Abstract
Autophagy is a fundamental eukaryotic process with multiple cytoplasmic homeostatic roles, recently expanded to include unique standalone immunological functions and interactions with nearly all parts of the immune system. Here, we review this growing repertoire of autophagy roles in innate and adaptive immunity and inflammation. Its unique functions include cell-autonomous elimination of intracellular microbes facilitated by specific receptors. Other intersections of autophagy with immune processes encompass effects on inflammasome activation and secretion of its substrates including IL-1β, effector and regulatory interactions with Toll-like and Nod-like receptors, antigen presentation, naïve T cell repertoire selection, and mature T cell development and homeostasis. Genome wide association studies in human populations strongly implicate autophagy in chronic inflammatory disease and autoimmune disorders. Collectively, the unique features of autophagy as an immunological process and its contributions to other arms of the immune system represent a new immunological paradigm.
Introduction
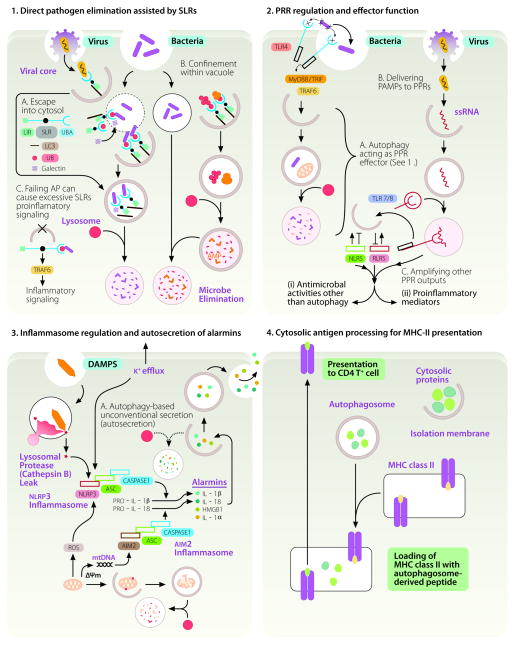
In this review, we cover the immunological roles of macroautophagy (1), a specific autophagic process that will be referred herein as the sensu stricto autophagy or simply autophagy. Autophagy is unique in its capacity to sequester, remove, or process bulk cytosol, cytoplasmic organelles (1), invading microbes, and immunological mediators (2) as depicted in Fig. 1. Another special property illustrated in Fig. 1 is that autophagy acts as a topological inverter - bringing molecules and objects from the cytosolic side to the lumenal side for degradation or processing, interaction with luminal receptors, or secretion from cells. Here, we cover the four principal manifestations of immunological autophagy (Fig. 1): (i) direct pathogen elimination assisted by sequestosome 1-like receptors (SLRs); (ii) regulation and effector functions of pattern recognition receptors (PRR); (iii) regulation of inflammasome activation and alarmin secretion; and (iv) cytoplasmic antigen processing for MHC II presentation and T cell homeostasis. We relate these processes to conventional immunological functions, defense against infectious agents, chronic inflammatory disorders and other immunological pathologies.
FIGURE 1.
