Abstract
Precise transcriptional networks drive the orchestration and execution of complex developmental processes. Transcription factors possessing sequence-specific DNA binding properties activate or repress target genes in a step-wise manner to control most cell lineage decisions. This regulation often requires the interaction between transcription factors and subunits of massive protein complexes that bear enzymatic activities towards histones. The functional coupling of transcription proteins and histone modifiers underscores the importance of transcriptional regulation through chromatin modification in developmental cell fate decisions and in disease pathogenesis.
Keywords: cancer, chromatin, development, differentiation, histone modifications, next-generation sequencing, pluripotency
Genome-wide changes in the mammalian epi-genome, including alterations in DNA methylation and in histone modification patterns, occur during the early stages of embryo development. Histone modifications influence chromatin-templated processes such as gene transcription, DNA repair and recombination. While chromatin states in mouse and human embryonic stem (ES) cells have been characterized extensively [1], little is currently known regarding histone modification changes and epigenetic regulation during the differentiation of tissues and organs in vivo. Histone modifications including methylation, acetylation, phosphorylation and ubiquitination are known to instruct transcriptional networks required for cellular differentiation. However, the mechanistic links that drive early-fate decisions as progenitor cells are pushed towards specific differentiated states are still unclear.
Striking similarities exist between the processes involved in tumor cell formation and the properties of untransformed stem cells. Defining molecular controls that underlie the high proliferation rates and increased plasticity of stem cells has helped to decipher analogous switches that drive normal cells to tumorigenic states [2]. For example, Trithorax group (TrxG) and Polycomb group (PcG) proteins dynamically regulate expression of Hox genes, which are involved in transcriptional networks that regulate cell proliferation and differentiation decisions in stem and progenitor cells that ultimately determine patterning during vertebrate development [3–5]. Aberrant Hox gene-expression patterns often correlate with altered activity of TrxG and/or PcG members in cancer cells, and contribute to malignancy in a context-dependent manner [6].
The chromatin environments that control these differing cell states require the orchestration of many factors, including histone modifications, DNA methylation, chromatin remodeling, nuclear organization, histone exchange and ncRNA molecules. Histone modifications control differentiation states mainly by coordinating gene expression programs, as they mark different sets of genes for either transcriptional activation or silencing. The chromatin of genes expressed in a specific cell lineage is marked with active histone modifications like H3K4 methylation and H4 acetylation, while the chromatin states of the same genes are enriched with repressive marks, like H3K27 methylation, in cell types in which they are silenced. Histone lysine methylation and acetylation are enzymatically reversible and are ‘written’ by lysine methyltransferases (KMTs) and lysine acetyltransferases (KATs), and ‘erased’ by lysine demethylases (KDMs) and histone deacetylases (HDACs), respectively. The enzymatic activity of KMTs, KATs, protein arginine methyltransferases (PRMTs), KDMs and HDACs is not necessarily restricted to histone proteins, as a growing number of nonhistone substrates are being identified. In addition to ‘writers’ and ‘erasers’ of histone marks, ‘reader’ proteins that contain specialized histone modification binding domains, recognize specific marks to elicit particular biological responses.
This review will focus on our current understanding of how histone-modifying enzymes coordinately regulate different histone modification patterns, and thus control cell fate decisions during mammalian embryonic development and tissue differentiation. We will then discuss how deregulation of these enzymes leads to tumorigenesis and developmental disorders.
Histone modifiers regulate early cell fate decisions
One crucial cell fate decision for mouse blastomeres is whether to establish pluripotent inner mass cells, from which ES cells are derived, or to differentiate into trophectoderm cells, which contribute to the formation of extraembryonic tissues. CARM1 (also known as PRMT4), an H3 arginine methyltransferase and transcriptional coactivator, appears to be involved in this decision as it influences 4-cell stage blastomeres to acquire a pluripotent fate; blastomeres with less H3 arginine methylation become tropho-ectoderm [7]. An important role for CARM1 epigenetic regulation in ES cells is further underscored by its regulation of the expression of the pluripotency factors OCT4, NANOG and SOX2. Similarly to its role in determining blastomere fate, CARM1-dependent H3 arginine methylation is required for maintenance of ES cell pluripotency, as direct binding of CARM1 at Oct4, Nanog and Sox2 promoters regulates H3R17 and H3R26 methylation and leads to subsequent increases in expression levels of these pluripotency factors [8]. A recent study by Parfitt and Zernicka-Goetz [9] shows a functional relationship between CARM1 methyltransferase activity and regulation of blastomeres’ cell polarity, offering an intriguing link between epigenetic modifications and cell fate specification events, like cell polarity and movement.
Transcriptional activation of pluripotency genes like Oct4 and Nanog is also regulated by histone marks that are observed on active gene promoters of differentiated somatic cells [10] including H3K4 methylation. TrxG chromatin modifiers that induce this modification generally act as transcriptional activators, and they antagonize repression mediated by members of the PcG family, which induce H3K27 methylation. H3K4 trimethylation of active pluripotency gene promoters is mediated by the MLL catalytic subunit of TrxG complexes [11], and this activity requires cofactors including ASH2L, WDR5 and RBBP5 [12]. A recent genome-wide protein localization analysis and expression profiling of ES cells identified WDR5 as an interacting partner of OCT4, and indicated that WDR5 is an indispensable regulator of pluripotency and self-renewal [11]. These findings offer a direct link between the WDR5 subunit of TrxG complex and regulation of OCT4 expression through H3K4me3, but further experiments are required for a deeper understanding of the role of the TrxG complex in early fate decisions.
