-cell responses seemed to be in the genotype group homozygous 359 wild type (359 wt/wt). *In this donor the number of spots was estimated to be > 1,000.
Discussion
Emerging evidence suggests that during cancer progression activation of the IDO pathway might act as an important modifier pathway for immune escape.12 Small molecule inhibitors of IDO and IDO2 heighten chemotherapeutic efficacy in mouse models of cancer in a nontoxic fashion.13 Hence, the targeting of these proteins offers promising ways to broaden the combinatorial attack on advanced cancers, where immune escape mechanisms likely provide pivotal support. Furthermore, it was recently shown that IDO2-specific T cells were present in the peripheral blood and that they were able to recognize and kill tumor cells.11 Interestingly, the coding region of the human IDO2 gene includes two nonsynonymous SNPs that strongly ablate enzymatic activity. The use of IDO2 as a target in the clinic will be restricted to patients having the active genotype. One SNP (R248W) has been associated with a reduction in IDO2 catalytic activity, whereas the other SNP (Y359stop) generates a premature stop codon abolishing activity completely.14
In the present study, we investigated the association between a specific cellular immune response and the functional status of the IDO2 protein as based on IDO2 SNP genotyping. Several HLA-A2-restricted IDO2-derived T-cell epitopes have been described. We examined the apparently most immunogenic epitope IDO2273–281, which are located before the Y359stop SNP, an additional epitope spanning the Y359stop SNP (IDO2354–362) and finally an epitope IDO2386–394 located down-stream the Y359stop SNP. We found that specific T cells against the three different HLA-A2-restricted peptides from IDO2 were restricted to individuals being either homozygous or heterozygous wildtype for the Y359stop SNP (359 wt/wt or 359 wt/stop). No T-cell responses were detected in individuals being homozygous for the 359 stop allele (359 stop/stop). Hence, these findings suggest that the induction of IDO2-specific T cells in peripheral blood requires the presence of a functional IDO2 protein and, consequently, is restricted to individuals that are not homozygous for the stop codon. Furthermore, there was a tendency of a higher number of T-cell responses within the homozygous wildtype genotype when compared with the heterozygous genotype. This could be due to a higher level of expression of IDO2 in these individuals.
Interestingly, when looking at the other SNP (R248W) we found that the immune responses seemed higher for the most immunogenic IDO2-derived epitope (IDO2273–281) in individuals homozygous for the SNP allele (248 low) giving reduction in IDO2 activity. Hence, spontaneous immune responses against IDO2 depend on the generation of a functional target protein but seem, in addition, to be associated with reduced enzymatic activity of IDO2. This could suggest that the enzymatic activity of IDO2 influences the systemic adaptive immune response and, furthermore, that the immune suppressive effects of IDO2 expression in the target cells suppress the cytotoxic T-cell responses generated against this cancer antigen.
We additionally analyzed the association between IDO2 genotype and T-cell responses toward virus antigens. The strongest CMV responses were detected in donors who were homozygous for the IDO2 359 wt/wt genotype, which could suggest a correlation between IDO2 and CMV responses. Although this needs to be verified, we have previously found that the presence of IDO-specific T-cell responses correlated with the presence of CMV-responses.
The number of IDO2-specific T-cell responses found in cancer patients and healthy donors did not differ significantly although responses appeared to be stronger among cancer patients. IDO2-specific T cells have been described to recognize and kill tumor cells. IDO2 might—especially considering the immune suppressive functions of IDO—be a useful target for anti-cancer immunotherapeutic strategies. However, since the spontaneous immune responses against IDO2, as described here, were depending on the IDO2 genotype, the patient IDO genotype might likewise influence the potential outcome of such clinical measures.
Materials and Methods
Donors
Peripheral blood mononuclear cells (PBMC) were collected from healthy individuals and cancer patients (renal cell carcinoma, melanoma and breast cancer). Blood samples from cancer patients were drawn a minimum of four weeks after termination of any kind of anti-cancer therapy. The majority of renal cell carcinoma patients had previously been treated with IL-2 and INFα, most melanoma patients had received high dose IL-2 and INFα, while the breast cancer patient was pre-treated with several kinds of chemotherapy, (e.g., epirubicin, docetaxel and cabecitabine), trastuzumab and/or endocrine therapy. PBMC were isolated using lymphoprep separation, HLA-typed (Department of Clinical Immunology, University Hospital, Copenhagen, Denmark) and frozen in FCS with 10% DMSO. Informed consent was obtained from the patients prior to any of these measures.