The four principal manifestations of immunological autophagy. 1. Direct pathogen elimination assisted by SLRs and DAMP receptors. (A) Invading microbes either escaping the endosomes or phagosome (thin outline) or remaining in phagosomes that can be partially permeabilized (dotted outline) are captured by galectins and sequestosome 1/p62-like receptors (SLR) that recognize tags such as ubiquitin (small red circles) or diacylglicerol and β-galactoside (not shown) on damaged host membranes. The captured microbes or co-captured with the earmarked membranes are delivered into autophagic organelles (thick outline) starting with phagophores (crescents) progressing through autophagosomes (full white circles) and ending in degradative autolysosomes (full pink circles). SLRs posses LC3 interacting region (LIR), phosphorylation sites (black dot, arbitrarily positioned), and tag recognition domain (UBA, depicted for p62). Galectins (hatched square), considered to be DAMP receptors, have carbohydrate recognition domains (not shown) that recognize sugars on glycans exposed on the endofacial lumenal membrane leaflet of permeabilized organelles. (B) Alternatively, autophagy can sequester cytosolic proteins such as ubiquitin and ribosomal proteins (pear shaped tan-colored objects, ribosomes) and digest them into antimicrobial peptides (AMPs) that can be delivered to pathogens confined in phagosomes. (C) SLRs can engage in pro-inflammatory signaling e.g. via TRAF6 (shown) or atypical PKC (not shown) or promote cell death by activating caspase-8 through aggregation (not shown). AP, autophagy. 2. PRR regulation and effector functions. A. Autophagy can be activated downstream of TLR signaling upon recognition of PAMPs (x-like objects). B. As a topological inverter device, autophagy can deliver cytosolic PAMPs to the lumen of endomembranous organelles where they can interact with the receptor portions of TLRs. Known functional interactions with NLRs and RLRs are summarized by positive (arrows) and negative (lines symbolizing inhibition) effects. 3. Inflammasome regulation and secretion of alarmins. Autophagy plays a dual role in controlling inflammasome output: it suppresses basal levels of inflammasome activation but also assists IL-1βand IL-18 release from the cells via an autophagy-dependent unconventional secretory pathway (A; autosecretion). Inflammasomes, heteromeric protein assemblages (consisting of ASC, caspase 1, and NLRP3 or AIM2) act as platforms for activation in response to K+ efflux or presence and action of DAMPs (silica, crystal like object; ROS, reactive oxygen species; mtDNA, mitochondrial DNA). ROS and mtDNA can be released as endogenous DAMPs by damaged mitochondria if they are not continuously removed by autophagy. This results in caspase-1 activation and proteolytic processing of pro-forms of proinflammatory cytokines (IL-1β). Whereas autophagy lowers the sources of endogenous DAMPs by disposing of depolarized (ΔΨm) or leaky mitochondria (sources of ROS and mtDNA), autophagy also enables secretion of the cytosolic IL-1β(and other alarmins such as HMGB1) during the very early stages of physiological inflammasome activation in response to exogenous DAMP sources (microbial or sterile). Autosecretion (autophagy-based unconventional secretion; see Fig. 2 and text for explanations) enables extracellular release of the cytosolic proteins such as IL-1β and HMGB1 per the illustrated process controlled by Atg factors and GRASP (see Fig. 2). Autosecretion occurs early in the process of stimulation and is swamped pre- or shortly post-stimulation by the anti-inflammatory effects of autophagy. The latter keep the tonic levels of inflammasome activation low and bring them back to the resting levels following stimulation. 4. Cytosolic antigen processing for antigen presentation. Autophagy assists as a topological inversion device in delivery of cytosolic (and nuclear) proteins to MHC II processing and presentation compartments. Explanations in the text include relationships to selection of naïve T cell repertoires and citrullination of antigens.
Autophagy pathway
The key morphological features of autophagy are endomembranous organelles, called autophagosomes (Fig. 1) whose formation is controlled by the Atg and additional factors comprehensively reviewed elsewhere (1). Briefly, the Atg system includes Ser/Thr kinases Ulk1 and Ulk2 (Atg1), Beclin 1 (Atg6; a subunit of the class III phosphatidylinositol 3-phosphate kinase (PI3K) hVPS34 complexes), Atg5-Atg12/Atg16L1 complex, and LC3s (multiple Atg8 orthologs), with LC3B being a commonly used marker for identification of autophagosomes (1). Ulk1/2 and Beclin 1-hVPS34 integrate upstream signals and direct the downstream Atg conjugation cascade, which involves Atg5-Atg12/Atg16L1 assembly as an “E3 enzyme” for LC3 lipidation. Lipidated LC3s in conjunction other factors assemble, elongate, and close nascent autophagic organelles. Autophagosomes interact with endosomal and lysosomal organelles to mature into autolysosomes (1), or promote unconventional secretion of cytoplasmic constituents, as first demonstrated in yeast (3) and recently shown to include immune mediators (4, 5). In addition to its immunological functions (2), autophagy plays a general cellular homeostatic role by supplying nutrients (e.g. amino acids) through cytosol autodigestion at times of starvation or growth factor withdrawal, and serves as a quality and quantity control mechanisms for intracellular organelles (1).