PcG proteins are also important in maintaining pluripotency of ES cells [13]. Members of both Polycomb repressive complex (PRC)1 and PRC2 are required for repression of a large number of transcription factors that function in organogenesis, morphogenesis, pattern specification and neurogenesis in ES cells [14]. The transcriptional repression of these genes is indispensable for the maintenance of ES cell pluripotency [13]. Genome wide localization of SUZ12, a subunit of the PRC2 complex, in human ES cells verified the repressive role of Polycomb-mediated H3K27 trimethylation in maintaining ES cell identity [15]. SUZ12 occupancy is diminished at the same developmental regulator genes in primary differentiated muscle cells, consistent with removal of PRC2-induced repressive marks during differentiation [15]. The removal of H3K27 methylation is regulated by UTX, a component of MLL2/3 complexes, with a JmjC-domain demethylating activity [16–18]. The recruitment of these MLL complexes to Hox gene promoters coordinates the addition of the H3K4me3 active mark through the MLL subunit and the removal of the H3K27me3 repressive mark through UTX, creating a switch that controls cell fate decisions. More recently the activity of the PRC2 complex has been shown to be fine-tuned both in ES cells and in early embryos by the presence of JARID2, which targets PRC2 to developmental genes, and counter-intuitively, represses PRC2 enzymatic activity [19–21]. This functional interplay highlights the importance of dynamic regulation of repressive histone marks, and specifically H3K27 methylation, in the transition between pluripotent and differentiated states.
RING1A/B, a subunit of the PRC1 complex with E3 ubiquitin ligase activity towards H2AK119, plays an important role in the repression of developmental genes in mouse ES cells. Recruitment of RING1A/B to target genes depends on OCT4, providing another functional link between PcG-dependent repression and the core transcriptional network in ES cells [22]. The presence of both active (H3K4me3) and repressive marks (H3K27me3) on some gene promoters creates a ‘bivalent’ transcriptional environment that allows for genes that are silent in ES cells to be promptly expressed in subsequent differentiation steps. RING1B-mediated H2A ubiquitination provides an additional regulatory step for bivalent promoters. RING1B keeps RNA polymerase (pol) II in check on bivalent promoters by enforcing a stem cell-specific RNA pol II conformation, preferentially phosphorylated at Ser5, which limits its processivity [23]. Although the current study relies on detecting the recruitment of RNA pol II on bivalent promoters by using antibodies specific for different phosphorylation states of RNA pol II, further structural analysis will be required to understand the mechanistic details of this regulation. Interestingly, the poised environment established on bivalent promoters regulated by both PRC2-mediated H3K4 trimethylation and PRC1-dependent H2A ubiquitination that is observed in ES cells does not occur in trophoblasts. Instead, transcriptional repression of bivalent genes in the trophoectoderm depends on SUV39H1-mediated H3K9 trimethylation in coordination with subsequent DNA methylation [24].
Maintenance of ES cell pluripotency also depends on the presence of two demethylases, JMJD1A and JMJD2C. Both demethylases are upregulated by Oct4 and maintain chromatin in a permissive state by removing repressive methylation marks (H3K9me) from the promoters of downstream pluripotency genes, like Nanog and Tcl1. When cells start to differentiate, Jmjd1a and Jmjd2c expression levels decrease, and the repressive methylation marks increase, resulting in downregulation of target genes [25]. The transcriptional shut-off of pluripotency-associated factors is mediated by another histone modifier, G9a, an H3K9 methyltransferase that inactivates Oct4 expression by inducing its heterochromatinization through H3K9 methylation, recruitment of chromodomain protein HP1, and de novo DNA methylation by DNMT3A/B [26,27].
In contrast to the extensive analysis of PcG proteins and the role of histone methylation–demethylation cycles in ES cell self-renewal and pluripotency, little is known about the role of histone acetylation–deacetylation in maintaining ES cell identity. Acetylation of H3 is permissive for transcriptional activation and has been shown to mark regions in ES cells that are required for subsequent cell differentiation [28]. The removal of methylation from H3K27 is not sufficient for derepression of these genes in ES cells; rather, subsequent acetylation of H3K27 by the acetyltransferases CBP and P300 is required for rapid activation of target genes during ES cell differentiation [16,29]. Both CBP and P300 are required for early mouse development [30], but our understanding of their role in ES cells is still minimal. Xu et al. uncovered an important role of P300 – mediated acetylation in the fate decision of ventral foregut endoderm embryonic mouse cells (embryonic day 8.25) that can differentiate towards a hepatic or pancreatic lineage [31]. P300-mediated acetylation was increased in differentiated embryonic hepatoblasts relative to undifferentiated endodermal cells, showing an important role of P300 at liver-specific regulatory elements.
Deacetylases are also important in early development. HDAC1 is expressed in the two-cell embryo and regulates levels of histone acetylation and gene-expression patterns until the pre-implantation stage [32]. HDAC1 and its paralog HDAC2 are components of multiprotein complexes with multiple histone modifying activities that play important roles in early mouse embryogenesis [33]. Two components of the nucleosome remodeling and deacetylase (NuRD) corepressor complex, MBD3 and P66A, are required for early cell fate decisions [34,35]. More specifically, MBD3 has a key role in the cell fate transition of inner cell mass cells to late epiblast after implantation [34] and is required for the maintenance of ES cells pluripotency by repressing the trophoectoderm cell lineage [36]. Except for NuRD and Sin3A complexes, the OCT4 and NANOG pluripotency factors are also found to interact with a stem cell specific repressive complex, NANOG and OCT4-associated deacetylase (NODE), which represses the expression of developmentally regulated genes in ES cells [37]. Conditional deletion of HDAC1, but not HDAC2, pushes cells towards mesodermal cell fate at the expense of endoderm [38].
More recently, HDAC1 has been implicated in the transcriptional repression of brachyury, a WNT-responsive developmental gene that is required for mesodermal differentiation. The repression of this gene in ES cells by HDAC1 highlights its importance for maintenance of ES cell self-renewal capacity [39]. Another HDAC, HDAC4, is repressed by OCT4 in mouse ES cells and is upregulated during differentiation [40]. These examples clearly indicate that HDACs exert their repressive functions on different genes during different developmental time points, adding another important level of regulation for ES cell pluripotency and plasticity.
