Abstract
A cancer immune signature implicating good prognosis and responsiveness to immunotherapy was described that is observed also in other aspects of immune-mediated, tissue-specific destruction (TSD). Its determinism remains, however, elusive. Based on limited but unique clinical observations, we propose a multifactorial genetic model of human cancer immune responsiveness.
Keywords: cancer genetics, cancer microenvironment, immune responsiveness, immunotherapy, melanoma
…answers preexist, and it is the question that needs to be discovered.
—Jonas Salk1
Introduction
Among the infinite permutations that deductive thinking applied to the current understanding of biology may conceive, only a few materialize because Nature adopts a handful to achieve its goals.2 For instance, a deductive approach could hypothesize an infinite number of limbs protruding from a mammalian body, but in reality only four are observed as evolution decided long ago that this is quite suitable for most creatures’ survival. An immunologic example is the realization by Hancock et al.3 that, “although almost every known chemokine and chemokine receptor is expressed at some stage during development of allograft rejection, mechanistic studies indicate that the actual key effector mechanisms are rather few. Thus, in vivo studies have alleviated concerns regarding possible biological redundancy and the pleiotropic effects of these molecules, and have resulted in a focus on CXCR3, CCR5 and their respective ligands as key mediators of host alloresponses, especially in acute rejection.” Interestingly, a bottom up, inductive approach applied to the study of human tissues undergoing rejection reached the same conclusion without need to test an infinite number of conceivable combinations.4-6 For this reason, we advocated that an inductive approach based on human observation may alleviate the daunting task of predicting in the laboratory what can be directly observed in nature.2 Following this observational approach, various groups concluded that human cancer biology can be simplified according to recurring immunologic themes.7 An observation pertinent to this manuscript suggests that some cancers display an effector immune phenotype associated with good prognosis.8 Concurrent with this observation, is the realization that a similar footprint is observable in cancers likely to regress in response to active-specific vaccination,9 treatment with systemic interleukin (IL)-210 or other types of immune therapy.7 Paradoxically, since these signatures are observed in growing cancers, they are not sufficient to clear entirely the organism of neoplastic cells but they sustain, as in chronic infections, a self-perpetuating inflammatory process that may slow cancer growth naturally and predispose to cancer rejection during immunotherapy.6 More broadly, the same signatures are observed in tissues undergoing other types of immune-mediated tissue-specific destruction (TSD) such as flares of autoimmunity, killing of pathogen-infected cells during acute infection, allograft rejection and graft-vs. host disease.5 The convergence of these phenomena into a common final pathway suggests that TSD is a conserved mechanism mediated by the activation of a specific set of genes that we named the immunologic constant of rejection (ICR).4 We suggested that the ICR includes at least four components: the activation of the signal transducer and activator of transcription (STAT)-1/interferon regulatory factor (IRF)-1 pathway resulting in the activation of interferon (IFN)-γ stimulated genes (ISGs), the expression of immune effector (IEFs) mechanisms and the CXCR3 and the CCR5 ligand chemokines.5 While the common pathway to rejection (how does rejection occur) is becoming progressively accepted, the factors determining its occurrence in individual cases (why does rejection occur) remain elusive. It is likely that while TSD results from a common pathway depending upon the activation of ICR genes, the upstream factors leading to TSD in individual cases are heterogeneous and disease-specific. Consequently, it is likely that the genetic and environmental factors predisposing to distinct immune pathologies are different and the genesis of TSD is multifactorial; conditions leading to TSD may be overlapping, mutually exclusive or synergist making the algorithm leading to rejection difficult to identify and unyielding to univariate comparisons as combinations rather that individual factors may be at the basis of the phenomenon. This view point will summarize our efforts to apply a discovery-driven, systematic approach to the identification of determinants of cancer immune responsiveness.
The Natural History of Cancers is Linked to its Immune Phenotype
The connotation of lymphocytic infiltrates as a favorable prognostic biomarker in cancer was originally reported by Cochran et al.11 in 1969; however, much progress has been made in the last decade in particular by Je´rôme Galon who characterized extensively the type and functional status of immune infiltrates associated with good prognosis in colon cancer.7,8 In particular, the expression of genes associated with a Th1 effector T-cell response was observed to bear superior prognostic accuracy compared with TNM staging; this lead to an effort to evaluate immune scoring as a new approach to complement the current classification of cancer.12 Others subsequently reported similar findings in melanoma, ovarian, breast and squamous cell cancer.7 Thus, it appears that the Th1 environment originally described for colon cancer is a widespread phenomenon related to the oncogenic process rather than the ontogeny of individual cancers. Although, a classification of cancer based on its immune phenotype has not been as yet broadly embraced for routine diagnostic and staging purposes, the confirmation by various groups including ours13 of these findings sheds a new light on the understanding of the immune biology of cancer. Thus, future work will be aimed at understanding the mechanism leading to this cancer phenotype whether this is dependent upon the genetic background of the host, the genetic makeup of individual cancers, environmental influences or a combination of them.