At the transcriptional level, regulation of autophagy is coupled to the lysosomal system via TFEB (transcription factor EB) (6) and other proteolytic systems via FoxO3A (7). However, autophagy is primarily a rapid-response remodeling of membranes that occurs in the cytoplasm under the control of the signaling systems faster than transcriptional changes. The classical nutritional/energy regulation of autophagy is via mTOR and AMPK inhibiting and activating, respectively, Ulk1/2 (1). This pathway merges with signaling via the inhibitor of NF-κB kinases (IKK), frequently involved in immune signaling. IKKα and IKKβ transduce the classical signal for autophagy induction – starvation (8) but this signaling is not based on nuclear NF-κB responses. Instead, IKK and AMPK signaling merge via TAK1 and its activators TAB2 and TAB3. Upon autophagy induction TAB2 and TAB3 dissociate from and thus activate Beclin 1 and also bind to and activate TAK1 (8), whereas TAK1 in turn phosphorylates and activates AMPK.
In T cells, autophagy is activated upon TCR engagement and CD28 co-stimulation and supports their effector functions and proliferation (9). Recently, class III PI3K hVPS34 was found to be dispensable for autophagy induction in T cells albeit required for T cell homeostasis via its regulation of receptor endocytosis (10) bringing up the possibility of alternative pathways in PI3P signaling, as suggested by the positive role of class I PI3K p110β(11).
Innate immune signaling can induce autophagy. TRAF6 downstream of TLR4 activates autophagy (12) Alarmins or damage associated molecular patterns (DAMP) induce autophagy (13, 14). HMGB1, an alarmin, undergoes translocation from the nucleus into the cytoplasm and then out of the cells by unconventional secretion (5, 15) or cell death-associated release, inducing autophagy at each stage: cytoplasmic through derepression of Beclin 1 by displacing its negative regulator Bcl-2, or extracellularly via RAGE signaling (13, 14). In addition to HMGB1, DAMPs such as ATP, IL-1β, and DNA complexes are known to induce autophagy (reviewed in (16)).
Autophagy in direct pathogen elimination
The evolutionarily most primal manifestation of immunological autophagy is direct capture and degradation of invading intracellular microbes by autophagy (Fig. 1, panel 1, left). This cell-autonomous defense function of autophagy is often countered by microbial adaptation mechanisms and a number of highly adapted pathogens can convert autophagic organelles into growth-supporting compartments (17). Autophagic capture of intracellular microbes is facilitated by autophagic adaptors, referred to as SLRs (sequestosome 1/p62-like receptors) (18). SLRs have LC3 interacting regions (LIR) and cargo-tag (e.g. ubiquitin) recognition domain and are modulated by protein kinases. Salmonella requires multiple SLRs (p62, NDP52, optineurin) (19, 20), phosphorylation of at least one of the SLRs (optienurin) with an IKK-related kinase, TBK-1 (20), and an intracellular DAMP receptor (galectin 8) (21). The SLRs p62 and NDP52 are also engaged in clearance of Shigella and Listeria (22–24), whereas Streptococci are affected by NDP52 (19). Sindbis virus interacts with p62 (25). Candidate E3 ligases contributing to target ubiquitination have been identified in some instances: SMURF1 for sindbis virus (26) and LRSAM1 as a candidate for Salmonella (27).