Differentiation & tissue development are controlled by histone-modifying enzymes
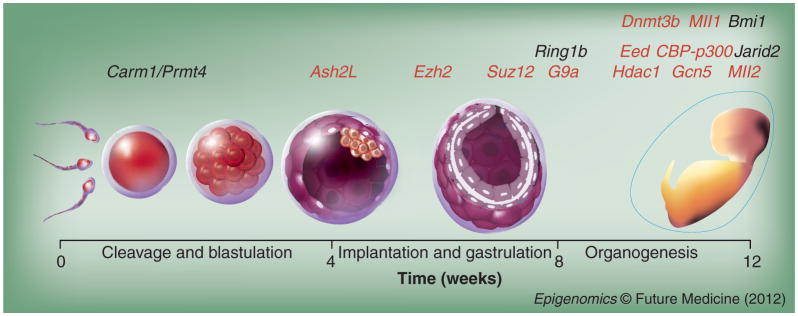
Early developmental stages are regulated by multiple histone-modifying enzymes, examples of which are described above. Most of them are indispensable to specific cell fate transitions, as they cause lethality in various embryonic stages when they are deleted (Figure 1). Later in development, multipotent progenitor cell populations control tissue development and maintenance through the execution of well-orchestrated programs. A common mechanism that undifferentiated progenitors employ to maintain their pluripotency is through the repression of differentiation factors using HDACs and PcG chromatin modifying complexes.
Figure 1. Examples of histone-modifying proteins implicated at different stages of mouse embryogenesis that have been analyzed in vivo.
The genes in red depict knockout models with embryonic lethality at the time points shown. The regulation of histone modifications between fertilization and blastulation is minimally characterized.
HDAC1 and HDAC2 play important roles in cardiac development as the deletion of both genes together in mouse cardiomyocytes causes a severe phenotype of cardiac arrhythmias, right and left ventricular dilation, and death at 14 days after birth [41]. Both deacetylases have been found to regulate genes important for the physiology of cardiomyocytes like Ca2+ channels and thin filaments components. When both deacetylases are absent, the expression of Ca2+ channel subunits is possibly increased and causes a pathological influx of Ca2+ into the cardiomyocytes, resulting in cardiac dysregulation [41]. In a different study, HDAC2 exerts its repressive effect on Inpp5f and regulates the constitutive activation of GSK3β by titrating AKT and PDK1 kinases away from the cell membrane [42]. Activated GSK3β inhibits proliferative signals that would lead to cardiac hypertrophy [42].
HDACs are broadly expressed in the brain [43], but their role in the development of the CNS remains largely unknown. Both HDAC1 and 2 are critical in the development of synaptic networks by controlling their excitatory drive [44]. A more recent study has evaluated the susceptibility of HDAC targets for drug treatment of progressive neurodegenerative disorders such as Huntington’s disease. Increased HDAC1 levels and decreased HDAC4, 5 and 6 correlate with disease progression in both mouse and human tissues, although the underlying regulatory mechanism remains to be discovered [45]. SAGA, a chromatin remodeling complex with both histone acetyltransferase and deubiquitinase activity, has also been implicated in the development of the neurodegenerative disease, spinocerebellar ataxia type 7 [46–48]. The etiology of spinocerebellar ataxia type 7 disease lies in polyglutamine expansions identified in the ATAXIN-7 protein, which cause toxicity in neuronal tissues, like the cerebellum and the retina, leading to abnormal gait, uncoordinated movements (ataxia) and visual loss [49,50]. Although the progression of the disease was initially thought to be caused by transcriptional deregulation of SAGAs downstream target genes, a recent study revealed genetic interactions between mutations in the GCN5 acetyltransfersase subunit and polyQ expansions in ATAXIN-7, which is a component of the SAGA deubiquitinase module, and pointed towards nontranscriptional effects of SAGA complex deregulation as a possible cause of neural degeneration [51].
In undifferentiated myoblasts, EZH2 is recruited to the promoters of the muscle-specific genes though interactions with YY1 and keeps these genes silenced in collaboration with HDAC1. Removal of the H3K27 repressive mark and recruitment of activators like SRF and MYOD are required for the promotion of muscle differentiation [52]. In additon, the ASH2L subunit of SET1/MLL complexes interacts with MEF2D to recruit H3K4 methyltransferase activity to activate expression of muscle-specific genes [53]. EZH2-mediated H3K27 trimethylation along with subsequent DNMT3B-mediated DNA methylation has been shown to repress myoblast proliferation and self-renewal [54] through the repression of the Notch-1 receptor. The regenerative potential of the myoblasts is severely compromised in Duchenne muscular dystrophy, and our understanding of the underlying mechanism creates new potential therapeutic approaches against degenerative diseases, as blocking the EZH2-mediated repression of Notch-1 receptor in dystrophic muscles could restore their regenerative potential [54].
PcG and TrxG proteins also regulate the proliferation and expansion of pancreatic β-cells both in humans and mice, showing an important role of epigenetic regulation in normal pancreatic functions and in the pathogenesis of diabetes. EZH2 is recruited to the promoters of p16INK4a and p19Arf, which act as negative regulators of the cell cycle and stop the proliferation of islet β-cells. EZH2, through H3K27 trimethylation, represses the expression of Ink4a/Arf, allowing the proliferation and expansion of pancreatic β-cells in neonate mice [55]. In aged islets, the increased expression of the negative regulators is associated with decreased Bmi1 recruitment and increased MLL1-mediated H3K4 trimethylation on the gene promoters [56], verifying the antagonistic role of the two complexes in epigenetic regulation of downstream targets. An important role of Ezh2 in maintaining proliferation of undifferentiated cells through regulation of Ink4a/Arf was also shown in embryonic epidermal progenitors [57]. Interestingly, this study showed that epidermal progenitor cells do not use ‘bivalent’ chromatin marks to maintain or relieve transcriptional repression. Instead, EZH2-mediated H3K27 trimethylation prevents the binding of key activators, like AP1, that are required for terminal differentiation [57].
PcG complexes also have important roles in neural stem cell specification and neuronal cell differentiation. PcG subunits RING1B and BMI1 promote neuronal stem cell self-renewal and maintain their undifferentiated state [58,59]. EZH2 regulates neurogenesis similarly to RING1B and BMI1 by maintaining stem cell populations, while it also controls cell fate decisions in later differentiation programs. Ezh2 is highly expressed in proliferating neural stem cells, and its expression decreases during neural stem cell differentiation and is lost after terminal differentiation to astrocytes. EZH2 expression persists in neural cells that differentiate into oligodendrocytes [60]. Hirabayashi et al. have uncovered a PcG role in neuronal differentiation that differs from PcG function in ES cells. They found that H3K27 trimethylation is increased at neuro-genin loci during neural stem cell differentiation and deletion of Ring1B in neural stem cells results in extended neurogenic capacity compared to the loss of neurogenic capacity observed in ES cells after deletion of Ring1B or Suz12 [61,62].