Peptides
Three synthetic HLA-A2 restricted IDO2-derived peptides were synthesized for ELISPOT (TAG Copenhagen, Copenhagen, Denmark. IDO2273–281 (VLHAFDEFL), IDO2354–362 (YLITAAAKA), IDO2386–394) and IDO2386–394 (AVMSFLKSV).Two HLA-A2 restricted virus peptides were used in ELISPOT: CMV pp65 (NLVPMVATV) and EBV BMLF1 (GLCTLVAML).
ELISPOT assay
The ELISPOT assay was used to quantify peptide-specific INFγ releasing effector cells as described.15 For analyses of IDO2 specific T-cell responses, PBMC were stimulated once in vitro with peptide prior to analysis as described16 to extend the sensitivity of the assay. After seven days in culture in X-vivo15TM (BioWhittaker) medium with 25 µg/ml peptide and 20 U/ml IL-2 (PeproTech), cells were counted and analyzed in ELISPOT. CMV and EBV-specific T cells were detected directly ex vivo with no prior stimulation and for these analyses PBMC were thawn and rested overnight before being counted and analyzed in ELISPOT. Briefly, nitrocellulose bottomed 96-well plates (MultiScreen MAIP N45; Millipore) were coated overnight with INFγ capture mAb (Mabtech). The wells were washed, blocked by X-vivo15 medium. Cells were added in duplicates at different cell concentrations, with (experimental wells) or without (control wells) 5 µg/ml peptide. The plates were incubated overnight. The following day, medium was discarded and the wells were washed prior to addition of appropriate biotinylated secondary mAb (Mabtech). The plates were incubated at room temperature (RT) for 2 h, washed, and Avidin-enzyme conjugate (AP-Avidin; Calbiochem/Invitrogen Life Technologies) was added to each well. Plates were incubated at RT for 1 h and the enzyme substrate NBT/BCIP (Invitrogen Life Technologies) was added to each well and incubated at RT for 5–10 min. Upon the emergence of dark purple spots, the reaction was terminated by washing with tap water. The spots were counted using the ImmunoSpot Series 2.0 Analyzer (CTL Analyzers). For indirect ELISPOT: positive responses were defined as (mean number of spots in experimental wells) - (mean number of spots in control wells) > 50 spots/2 × 105 PBMC as previously.11 In general, direct ELISPOT did not give any background spots.
Genotyping
Genotyping was performed by polymerase chain reaction (PCR) and subsequent sequencing. We isolated genomic DNA using the NucleoSpin© tissue kit (Macherey-Nagel, Germany) and performed PCR amplifications of the Y359Stop (forward primer: 5′-TAGAAGACATCCACTCAGCACC-3′, reverse primer: 5′-CGTGGGTGAAGGATTGACTC-3′) and R248W (forward primer: 5′-TGCAGCTTTCATTCCAGGTCTCCA-3′ and the reverse primer: 5′-CTGAGAGTGGATCCCTAGCAAGAGT-3′) polymorphisms.9 Amplifications were performed in a total volume of 15 µl containing 1× PCR buffer [50 mM KCl, 20 mM Tris pH 8.4, 2.0 mM MgCl2, 0.2 mM cresol red, 12% sucrose, 0.005% (wt/v) BSA (Boehringer-Mannheim)], 5 pmol of each primer, 40 mM dNTPs (Pharmacia LKB, Sweden) and 1.25 U of AmpliTaq polymerase (Perkin Elmer Cetus Corporation). Conditions used for amplification were 40 cycles at 94°C for 2 min, 94°C for 30 sec, 65°C for 60 sec, and 72°C for 60 sec. Sequencing was performed using the BigDye® Terminator v3.1 Cycle Sequencing Kit (sequencing primer for Y359stop: 5′-ACGTGGGTGAAGGATTGACTC-3′ and R248W: 5′-TGCAGCTTTCATTCCAGGTCTCCA-3′ (forward primer)) and analyzed by a 3500 Genetic Analyzer (Applied Biosystems).
Statistics
Fisher’s exact test was used for comparing categorical variable ie genotypes and ELISPOT responses. When analyzing genotype as an ordinal variable spearman rank correlation was used.
Due to the sample size exact or Monte Carlo methods were used for calculating p values.