The most recent player in these processes is galectin 8, a cytosolic lectin binding to β-galactoside glycans. The membrane glycans are normally present only on the lumenal side of parasitophorous vacuoles. However, upon membrane damage the glycans come in contact with the cytosol and thus become recognized by cytosolic gelectins (21) (Fig. 1, panel 1, hatched square). Galectin 8 is important to restrict Salmonella proliferation, and plays an early role until supplanted by a phase dominated with ubiquitin and ubiquitin-recognizing SLR – NDP52. It appears that the phases and the sequence of recognizing membrane damage could be ushered by the appearance of diacylglycerol (28), followed by galectin-β-galactoside recognition, followed by NDP52-ubiquitin recognition. Since galectin 8 and NDP52 interact, a sequential action is doubly ensured. Galectin 8 is important for the recruitment of NDP52, since the requirement for galectin 8 to restrict Salmonella proliferation could be bypassed by expressing a fusion hybrid between galectin 3 and NDP52. Galectin 3 per se is not required for restriction although it is found on Salmonella vacuoles, primarily since it – unlike galectin 8 - cannot interact with NDP52. Galectin 8 recognizes host membrane glycans and not directly Salmonella carbohydrates, albeit it can directly recognize blood-group-B-positive E. coli O86. Galectin 8 is important also for Shigella, Listeria and even recognized sterile damage to endosomes and lysosomes.
SLRs can also act in a completely different manner to promote autophagic killing of intracellular microbes (Fig. 1, panel 1, right). They gather cytoplasmic proteins (e.g. ubiquitin and ribosomal proteins) to be converted in autolysosomes into anti-microbial products that upon delivery to cytoplasmic compartments harboring microbes transform them into autophagolysosomes, organelles with enhanced antimicrobial capacities relative to conventional phagolysosomes (29–31).
Autophagy and pattern recognition receptors
Autophagy interacts with classical pattern recognition receptors (PRR), including Toll-like receptors (TLR), Nod-like receptors (NLR), and RIG-I like receptors (RLR). TLRs and autophagy intersect in two ways illustrated in Fig. 1, panel 2. Firstly, autophagy is an effector mechanism (e.g. elimination of microbes illustrated in Fig. 1, panel 1) downstream of TLR activation. TLR4 triggers autophagy via TRAF6 E3 ligase, ubiquitination of Beclin 1, and Bcl-2 dissociation from the BH3 domain of Beclin 1 (12). Secondly, autophagy as a topological inverter devise can bring cytosolic pathogen associated molecular pattern (PAMP) molecules into the lumen where they can bind the ligand recognition side of the TLR receptor. This has been demonstrated for TLR7 (32), TLR4 ligands (33) and TLR9 in the context of B cell receptor signaling (34).
NLR and autophagy interactions are evolutionarily conserved from Drosophila (35) to humans (16). Nod1 and Nod2 interact with Atg16L1 (36, 37), of significance for Crohn’s disease (CD) since Nod2 and Atg16L1 are risk loci for CD (38). NLRC4 (Ipaf) and NLRP4 inhibit autophagy (39) and are found in macromolecular complexes with Beclin 1. RLRs activate autophagy with biologically important effects (40) but thus far more attention has been given to negative regulation of RLR signaling by autophagy factors Atg5-Atg12 (41) and Atg9 (42). Atg9 negatively regulates trafficking and activation of TBK1 in the type I interferon response to double stranded DNA (42).
Autophagy and inflammasome
Autophagy and inflammasomes interact in two ways (Fig. 1, panel 3). All reports thus far (5, 43–46) agree on the observation that autophagy plays a negative role in inflammasome activation. Autophagy lowers basal level of inflammasome activation by continually removing endogenous irritants (43, 44). For example, autophagy prevents spurious inflammasome activation by eliminating defunct mitochondria that otherwise represent endogenous sources of inflammasome agonists such as ROS and mitochondrial DNA (43, 44) (Fig. 1, panel 3). In the absence of basal autophagy, endogenous factors lead to inflammasome activation and increased IL-1β processing and represent sources of sterile inflammation. This explains how loss Atg16L1 elevates IL-1β levels in a murine model of CD (47).