The functional interplay between PcG and TrxG proteins in various developmental processes and the different nuances of fine-tuning of their opposing roles in cell fate decisions has greatly enhanced our understanding of tumorigenesis and cancer progression in different tissues and organs. A great number of genetic aberrations on PcG and TrxG complex subunits, as well as on other chromatin modifiers, have been identified, and great effort is directed towards defining the molecular events that lead to tumor development.
Mutations in histone-modifying enzymes cause human diseases
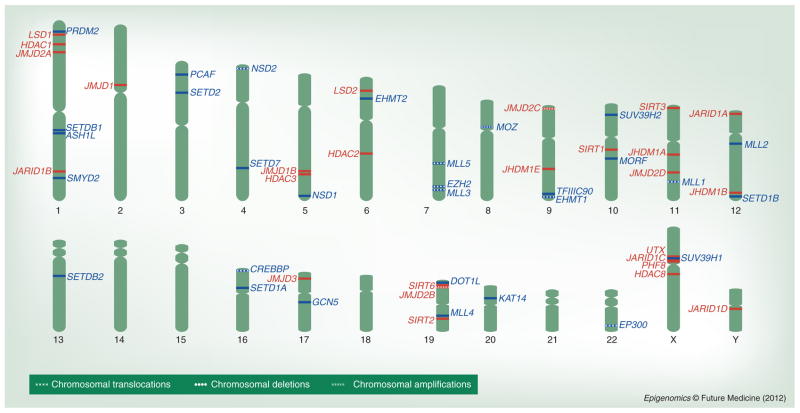
Genetic, cytogenetic and molecular approaches have identified many chromosomal translocations, deletions, and amplification events that link histone-modifying enzymes to human diseases. As Figure 2 illustrates, histone-modifying enzymes with opposing functions, such as KMTs and KDMs, are often positioned near one another in the chromosomes, and therefore may be affected by the same genomic insult associated with a particular disease (Figure 2). For instance, genes encoding the TrxG and PcG members MLL3/KMT2C, MLL5/KMT2E, and EZH2/KMT6 are located in regions 7q36.1, 7q22.1 and 7q35-36, respectively, that are often deleted in myeloid malignancies [63,64]. Likewise, the genes encoding the H3K4 demethylase JARID1C/SMCX/KDM5B [65] and the H3K9 demethylase PHF8 [66] are located near one another in the pericentric region of the X chromosome, and both genes are enriched in point mutations that give rise to X-linked mental retardation [67]. Mutations that occur within histone modifiers possessing similar functions may also give rise to a particular disease. For example, translocations involving the KATs CBP and p300, whose genes reside in chromosomes 16 and 22, respectively, are observed in myeloid cancers [68] (Figure 2). Furthermore, some chromosomes encode multiple histone H3 lysine modifiers, like chromosome 1 and chromosome X (Figure 2), suggesting that disrupted modification of histone H3 may underlie the pathogenesis of human diseases involving gains, losses and gross chromosomal rearrangements of these chromosomes.
Figure 2. Chromosomal distribution of the genes encoding histone H3 modifying enzymes.
The chromosomal locations of genes encoding enzymes known to modify lysine residues within histone H3 are depicted. The gene names of writers are shown in blue to the right of the chromosome, and erasers are shown in red to the left. The chromosomal locations were verified using the NCBI Gene and Ensembl Databases.
Next-generation whole-genome sequencing (WGS) has become an invaluable tool used to determine more subtle genetic events underlying disease states. Whole-exome sequencing (WES) is quickly emerging as a popular variation of WGS to discover causative mutations that underlie cancer and rare genetic diseases and this is mostly due to its lower cost, easier data management and the ability to detect non-synonymous genetic variants within a small sample population [69–71]. A number of recent WGS and WES studies designed to profile genetic mutations in human cancers and developmental disorders have highlighted an enrichment of novel point mutations, many causative, in proteins involved in chromatin modification. The mutated proteins belong to all classes of chromatin modifiers: writers, erasers, readers and remodelers, underscoring the significance of the idea that complex diseases often arise from multiple mutations that occur within a common biological pathway [72].
Novel mutations discovered in histone modifiers are involved in hematological malignancies
The important roles of TrxG and PcG complexes in regulating developmental processes foreshadows the involvement of mutations in these factors in human cancers that possess features of undifferentiated cells. For example, reciprocal chromosomal translocation events involving 11q23 causally link the founding member of the MLL gene family, MLL1/KMT2A, to acute myeloid and acute lymphoid leukemias (Figure 2) [73]. Although the mechanisms that drive MLL-mediated leukemogenesis remain unclear, many of the resulting oncogenic MLL fusion proteins act dominantly to promote tumorigenesis [74]. For instance, the MLL-AF4 oncoprotein colocalizes with aberrant H3K4 and H3K79 methylation at genes that mediate hematopoietic stem cell self-renewal [75], implying that mistargeted MLL complexes contribute to a transcriptional program that promotes stem cell propagation rather than differentiation.
WES sequencing helped to identify inactivating mutations within or surrounding the functional PHD, FYRN and SET domains of MLL2/KMT2B that are enriched in non-Hodgkin’s lymphoma patient samples [76,77]. Importantly, these studies implicate disruption of MLL2 in the pathogenesis of hematological malignancies for the first time. Furthermore, gene ontology analysis performed with all candidate genes revealed that chromatin modification and transcriptional regulation were among the top biological processes in which mutations are enriched in the analyzed lymphomas [76,77].
Clinical observations combined with the hematopoietic defects observed in MLL-deficient mice [78,79] support the role of MLL family members as tumor suppressors in hematopoietic cells. However, the precise mechanisms involving these enzymes in differentiation and proliferation decisions remain largely unknown. Determining whether MLL family members regulate H3K4 methylation at overlapping or distinct sets of target genes, how these enzymes are targeted to gene loci, and whether these enzymes methylate nonhistone substrates will shed light on the oncogenic mechanisms used by MLL family members in hematopathologies.