All p values were two-sided and p-values below 0.05 were considered significant. All calculation were done with R statistical software version 2.12.1 (R Foundation for Statistical Software, 2011). Calculations using exact and Monte Carlo methods were done with the “coin” package.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We would like to thank James DuHadaway and George C. Prendergast for kindly providing us with primer sequences for genotyping of the SNP R248W. In addition, we thank Merete Jonassen and Tina Seremet for excellent technical assistance. Grant support: The Danish Cancer Society, Novo Nordisk Foundation, Danish Medical Research Council, Lundbeck Foundation, and Herlev Hospital.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/19654
References
- 1.Prendergast GC. Immune escape as a fundamental trait of cancer: focus on IDO. Oncogene. 2008;27:3889–900. doi: 10.1038/onc.2008.35. [DOI] [PubMed] [Google Scholar]
- 2.Prendergast GC, Metz R, Muller AJ. IDO recruits Tregs in melanoma. Cell Cycle. 2009;8:1818–9. doi: 10.4161/cc.8.12.8887. [DOI] [PubMed] [Google Scholar]
- 3.Muller AJ, Prendergast GC. Indoleamine 2,3-dioxygenase in immune suppression and cancer. Curr Cancer Drug Targets. 2007;7:31–40. doi: 10.2174/156800907780006896. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee T, Duhadaway JB, Gaspari P, Sutanto-Ward E, Munn DH, Mellor AL, et al. A key in vivo antitumor mechanism of action of natural product-based brassinins is inhibition of indoleamine 2,3-dioxygenase. Oncogene. 2008;27:2851–7. doi: 10.1038/sj.onc.1210939. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Jaller D, Patel B, LaLonde JM, DuHadaway JB, Malachowski WP, et al. Structure based development of phenylimidazole-derived inhibitors of indoleamine 2,3-dioxygenase. J Med Chem. 2008;51:4968–77. doi: 10.1021/jm800512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S, Malachowski WP, DuHadaway JB, LaLonde JM, Carroll PJ, Jaller D, et al. Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based inhibitors. J Med Chem. 2008;51:1706–18. doi: 10.1021/jm7014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metz R, Duhadaway JB, Rust S, Munn DH, Muller AJ, Mautino M, et al. Zinc protoporphyrin IX stimulates tumor immunity by disrupting the immunosuppressive enzyme indoleamine 2,3-dioxygenase. Mol Cancer Ther. 2010;9:1864–71. doi: 10.1158/1535-7163.MCT-10-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- 9.Metz R, Duhadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res. 2007;67:7082–7. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- 10.Witkiewicz AK, Costantino CL, Metz R, Muller AJ, Prendergast GC, Yeo CJ, et al. Genotyping and expression analysis of IDO2 in human pancreatic cancer: a novel, active target. J Am Coll Surg. 2009;208:781–7, discussion 787-9. doi: 10.1016/j.jamcollsurg.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sørensen RB, Køllgaard T, Andersen RS, van den Berg JH, Svane IM, Straten P, et al. Spontaneous cytotoxic T-Cell reactivity against indoleamine 2,3-dioxygenase-2. Cancer Res. 2011;71:2038–44. doi: 10.1158/0008-5472.CAN-10-3403. [DOI] [PubMed] [Google Scholar]
- 12.Prendergast GC, Metz R, Muller AJ. Towards a genetic definition of cancer-associated inflammation: role of the IDO pathway. Am J Pathol. 2010;176:2082–7. doi: 10.2353/ajpath.2010.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–9. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 14.Witkiewicz AK, Costantino CL, Metz R, Muller AJ, Prendergast GC, Yeo CJ, et al. Genotyping and expression analysis of IDO2 in human pancreatic cancer: a novel, active target. J Am Coll Surg. 2009;208:781–7, discussion 787-9. doi: 10.1016/j.jamcollsurg.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen MH, Pedersen LO, Becker JC, Straten PT. Identification of a cytotoxic T lymphocyte response to the apoptosis inhibitor protein survivin in cancer patients. Cancer Res. 2001;61:869–72. [PubMed] [Google Scholar]
- 16.McCutcheon M, Wehner N, Wensky A, Kushner M, Doan S, Hsiao L, et al. A sensitive ELISPOT assay to detect low-frequency human T lymphocytes. J Immunol Methods. 1997;210:149–66. doi: 10.1016/S0022-1759(97)00182-8. [DOI] [PubMed] [Google Scholar]