On the flip side, autophagy plays a positive (but only acute, short term) role in delivering outside of the cell the effector products of inflammasome activation, such as IL-1β and potentially other alarmins, in a process referred to as the unconventional secretion of IL-1β(5). Although IL-1β and IL-18 do not have signal peptides to deliver them into the lumen of the organelles of the conventional secretory pathway (ER-Golgi-plasma membrane), they are released extracellularly upon inflammasome activation. This is at least in part supported by the topological inversion properties of autophagy, ferrying molecules from cytosolic side into lumen of putative secretory vesicles. However, this effect wanes quickly with time and the downregulation of inflammasome by autophagy becomes dominant once again (46). Thus, autophagy controls negatively inflammasome activation (5, 43–46) and positively IL-1β secretion per se (5). The topological inversion action and positive role of autophagy in secretion of alarmins is not limited to IL-1β and extends to HMGB1 (5).
Autophagy in antigen presentation and T cell homeostasis
The role of autophagy as a topological inverter (transport from cytosol to lumen) and its other functions contribute to MHC II presentation of endogenous cytosolic antigens (33, 48, 49) (Fig. 1, panel 4). The physiological role of this is manifested in immune surveillance of viral infections (48) and inhibition of this process by HIV-1 (49). Autophagy-dependent presentation of endogenous antigens plays a role in positive and negative selection of naïve T cells repertoires in the thymus (50). It has been hypothesized that peripheral tissue autophagic activities may have to be matched by central tolerance mechanisms dependent on autophagy in the thymus to prevent autoimmunity (50). Autophagy plays a role in mature T cell homeostasis, and is essential for T cell survival following exit from the thymus in part based on the requirement for autophagy to physiologically reduce the mitochondrial and ER content in maturing T cells (51–53).
Autophagy in chronic inflammatory and autoimmune diseases
Genetic links between autophagy and chronic inflammatory disorders and autoimmune diseases continue to be uncovered by genome-wide association studies (GWAS). Genetic variations in the PRDM1-ATG5 intergenic region have been associated with rheumatoid arthritis (RA) (54). Autophagy specifically favors presentation of citrullinated proteins, which may contribute to autoimmune disorders such as RA (55). The initial GWAS linking of ATG16L1 and IRGM (a modulator of autophagy (56, 57)) with CD (38) have been replicated in nearly 50 independent population studies. Polymorphisms in another autophagy gene, ULK1, are also associated with CD (58). Genetic associations of CD with IRGM have been extended to IRGM copy number variants in human populations (59). IRGM has furthermore been linked to systemic lupus erythematosus (SLE) in a recent meta-analysis of autoimmune diseases (60). GWAS in different populations link ATG5 variants to SLE (61, 62). This genetic evidence and other studies implicate autophagy in chronic inflammatory diseases and autoimmunity disorders.
Conclusions
The initial sporadic observations that autophagy can play a role in cell-autonomous defense against intracellular bacteria such as Mycobacterium tuberculosis (63) and streptococci (64) have been extended in past several years to various facets of immunity. The connections of autophagy with normal function of innate and adaptive immunity at almost every level, genetic and functional associations with immunological disorders, and unique, specialized mechanisms of autophagy as standalone immune processes reviewed here and elsewhere (2) are consistent with the thesis of this review that autophagy represents a new and growing immunological paradigm.
Acknowledgments
Funding support: AI042999, AI069345, and ARRA RC1AI086845 from National Institutes of Health, Crohn’s & Colitis Foundation of America CCFA2053, and Bill and Melinda Gates Grand Challenge Explorations grant.
The author apologies to the colleagues for omissions imposed by space limitations, including microbial defenses against autophagy, non-autophagic functions of the ATG factors, and roles of autophagy processes other than macroautophagy. The author is grateful to Carolyn Mold for comments on the text and to Dara Elerath for graphic design.