As described earlier, the PcG enzyme EZH2 is critical for maintaining the proliferative capacity of undifferentiated cells in developing tissues by controlling the expression of cell cycle regulators, such as p16INK4a and p19Arf [55]. Similarly, EZH2 promotes the normal maturation of naive B cells into highly proliferative germinal center B cells by repressing the expression of genes involved in cell proliferation and differentiation, including p16INK4a [80]. Increased EZH2-mediated silencing of tumor suppressor genes may play a role in tipping the balance from regulated to uncontrolled proliferation of germinal center B cells, resulting in diffuse large B-cell lymphoma (DLBCL). Activating and inactivating mutations within EZH2 were recently reported in hematological cancers [81]. A specific heterozygous gain-of-function mutation within the EZH2 catalytic SET domain was identified in follicular lymphoma and DLBCL patient samples [82]. This EZH2 mutant has enhanced catalytic activity and cooperates with wild-type EZH2, thus mimicking the EZH2 over-expression phenotype observed in solid tumors [83]. Knockdown of EZH2 in DLBCL cells results in upregulation of the tumor suppressor genes and cell cycle arrest, further supporting an oncogenic role for EZH2 [80].
Conversely, a spectrum of missense and nonsense mutations that truncate EZH2 were found in acute and chronic myeloid malignancies [84–88]. Furthermore, EZH2 mutations correlate with decreased overall survival of chronic myelomonocytic (CMML) patients, indicating that EZH2 mutational status likely holds prognostic value in CMML [88]. The fact that both gain and loss of function mutations within EZH2 contribute to oncogenesis in cells of different hematopoietic lineages suggests that additional cell type-specific factors may be involved in regulating EZH2 expression or targeting of EZH2 activity, and that altering the balance of these activities promotes leukemogenesis.
As described above, UTX/KDM6A opposes EZH2 function to regulate cell fate decisions by removing methyl groups from histone H3K27 [17,18]. Inactivating mutations in UTX were found in a number of cancer tissues, and were the first cancer-associated mutations identified in a histone demethylase [89]. Somatic mutations that often truncate UTX prior to the catalytic jumonji-C domain were also observed in CMML, although not in conjunction with EZH2 mutations [87], suggesting independent mechanisms that promote transformation.
Mutations in UTX were identified in a screen of multiple myeloma patient samples along with mutations in TrxG genes, MLL1, MLL2, MLL3 and the H3K9 demethylase, KDM3B [90]. Multiple myeloma arises from clonal expansion of plasma B cells, therefore inappropriate transcriptional regulation of genes involved in cell proliferation likely plays a role in transformation. Indeed, increased expression of the MLL target gene HoxA9 was observed along with decreased global H3K27 methylation in multiple myeloma cell lines, giving the cells a competitive advantage [90]. In addition, novel deletion and missense mutations were recently identified in the H3K36 methyltransferase NSD2/WHSC1/MMSET, which has been linked previously to multiple myeloma via translocations (Figure 2) [91]. NSD2 has also been implicated in regulating genes involved in cell growth, apoptosis and cell adhesion [92]. Although MLL family members and NSD2 methylate histones in genes involved in cell proliferation, the mechanisms by which mutations in these enzymes contribute to multiple myeloma have not been elucidated. Functional testing of the disease-associated mutant enzymes is required to determine their biological relevance.
Translocations within CBP/CREBBP/KAT3A and p300/EP300/KAT3B that generate fusion proteins have long been observed in acute myeloid leukemia and treatment-related hematological disorders (Figure 2) [68]. Recently, WGS analysis revealed point and deletion mutations within CBP in relapsed acute lymphoid leukemia patient samples [93], further underscoring that inactivation of CBP plays a role in oncogenic transformation.
A separate WES sequencing study of DLBCL patient samples conducted by Pasqualucci et al. uncovered a number of deleterious mutations within the genes encoding CBP and p300 that occur in a monoallelic fashion [94]. Many of the identified CBP mutations cluster around the HAT domain, and although effects on histone acetylation levels were not reported in this study, acetyltransferase activity toward nonhistone substrates of CBP, p53 and BCL-6, was compromised resulting in reduced tumor suppressor activity and increased oncoprotein activation, respectively. It will be interesting to determine whether global or gene specific histone acetylation patterns are also affected by these pathogenic mutations within CBP and p300.
Examples of mutations in histone-modifying enzymes linked with hematological malignancies are abundant in the literature, as evidenced above. This may reflect the necessity of precise regulation of chromatin modifications during normal hematopoiesis, which requires many cell-fate decisions during embryonic development as well as throughout adulthood. The developmental and hematopoietic defects observed in MLL and CBP-deficient mouse models support this hypothesis [78,79,95]. Blood is often more readily available from patients than solid tumor tissue, and this likely has facilitated the discovery of mutations in chromatin modifiers in blood malignancies. Nevertheless, the advent of WGS methods is now facilitating the discovery of disease-causing mutations in solid tumors and developmental disease states where acquisition of affected tissue from patients is often limiting.
Histone-modifying enzymes emerge as potential therapeutic targets in solid tumors
The recent discovery of somatic mutations in MLL2 and MLL3 in adult and pediatric medulloblastoma (MB) samples reveals that these enzymes function as tumor suppressors in tissues other than blood [96]. The high percentage of truncating mutations identified in MLL2 compared with the overall abundance of truncating mutations observed in the MB samples classifies these mutations as driver rather than passenger mutations, and suggests that MLL2 functions as a tumor suppressor gene in MB [96]. All of the identified missense and nonsense mutations are predicted to eliminate the conserved FYRN, FYRC and SET functional domains. The mutations identified in MLL3 would leave the PHD domains intact, suggesting that the presence of these domains in the absence of the catalytic SET domain confers a cell survival or growth advantage. Although mechanisms have yet to be elucidated, it will be interesting to see whether the truncated MLL2 and MLL3 proteins are actually expressed, and if so, whether they promote tumor growth in MB through H3K4me-dependent or independent pathways. Loss of function mutations within MLL2 and MLL3 suggest that these histone modifiers might provide new therapeutic targets for pediatric and adult MB patients [96].