The Effector Immune Phenotype of Cancer also Predicts Immune Responsiveness
Concurrent to the previous observations, others observed that the expression of ISGs, IEFs and CXCR3 and CCR5 ligand chemokines was predictive of tumor responsiveness to immunotherapy such as vaccination with antigenic peptides, dendritic cell-based vaccines and systemic cytokine therapy.7 About a decade ago, we observed that melanoma metastases likely to regresss following immune therapy with active specific vaccination and systemic IL-2 therapy, displayed before treatment an activate immune microenvironment.14 We were recently able to confirm this finding in a small prospective study in which patients with metastatic melanoma were treated with systemic IL-2 and their metastases were serially biopsied with fine needle aspiration biopsies (FNA) before and during treatment.10 This approach directly links the biology of individual lesions to their responsiveness to treatment.14-16 In two patients, pre-treatment biopsies clearly identified lesions destined to respond, which expressed the ICR signature.10 A more extensive unpublished study in which patients with melanoma or lung cancer were treated with a MAGE-A3 peptide plus adjuvant confirmed these signatures as harbinger of good outcome.9 We recently summarized these and similar observations in a more comprehensive review beyond the purposes of this article.7
The Immune Responsive Phenotype is Enhanced in Responsive Lesions During Treatment
Immunologic patterns that define the good prognostic phenotype are qualitatively similar to those observed during TSD.5,6 By obtaining serial FNA biopsies of melanoma metastases during and/or following therapy, we observed a decade ago14 that lesions undergoing rejection displayed enhanced expression of IRF-1, which later became recognized as the master regulatory transcription factor modulating the switch from chronic to acute inflammation during TSD.17 The observation was expanded subsequently to a complete set of genes associated to the ICR; in a double blind randomized placebo control study, we serially biopsied basal cell carcinomas undergoing TSD following treatment with the Toll-like receptor agonist Imiquimod and identified a rejection-specific signature.18,19 The set of genes associated with the rejection of basal cell carcinoma in patients receiving Imiquimod, was then observed in several other types of TSD following different treatments, in different diseases and even in different mammalian species5,7,20 confirming the breath of the phenomenon. Most recently, by prospectively following serially biopsied melanoma metastases in patients treated with high dose systemic IL-2, we could document the switch from a moderate activation of ICR genes in pre-treatment lesions to a fully blown activation during treatment only in lesions that eventually underwent complete regression.10 Since the signatures observed pre-treatment in responsive lesions were qualitatively similar to those observed during treatment, we postulate that the biology underlining immune responsiveness is a necessary pre-existent requirement that is however not sufficient for tumor rejection. Thus, immune-mediated rejection of tumors reflects a continuum that spans from an indolent immune recognition that may slow but not stop in natural conditions tumor growth to a potent activation of acute inflammation that embraces the complete set of effector components necessary for tissue rejection during immune stimulation. Similarly, comparison of metastases from two patients who experienced a mixed response following immunotherapy identified the activation of ICR genes in the responding but not in the non responding lesions.16
The Activation of ICR Genes During Cancer Rejection is Shared by Other Immune Pathologies Suggesting that Cancer Rejection is a Form of Autoimmunity
The signatures identified in immune responsive lesions before treatment and that are amplified during and/or following treatment are similar to those observed during other forms of TSD such as allograft rejection, graft vs. host disease, clearance of pathogen and flares of autoimmunity.4,5,21 Among them, autoimmunity is the one that most closely resembles tumor rejection as it is the only one directed toward self. Thus, we suggest that tumor rejection whether occurring spontaneously or induced by immune manipulation is a form of tissue-specific autoimmunity directed against the neoplastic organ. Indeed, immune responsiveness of melanoma to various treatments including systemic IL-2 administration or IFNα, has been associated with various forms of autoimmunity including vitiligo,22 thyroiditis,23 autoimmune rethynopathy24 and development of antibodies against self.25 While the association between vitiligo and rejection of melanoma may represent the immune response against antigens shared by cells of the melanocytic lineage, the reason for the association with other autoimmune diseases remains to be clarified as it could represent a response to still unknown shared antigens or reflect a status of hyper immune activation. Among the various manifestations of autoimmunity, a quite aggressive is systemic lupus erythematosus (SLE). A clear link between SLE and responsiveness to immunotherapy has never been described. This could be partly explained by the preferential exclusion of patients with severe autoimmunity from immunotherapy trials. It was, however, observed that symptoms related to SLE could be exacerbated by immunotherapy in cases when patients were enrolled.26 Moreover, patterns associated with tumor rejection are similar to SLE as they share a relatedness in ISG activation;27,28 this observation lead us to study whether genetic traits associated to the development of SLE could also apply to cancer immune responsiveness.