Abbreviations used in this article
- AMPK
AMP-activated protein kinase
- ATG
autophagy related genes
- CD
Crohn’s disease
- DAMP
danger/damage associated molecular patterns
- GWAS
genome-wide association studies
- HMGB1
high-mobility group protein B1
- hVPS34
human vacuolar protein sorting 34
- IKK
inhibitor of NF-κB kinases
- IRGM
immunity related GTPase M
- LC3
microtubule-associated protein light chain 3
- LIR
LC3-interacting region
- LRSAM1
leucine rich repeat and sterile alpha motif containing 1
- mTOR
mammalian target of rapamycin
- NDP52
Nuclear domain 10 protein / antigen nuclear dot 52 kDa protein
- NLR
nucleotide binding and oligomerization domain-like receptors
- PAMP
pathogen associated molecular patterns
- PI3K
phosphatidylinositol 3-phosphate kinase
- PRR
pattern recognition receptors
- RAGE
receptor for advanced glycation endproducts
- RLR
RIG-I-like receptors
- SLE
systemic lupus erythematosus
- SLR
sequestosome 1/p6-like receptors
- ROS
reactive oxygen species
- SMURF1
SMAD specific E3 ubiquitin protein ligase 1
- TAB2 and TAB3
TGFβ–Activated Kinase 1 (TAK1)-Binding Proteins 2 and 3
- TAK1
Transforming growth factor β activated kinase 1
- TBK-1
TANK-binding kinase 1
- TCR
T cell receptor
- TFEB
transcription factor EB
Footnotes
DISCLOSURES
The author has no financial conflicts of interest to disclose.
References
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, Rello-Varona S, Tailler M, Menger L, Vacchelli E, Galluzzi L, Ghiringhelli F, di Virgilio F, Zitvogel L, Kroemer G. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 5.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. Embo J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, Rain JC, Ninomiya-Tsuji J, Fuentes JM, Lavandero S, Galluzzi L, Maiuri MC, Kroemer G. Inhibition of autophagy by TAB2 and TAB3. Embo J. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. J Immunol. 2011;187:5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, Yue Z, Lin RZ, Zong WX. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol. 2010;191:827–843. doi: 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, Zeh HJ, Lotze MT. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller M, Ruegg A, Werner S, Beer HD. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 16.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Op Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 20.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 Proteins Target Intracytosolic Shigella and Listeria to Different Autophagy Pathways. J Biol Chem. 2011;286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 25.Orvedahl A, Macpherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy Protects against Sindbis Virus Infection of the Central Nervous System. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng AC, Eisenberg JM, Heath RJ, Huett A, Robinson CM, Nau GJ, Xavier RJ. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4631–4638. doi: 10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HWt, Kyei GB, Johansen T, Vergne I, Deretic V. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 32.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 33.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28:799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 37.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 38.Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jounai N, Kobiyama K, Shiina M, Ogata K, Ishii KJ, Takeshita F. NLRP4 negatively regulates autophagic processes through an association with beclin1. J Immunol. 2011;186:1646–1655. doi: 10.4049/jimmunol.1001654. [DOI] [PubMed] [Google Scholar]