While inactivating mutations in MLL2 and MLL3 imply methylation changes involving H3K4, regulators of H3K9 methylation emerged as commonly mutated targets in a separate MB patient study. Homozygous deletion of chromosome 9q was detected in two primary tumor samples taken from pediatric patients suffering from MB [97]. In accordance, decreased expression of the histone H3K9 methyltransferase EHMT/GLP/KMT1D and decreased levels of H3K9me2 were detected in an MB tissue array. Chromosomal deletions affecting the H3K9 readers, L3MBTL2 and L3MBTL3, as well as amplification of the H3K9 demethylases JMJD2C and JMJD2B and the H3K9 KAT MOZ were also identified in patient samples (Figure 2) [97]. Notably, mutations affecting these H3K9 methylation and acetylation regulators were identified as mutually exclusive events [97], which further underscores the significance of the downstream consequence for any of these individual mutations. It will be interesting to determine if mutations affecting the methylation status of H3K4 or H3K9 converge to disrupt the expression of the same target gene sets, which would then contribute to the pathogenesis of MB.
Missense, nonsense and frameshift mutations in MLL1 and MLL3 were identified by exome sequencing of transitional cell carcinoma (TCC) of the bladder patient samples [98]. The majority of mutations within MLL3 are predicted to result in loss of function by eliminating the catalytic SET domain, suggesting MLL3 functions as a tumor suppressor in TCC [98]. The identification of truncating mutations within MLL1 suggests that it may also serve as a tumor suppressor in TCC, although the majority of MLL1 mutations discovered in these samples are missense mutations [98] and additional biochemical analyses are required to determine their functional impact. Truncating mutations within UTX prior to the jumonji-C domain are observed in TCC as well [98], indicating that mutations occur on both sides of the TrxG/PcG axis which would predict a loss of transcriptional activating marks and gain of repressive marks in this disease. In further support of the formation of a repressive chromatin structure in TCC, deleterious mutations were identified that are predicted to compromise the transcriptional activation properties of the KATs CBP and p300 [98].
Somatic mutations in enzymes that regulate methyl marks at H3K4 (JARID1C), H3K27 (UTX) and H3K36 (SETD2/KMT3A), were identified in clear cell renal cell carcinoma (ccRCC) patient samples [89,99,100]. As JARID1C and UTX regulate H3 marks with opposing transcriptional roles, it is possible that inactivating mutations in these demethylases affect the expression of different target genes. Although, corresponding expression changes were observed for a subset of genes in samples with either JARID1C or UTX mutations, suggesting some overlapping transcriptional regulation [99]. It is also possible that these demethylases share common nonhistone substrates that play a role in ccRCC pathogenesis. Further molecular and biochemical analyses are required to define the functional significance of these mutations in promoting ccRCC.
Taken together, these studies reveal new pathogenic roles for histone-modifying enzymes in solid tumors in which no previous associations had been made. Defining the molecular mechanisms by which mutations in these enzymes contribute to transformation will require functional testing. Many predictions on how the mutations would affect cell growth or differentiation are based on the known roles for these enzymes in transcriptional regulation and during embryo development. It will also be important to consider how these mutations may impact transcription-independent functions of these enzymes, such as protein–protein interactions or modification of nonhistone substrates.
Mutations in histone modifiers underlie developmental disorders & cancer
A growing number of mutations in genes encoding histone-modifying enzymes contribute to developmental disorders when they occur in the germline but give rise to cancer when acquired as somatic mutations (Table 1). The identified germline and somatic mutations in any given enzyme are not necessarily identical, however, they likely result in similar cellular consequences, such as deregulated proliferation and differentiation.
Table 1.
Histone-modifying enzymes involved in developmental disorders and cancer.
| Histone modifier | Molecular function | Mutations | Associated human disease | Ref. |
|---|---|---|---|---|
| p300 | Writer (KAT) | Germline | Rubinstein–Taybi syndrome | [102] |
| Somatic | Diffuse large B-cell lymphoma, transitional cell carcinoma of the bladder | [76,94,98] | ||
|
| ||||
| CBP | Writer (KAT) | Germline | Rubinstein–Taybi syndrome | [101] |
| Somatic | Relapsed acute lymphoblastic leukemia, diffuse large B-cell lymphoma, transitional cell carcinoma of the bladder | [76,93,94,98] | ||
|
| ||||
| MLL2 | Writer (KMT) | Germline | Kabuki syndrome | [113,114] |
| Somatic | Non-Hodgkin lymphoma, medulloblastoma | [76,77,96] | ||
|
| ||||
| NSD2 | Writer (KMT) | Germline | Wolf–Hirschhorn syndrome | [91] |
| Somatic | Multiple myeloma | [90,91] | ||
|
| ||||
| GLP | Writer (KMT) | Germline | Kleefstra syndrome | [107] |
| Somatic | Medulloblastoma, ganglioglioma | [97,109] | ||
|
| ||||
| JARID1C | Eraser (KDM) | Germline | X-linked mental retardation | [110] |
| Somatic | Clear cell renal cell carcinoma | [99] | ||
KAT: Lysine acetyltransferase; KDM: Lysine demethylase; KMT: Lysine methyltransferase.
Somatic mutations in CBP give rise to hematological malignancies and solid tumors as described above, while germline mutations that result in haploinsufficiency or dominant negative functions of CBP cause the multiple malformation disorder Rubinstein–Taybi syndrome (RSTS). Clinical features of the disorder include moderate to severe mental retardation, dysmorphic facial features and skeletal abnormalities, including broad digits [101–103]. CBP-heterozygous mice share some of the clinical features of RSTS, highlighting the required function of CBP during and postembryonic development [104]. Point mutations occurring in the related KAT, p300, have been identified in a small percentage of RSTS patients that do not carry mutations in CBP [105], implicating both enzymes in the pathogenesis of this developmental disorder. Patients diagnosed with RSTS also have an increased risk for developing cancer, especially neural tumors and, not surprisingly, hematological malignancies [103,106]. Given the mutation spectrum of CBP and p300 observed in RSTS and in human malignancies, it is clear that appropriate expression and function of CBP and p300 is required for cell division, proliferation and differentiation. Perhaps tissues that rely on a pool of well-defined progenitor cells for homeostasis, such as the neural and hematopoietic systems, require the continued function of CBP and p300 postdevelopmentally.