The Immune Phenotype Associated with Immune Responsiveness is Partly Dependent upon the Genetics of the Host
As discussed in the previous paragraph, if autoimmunity is associated with cancer immune responsiveness, polymorphisms of genes related to the former might be associated with the latter. We studied recently patients enjoying better overall survival following treatment with IFNα. We observed that patients carrying either DRB1*15 or HLA-Cw7 suffered worse overall survival while patients with either HLA-Cw6 or HLA-B44 enjoyed a better one. In addition to HLA, CTLA4 polymorphism was identified as a marker associated with shorter overall survival. Importantly, multivariate analysis revealed that a five-marker genotyping signature bore even better prognostic significance. A multivariate Cox regression model demonstrated that the presence of HLA-B38, HLA-C15, HLA-C3, DRB1*15 and the CTLA4 polymorphisms CT60*G/G (0.081) were significantly associated with overall survival suggesting that genetic factors predisposing to immune responsiveness may be multiple and analysis of them in combination may be more informative than studying each separately.29 Others have looked at the Δ32 variant of CCR that induces the expression of a non functional protein; patients with melanoma carrying this polymorphism were observed to score worse after immunotherapy.30 Perhaps, the most striking link between the genetic characteristic of the host and immune responsiveness came from a recent study that we performed in patients with melanoma treated with the adoptive transfer of ex vivo activated tumor infiltrating lymphocytes. Analysis of IRF-5 polymorphisms previously described to be associated with protection from developing SLE demonstrated that the same where highly predictive of non-responsiveness to melanoma (Uccellini et al., manuscript in preparation). Importantly, we observed that metastases from patients carrying this variant of IRF-5 displayed a distinct transcriptional pattern. Moreover, melanoma cell lines derived from the same metastases displayed in vitro distinct transcriptional patterns between the two genotypes that could be used to impute immune responsiveness of the parental metastases. This observation suggests that germline variants can affect directly the intrinsic biology of cancer cells besides affecting the behavior of host’s cells.
The Immune Phenotype Associated with Immune Responsiveness is Partly Dependent Upon the Genetics of the Cancer
The most striking evidence that immune responsiveness is, at least in part, related to acquired alterations of cancer cell genetics is the phenomenon of the mixed responses. A proportion of cancer patients experience a mix response to therapy characterized by the simultaneous regression of some metastases, while other progress. These relatively rare cases are precious since they single out the tumor’s aspects of immune responsiveness excluding from the equation the genetic background of the patient or external factors affecting the effectiveness of treatment. We recently performed a comparative gene expression analysis of 15 metastases (10 regressing and 5 progressing) obtained from two melanoma patients experiencing a mixed response following immunotherapy.16 Transcriptional analysis indicated that regression of melanoma metastases is associated with acute inflammation mediated by the upregulation of genes involved in antigen presentation, which in turn are activated downstream of the STAT-1/IRF-1 pathway. The molecular signature obtained in regressing metastases was similar to that observed in other types of TSD supporting the ICR hypothesis. This limited but unique study provides a striking demonstration that within the same genetic background and identical therapeutic conditions, tumors can behave differently and, therefore, that tumor rejection is at least in part dependent upon tumor biology.