- 40.Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, Mulero F, Piris MA, Dash R, Barral PM, Rodriguez-Peralto JL, Ortiz-Romero P, Tuting T, Fisher PB, Soengas MS. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell. 2009;16:103–114. doi: 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 44.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris J, Hartman M, Roche C, Zeng SG, O’Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1βsecretion by targeting pro-IL-1βfor degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 48.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 49.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, Schwartz O, Deretic V, Piguet V. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 51.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 53.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Burtt NP, Chang M, Coblyn J, Costenbader KH, Criswell LA, Crusius JB, Cui J, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Kurreeman FA, Lee AT, Liu X, Li Y, Martin P, Morgan AW, Padyukov L, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth P, Altshuler D, Karlson EW, Toes RE, de Vries N, Begovich AB, Siminovitch KA, Worthington J, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Expe Med. 2011;208:2625–2632. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, Komatsu M, Deretic V. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 58.Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K, Rutgeerts P, Vermeire S. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm Bowel Dis. 2011;17:1392–1397. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 59.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, Holmes C, Marchini JL, Stirrups K, Tobin MD, Wain LV, Yau C, Aerts J, Ahmad T, Andrews TD, Arbury H, Attwood A, Auton A, Ball SG, Balmforth AJ, Barrett JC, Barroso I, Barton A, Bennett AJ, Bhaskar S, Blaszczyk K, Bowes J, Brand OJ, Braund PS, Bredin F, Breen G, Brown MJ, Bruce IN, Bull J, Burren OS, Burton J, Byrnes J, Caesar S, Clee CM, Coffey AJ, Connell JM, Cooper JD, Dominiczak AF, Downes K, Drummond HE, Dudakia D, Dunham A, Ebbs B, Eccles D, Edkins S, Edwards C, Elliot A, Emery P, Evans DM, Evans G, Eyre S, Farmer A, Ferrier IN, Feuk L, Fitzgerald T, Flynn E, Forbes A, Forty L, Franklyn JA, Freathy RM, Gibbs P, Gilbert P, Gokumen O, Gordon-Smith K, Gray E, Green E, Groves CJ, Grozeva D, Gwilliam R, Hall A, Hammond N, Hardy M, Harrison P, Hassanali N, Hebaishi H, Hines S, Hinks A, Hitman GA, Hocking L, Howard E, Howard P, Howson JM, Hughes D, Hunt S, Isaacs JD, Jain M, Jewell DP, Johnson T, Jolley JD, Jones IR, Jones LA, Kirov G, Langford CF, Lango-Allen H, Lathrop GM, Lee J, Lee KL, Lees C, Lewis K, Lindgren CM, Maisuria-Armer M, Maller J, Mansfield J, Martin P, Massey DC, McArdle WL, McGuffin P, McLay KE, Mentzer A, Mimmack ML, Morgan AE, Morris AP, Mowat C, Myers S, Newman W, Nimmo ER, O'Donovan MC, Onipinla A, Onyiah I, Ovington NR, Owen MJ, Palin K, Parnell K, Pernet D, Perry JR, Phillips A, Pinto D, Prescott NJ, Prokopenko I, Quail MA, Rafelt S, Rayner NW, Redon R, Reid DM, Renwick, Ring SM, Robertson N, Russell E, StClair D, Sambrook JG, Sanderson JD, Schuilenburg H, Scott CE, Scott R, Seal S, Shaw-Hawkins S, Shields BM, Simmonds MJ, Smyth DJ, Somaskantharajah E, Spanova K, Steer S, Stephens J, Stevens HE, Stone MA, Su Z, Symmons DP, Thompson JR, Thomson W, Travers ME, Turnbull C, Valsesia A, Walker M, Walker NM, Wallace C, Warren-Perry M, Watkins NA, Webster J, Weedon MN, Wilson AG, Woodburn M, Wordsworth BP, Young AH, Zeggini E, Carter NP, Frayling TM, Lee C, McVean G, Munroe PB, Palotie A, Sawcer SJ, Scherer SW, Strachan DP, Tyler-Smith C, Brown MA, Burton PR, Caulfield MJ, Compston A, Farrall M, Gough SC, Hall AS, Hattersley AT, Hill AV, Mathew CG, Pembrey M, Satsangi J, Stratton MR, Worthington J, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand W, Parkes M, Rahman N, Todd JA, Samani NJ, Donnelly P. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramos PS, Criswell LA, Moser KL, Comeau ME, Williams AH, Pajewski NM, Chung SA, Graham RR, Zidovetzki R, Kelly JA, Kaufman KM, Jacob CO, Vyse TJ, Tsao BP, Kimberly RP, Gaffney PM, Alarcon-Riquelme ME, Harley JB, Langefeld CD. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 2011;7:e1002406. doi: 10.1371/journal.pgen.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 63.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 64.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]