EHMT1 haploinsufficiency, causes Kleefstra syndrome (previously referred to as 9q34 sub-telomeric deletion syndrome) [107]. Intragenic mutations within EHMT1, or microdeletions occurring within the EHMT1 locus on chromosomal region 9q34 (Figure 2), are common genomic aberrations that are shared by Kleefstra syndrome patients. Common clinical features of Kleefstra syndrome include severe mental retardation, craniofacial abnormalities and hypotonia, while some patients exhibit impaired social interaction and behavioral problems that may worsen with age [107,108]. Historically, Kleefstra syndrome patients have not shown an increased risk for developing malignancies, with only one recently identified patient having developed ganglioglioma [109].
The discovery of inactivating mutations in the histone H3K4 demethylase JARID1C first linked histone H3K4 methylation with X-linked mental retardation [65,110,111]. JARID1C binds to H3K9me3 through its PHD domain [65], and functions with HDACs, REST and G9a as a complex to repress the expression of genes important for neuronal development [112]. Mutations occurring within the PHD and catalytic domains of JARID1C were found to be detrimental to H3K9me3 binding as well as demethylase activity toward H3K4, respectively, suggesting that functions mediated by both of these domains are relevant to disease [65].
A number of mutations throughout MLL2 were recently identified by exome sequencing in patients diagnosed with Kabuki syndrome [113–116]. Kabuki syndrome is a rare multiple malformation disorder clinically diagnosed by the presence of multiple phenotypes including distinct facial anomalies, short stature, cardiac and renal abnormalities, and mental retardation [117,118]. Kabuki syndrome patients with MLL2 mutations are more likely to have renal abnormalities than those without MLL2 mutations [115,116]. The majority of the identified MLL2 mutations are nonsense and frameshift mutations that are located in regions that do not encode characterized functional domains, although they are predicted to result in a loss-of-function allele by truncating MLL2 upstream of the catalytic SET domain [113–116]. Interestingly, a number of mutations cluster within the region that encodes the FYRN and FYRC domains [113,115]. While the functions of these domains within MLL2 have not been defined, the FYR domains of MLL1 were recently implicated in mediating heterodimerization between the MLL1–AF4 fusion oncoprotein and wild-type MLL1, which promotes leukemogenesis [119]. Perhaps the Kabuki syndrome mutations that disrupt the MLL2 FYR domains interrupt protein interactions that are critical to maintain appropriate gene-expression programs that regulate neural, skeletal and muscular development. The identification of MLL2 genomic targets, nonhistone substrates, as well as interacting proteins will help define its pathogenic role in Kabuki syndrome.
Clearly, next-generation sequencing technologies will enhance the set of genetic testing methods currently used in the diagnosis and treatment of genetic diseases [69,71]. Importantly, WGS and WES offer unbiased approaches for discovering new candidates involved in pathogenesis. For instance, these techniques allow for discovery of mutations within multiple enzymes/proteins involved in a process by not limiting the inquiry to a particular chromosomal region or set of genes. Also, the sensitivity of next-generation sequencing allows for discovery of mutations that may not otherwise be easily detectable due to tumor heterogeneity. Finally, the powerful bioinformatics tools used to analyze WGS and WES data can identify causative mutations and correlate mutations within and between datasets to reveal common themes that emerge in a particular disease state.
Conclusion
Histone modifications, mediated by a plethora of histone modifying complexes, constitute an important epigenetic mechanism that regulates cell fate decisions. Deregulation of this mechanism contributes to tumorigenesis as indicated by the growing number of mutations that have been identified in chromatin modifiers in affected tissues. Regulation of downstream genes often involves cooperation between multiple chromatin and histone modifying activities. Crosstalk between chromatin modifiers may explain the haploinsufficiency caused by most of the gene mutations, as partially impaired function of histone-modifying enzymes is likely enough to tilt the balance towards ‘hyperactivation’ or ‘hyporepression’ of the downstream genes, with detrimental consequences for the physiology of the affected tissue.
Future perspective
High-throughput sequencing methods provide an invaluable window to survey the entire genome for subtle genetic alterations that contribute to human diseases. The development of efficient, low cost and robust assays to validate the functional significance of the identified mutations will be critical in order to keep up with the wealth of information generated by WGS or WES screens. Histone modification patterns associated with transcription have been extensively characterized and are often used as a measure of histone modifying activity. However, it is important to remember that histone modifications play a role in additional chromatin-templated processes, such as DNA repair and recombination, which are crucial for the maintenance of genome stability. Also, many of the enzymes that modify histones have nonhistone substrates that are involved in maintaining cellular homeostasis. Moving forward, it will be necessary to include consideration of these functions when investigating potential disease-causing mechanisms. Mutations affecting several histone modifiers are often identified in one disease. Defining mutational patterns within chromatin modifying networks that result in impaired developmental processes or that drive oncogenesis will shed light on the underlying mechanism of a particular disease state, and will ultimately enhance the development of personalized treatment approaches for patients suffering from these diseases.
Executive summary.
Histone modifiers regulate early cell fate decisions
Early cell fate decisions, like blastomere formation, are regulated by the presence of H3 arginine methylation.
The interplay between histone methylation active marks and repressive marks has been extensively analyzed during embryonic development, but the role of histone acetylation is just starting to emerge.
Differentiation & tissue development are controlled by histone-modifying enzymes
A common pattern observed in progenitor cells later in development is the repression of differentiation genes via repressive histone marks that can be removed during terminal differentiation.
The functions of different histone-modifying enzymes have been studied during tissue formation, providing insights in the molecular mechanisms that are deregulated in various developmental diseases.
Mutations in histone-modifying enzymes cause human diseases
Histone-modifying enzymes are commonly the targets of genomic aberrations, such as translocations, deletions and amplifications that give rise to human diseases.
Next-generation sequencing technologies, including whole-genome sequencing and whole-exome sequencing, are powerful methods to define causative mutations underlying genetic diseases.
Recent whole-genome sequencing and whole-exome sequencing studies have highlighted histone-modifying enzymes as a functional group enriched for mutations in cancer and developmental disorders.