A Proposed Algorithm Governing Immune Responsiveness of Human Cancers that Includes the Genetic Background of the Host and Somatic Alterations of Cancer
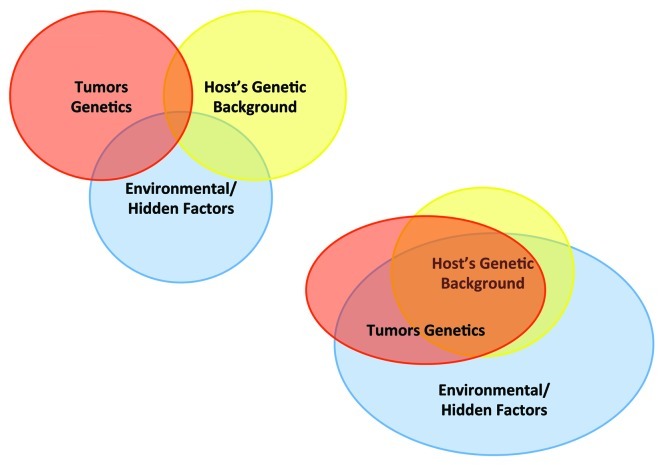
The last two paragraphs are in apparent contrast; on one side it appears that the genetic background of the host’s bears significantly on immune responsiveness, on the other it appears that tumor can behave differently within the same genetic background. This apparent paradox can only be explained by a multi-factorial model of cancer immune responsiveness. It should be emphasized that host and cancer genetics are largely overlapping since cancer cells carry the majority of the host’s genetics (Fig. 1). Thus, inherited genetic factors may affect the biology of cancer cells besides that of normal cells. Thus, it could be postulated that some patients carry a genetic background that make them resistant to immunotherapy by effecting either the biology of the immune response, the biology of the cancer cells or both. On the other hand, “an immune-responsive genotype” may still be limited by the genetics of the tumors: in other words, although the patient may be predisposed to cancer rejection the tumor lacks additional properties necessary for its recognition by the immune response (Fig. 2). In this model, a favorable genetic background of the host is necessary but not sufficient for tumor rejection as the possession of a shotgun is necessary to shoot a duck but at the same time a skill in shooting is required. A good example is provided by the analysis of patients with IRF-5 polymorphism; the “immune resistant phenotype” appears to almost exclusively preclude cancer rejection during adoptive therapy with tumor infiltrating lymphocytes; however, “the immune responsive phenotype” can be segregated into two categories; one enriched in patients responding to therapy and the other of non-responding. Although, other host’s genetic factors could be responsible for this sub-classification, it is also possible that, given a favorable genetic background, the genetics of the tumor may become the determining factor.
Figure 1. Interplay among categorical modifiers of responsiveness. (A) Classical view of the relationship between host genetic background, tumor genetics and environmental factors. (B) A more likely scenario integrating the large overlap between the genetics of the host and the tumor and the over-reaching effect of environmental factors on both of them.
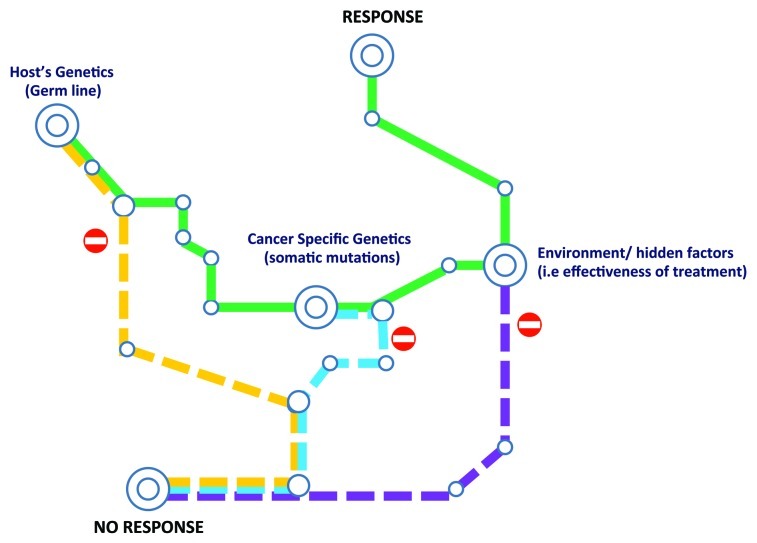
Figure 2. Check point regulating the winding road to cancer immune responsiveness. The host’s genetic may be the first limiting factor and a genetic predisposition to immune response may be necessary but not sufficient to allow cancer response to immunotherapy. Subsequently, genetic alteration of cancer cells may allow escape from immune recognition in spite of a favorable genetic background. Finally, quality and intensity of treatment and a myriad of hidden external factors may determine the final outcome.
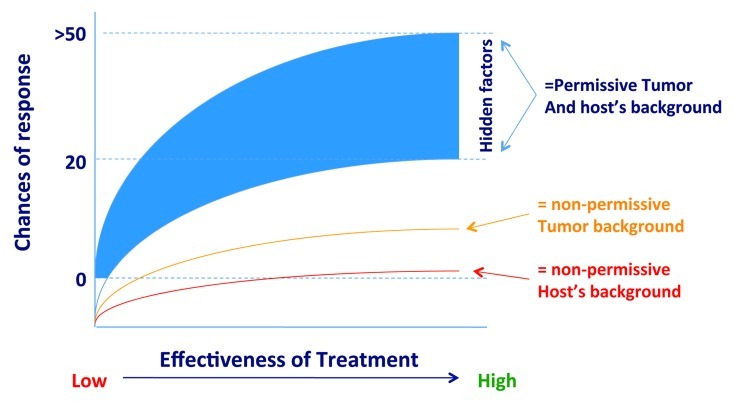
We recognize that this classification of factors that may influence immune responsiveness may be too rigid. In reality, immune responsiveness may depend upon a continuum determined by the interaction of a multitude of factors that for simplicity can be separated into broad categories depending upon the host’s genetic background, somatic mutations, and external factors such as intensity and effectiveness of treatment, general condition of the patient and a multitude of other hidden co-factors (Fig. 3).
Figure 3. A quantitative continuum determining the responsiveness of tumors according to genetic background of the host, genetics of cancer cells, effectiveness of treatment and other “hidden” external factors that may affect the final outcome.
We acknowledge that these conclusions are speculative but we hope that this commentary may stimulate an integrated approach to the study of cancer patients receiving immunotherapy that includes the analysis of germline variants, tumor genetics and other post-genomic modifications.
Glossary
Abbreviations:
- FNA
fine needle aspiration biopsy
- ICR
immunologic constant of rejection
- IEFs
immune effector genes
- IFN
interferon
- IL
interleukin
- IRF
interferon regulatory factor
- ISGs
interferon-stimulated genes
- SLE
systemic lupus erythematosus
- STAT-1
signal transducer and activator of transcription
- TSD
immune-mediated tissue-specific destruction
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/19531
References
- 1.Salk J. Immunological paradoxes: theoretical considerations in the rejection or retention of grafts, tumors, and normal tissue. Ann N Y Acad Sci. 1969;164:365–80. doi: 10.1111/j.1749-6632.1969.tb14051.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang E, Marincola FM. Bottom up: a modular view of immunology. Immunity. 2008;29:9–11. doi: 10.1016/j.immuni.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock WW, Wang L, Ye Q, Han R, Lee I. Chemokines and their receptors as markers of allograft rejection and targets for immunosuppression. Curr Opin Immunol. 2003;15:479–86. doi: 10.1016/S0952-7915(03)00103-1. [DOI] [PubMed] [Google Scholar]
- 4.Wang E, Worschech A, Marincola FM. The immunologic constant of rejection. Trends Immunol. 2008;29:256–62. doi: 10.1016/j.it.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Immunologic Signatures of Rejection. New York, NY: Springer, 2010. [Google Scholar]
- 6.Spivey TL, Uccellini L, Ascierto ML, Zoppoli G, De Giorgi V, Delogu LG, et al. Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J Transl Med. 2011;9:174. doi: 10.1186/1479-5876-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ascierto ML, De Giorgi V, Liu Q, Bedognetti D, Spivey TL, Murtas D, et al. An immunologic portrait of cancer. J Transl Med. 2011;9:146. doi: 10.1186/1479-5876-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagès F, Galon J, Dieu-Nosjean MC, Tartour E, Sautès-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 9.Louahed J, Grusell O, Gaulis S, et al. Expression of defined genes indentifed by pre-treatment tumor profiling: association with clinical response to GSK MAGE A-3 immunetherapeutic in metastatic melanoma patients. J Clin Oncol. 2008;26:A9045. [Google Scholar]
- 10.Weiss G, Grosh WW, Chianese-Bullock KA, et al. Molecular insights on the peripheral and intra-tumoral effects of systemic high dose rIL-2 (Aldesleukin) administration for the treatment of metastatic melanoma. Clin Cancer Res. 2011;17:7440–50. doi: 10.1158/1078-0432.CCR-11-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochran AJ. Histology and prognosis in malignant melanoma. J Pathol. 1969;97:459–68. doi: 10.1002/path.1710970305. [DOI] [PubMed] [Google Scholar]
- 12.Galon J, Pagès F, Marincola FM, Thurin M, Trinchieri G, Fox BA, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med. 2012;10:1. doi: 10.1186/1479-5876-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ascierto ML, Kmieciak M, Idowo MO, et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2012;131:871–80. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang E, Miller LD, Ohnmacht GA, Mocellin S, Perez-Diez A, Petersen D, et al. Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2002;62:3581–6. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang E, Marincola FM. A natural history of melanoma: serial gene expression analysis. Immunol Today. 2000;21:619–23. doi: 10.1016/S0167-5699(00)01724-2. [DOI] [PubMed] [Google Scholar]
- 16.Carretero R, Wang E, Rodriguez AI, Reinboth J, Ascierto ML, Engle AM, et al. Regression of melanoma metastases after immunotherapy is associated with activation of antigen presentation and interferon-mediated rejection genes. Int J Cancer. 2011 doi: 10.1002/ijc.26471. Epub ahead of press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang E, Marincola FM. From the “delayed allergy reaction” to the “immunologic constant of rejection”. In: Wang E, Marincola FM, eds. Immunologic Signatures of Rejection. New York, NY: Springer, 2010:3-8. [Google Scholar]
- 18.Panelli MC, Stashower ME, Slade HB, Smith K, Norwood C, Abati A, et al. Sequential gene profiling of basal cell carcinomas treated with imiquimod in a placebo-controlled study defines the requirements for tissue rejection. Genome Biol. 2007;8:R8. doi: 10.1186/gb-2007-8-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deonarine K, Panelli MC, Stashower ME, Jin P, Smith K, Slade HB, et al. Gene expression profiling of cutaneous wound healing. J Transl Med. 2007;5:11. doi: 10.1186/1479-5876-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worschech A, Chen N, Yu YA, Zhang Q, Pos Z, Weibel S, et al. Systemic treatment of xenografts with vaccinia virus GLV-1h68 reveals the immunologic facet of oncolytic therapy. BMC Genomics. 2009;10:301. doi: 10.1186/1471-2164-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedognetti D, Wang E, Sertoli MR, Marincola FM. Gene-expression profiling in vaccine therapy and immunotherapy for cancer. Expert Rev Vaccines. 2010;9:555–65. doi: 10.1586/erv.10.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan GQ, Attia P, Steinberg SM, White DE, Rosenberg SA. Factors associated with response to high-dose interleukin-2 in patients with metastatic melanoma. J Clin Oncol. 2001;19:3477–82. doi: 10.1200/JCO.2001.19.15.3477. [DOI] [PubMed] [Google Scholar]
- 23.Gogas H, Ioannovich J, Dafni U, Stavropoulou-Giokas C, Frangia K, Tsoutsos D, et al. Prognostic significance of autoimmunity during treatment of melanoma with interferon. N Engl J Med. 2006;354:709–18. doi: 10.1056/NEJMoa053007. [DOI] [PubMed] [Google Scholar]
- 24.Chan C, O’Day J. Melanoma-associated retinopathy: does autoimmunity prolong survival? Clin Experiment Ophthalmol. 2001;29:235–8. doi: 10.1046/j.1442-9071.2001.00425.x. [DOI] [PubMed] [Google Scholar]
- 25.Maire C, Vercambre-Darras S, Devos P, D’Herbomez M, Dubucquoi S, Mortier L. Metastatic melanoma: spontaneous occurrence of auto antibodies is a good prognosis factor in a prospective cohort. J Eur Acad Dermatol Venereol. 2011 doi: 10.1111/j.1468-3083.2011.04364.x. [DOI] [PubMed] [Google Scholar]
- 26.Kähler KC, Hauschild A. Treatment and side effect management of CTLA-4 antibody therapy in metastatic melanoma. J Dtsch Dermatol Ges. 2011;9:277–86. doi: 10.1111/j.1610-0387.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 27.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–3. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 28.Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol. 2006;18:676–82. doi: 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Wang E, Zhao Y, Monaco A et al. A multi-factorial genetic model for prognostic assessment of high risk melanoma patients receiving adjuvant interferon. Submitted 2012. [DOI] [PMC free article] [PubMed]
- 30.Ugurel S, Schrama D, Keller G, Schadendorf D, Bröcker EB, Houben R, et al. Impact of the CCR5 gene polymorphism on the survival of metastatic melanoma patients receiving immunotherapy. Cancer Immunol Immunother. 2008;57:685–91. doi: 10.1007/s00262-007-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]