Novel mutations discovered in histone modifiers are involved in hematological malignancies
Next-generation sequencing led to the discovery of novel deleterious point mutations in histone acetyltransferases and methyltransferases, which were previously implicated in hematological malignancies by translocations or deletions.
Mutations in histone modifiers may affect their activity toward histone or nonhistone substrates.
Histone-modifying enzymes emerge as potential therapeutic targets in solid tumors
Next-generation sequencing studies carried out in several types of solid tumors reveal novel pathogenic roles for histone-modifying enzymes.
Often, mutations were discovered in genes encoding several histone-modifying enzymes in a particular disease state, suggesting multiple mechanisms for disease progression that may involve histone modifications.
Mutations in histone modifiers underlie developmental disorders & cancer
Germline mutations in some histone-modifying enzymes cause developmental disorders, while acquired somatic mutations in the same enzyme cause cancer.
Mutations occurring in several different histone-modifying enzymes cause developmental disorders that share similar phenotypes, including mental retardation, musculoskeletal defects and facial anomalies.
Acknowledgments
The authors would like to thank H Graham and B Atanassov for helpful comments on the manuscript. The authors would also like to thank Y Geng and L Gann from the Research Medical Library at the University of Texas MD Anderson Cancer Center (TX, USA) for their assistance with literature searches.
Footnotes
Financial & competing interests disclosure
This work was supported by the Cancer Prevention Research Institute of Texas (CPRIT) RP100429 and by the NIH GM067718 (to SYR Dent). A Schibler is supported by the Schissler Fellowship in Genetics of Human Disease. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills AA. Throwing the cancer switch. reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 4.Soshnikova N, Duboule D. Epigenetic regulation of vertebrate Hox genes: a dynamic equilibrium. Epigenetics. 2009;4(8):537–540. doi: 10.4161/epi.4.8.10132. [DOI] [PubMed] [Google Scholar]
- 5.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344(1):7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 7▪▪.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445(7124):214–218. doi: 10.1038/nature05458. Dissects the function of H3 arginine methyltransferase in cell fate determination of a four-cell stage mouse embryo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q, Bruce AW, Jedrusik A, et al. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27(11):2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parfitt DE, Zernicka-Goetz M. Epigenetic modification affecting expression of cell polarity and cell fate genes to regulate lineage specification in the early mouse embryo. Mol Biol Cell. 2010;21(15):2649–2660. doi: 10.1091/mbc.E10-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’neill LP, Vermilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38(7):835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 11.Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou Y, Milne TA, Ruthenburg AJ, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 13.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 14.Conerly MI, Macquarrie KL, Fong AP, Yao Z, Tapscott SJ. Polycomb-mediated repression during terminal differentiation: what don’t you want to be when you grow up? Genes Dev. 2011;25(10):997–1003. doi: 10.1101/gad.2054311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 17.Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 18.Lan F, Bayliss PE, Rinn JL, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(7163):689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 19.Peng JC, Valouev A, Swigut T, et al. Jarid2/ Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasini D, Cloos PA, Walfridsson J, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 21.Shen X, Kim W, Fujiwara Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endoh M, Endo TA, Endoh T, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135(8):1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 23.Stock JK, Giadrossi S, Casanova M, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9(12):1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 24.Alder O, Lavial F, Helness A, et al. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137(15):2483–2492. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21(20):2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8(2):188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 27.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15(11):1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol Cell Biol. 2005;25(5):1804–1820. doi: 10.1128/MCB.25.5.1804-1820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasini D, Malatesta M, Jung HR, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38(15):4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao TP, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 31▪▪.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin ‘prepattern’ and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332(6032):963–966. doi: 10.1126/science.1202845. Highlights the importance of chromatin patterns in the fate decision between liver and pancreas by isolating ventral foregut endoderm cells from mouse embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319(1):110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53(2–3):275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- 34.Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134(6):1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 35.Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66α are embryonic lethal. PLoS ONE. 2007;2(6):e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu D, Fang J, Li Y, Zhang J. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS ONE. 2009;4(11):e7684. doi: 10.1371/journal.pone.0007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 38.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107(18):8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyu J, Jho EH, Lu W. Smek promotes histone deacetylation to suppress transcription of Wnt target gene brachyury in pluripotent embryonic stem cells. Cell Res. 2011;21(6):911–921. doi: 10.1038/cr.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Addis RC, Prasad MK, Yochem RL, et al. OCT3/4 regulates transcription of histone deacetylase 4 (Hdac4) in mouse embryonic stem cells. J Cell Biochem. 2010;111(2):391–401. doi: 10.1002/jcb.22707. [DOI] [PubMed] [Google Scholar]
- 41.Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trivedi CM, Luo Y, Yin Z, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3β activity. Nat Med. 2007;13(3):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 43.Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31(1):47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 44.Akhtar MW, Raingo J, Nelson ED, et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29(25):8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinti L, Chopra V, Rotili D, et al. Evaluation of histone deacetylases as drug targets in Huntington’s disease models. Study of HDACs in brain tissues from R6/2 and CAG140 knock-in HD mouse models and human patients and in a neuronal HD cell model. PLoS Curr. 2010;2 doi: 10.1371/currents.RRN1172. pii: RRN1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helmlinger D, Hardy S, Eberlin A, Devys D, Tora L. Both normal and polyglutamine-expanded ataxin-7 are components of TFTC-type GCN5 histone acetyltransferase – containing complexes. Biochem Soc Symp. 2006;73:155–163. doi: 10.1042/bss0730155. [DOI] [PubMed] [Google Scholar]
- 47.Mcmahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci USA. 2005;102(24):8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palhan VB, Chen S, Peng GH, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci USA. 2005;102(24):8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.David G, Abbas N, Stevanin G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17(1):65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- 50.David G, Durr A, Stevanin G, et al. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7) Hum Mol Genet. 1998;7(2):165–170. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- 51.Chen YC, Gatchel JR, Lewis RW, et al. Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Hum Mol Genet. 2012;21(2):394–405. doi: 10.1093/hmg/ddr474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caretti G, Di Padova M, Micales B, Lyons Ge, Sartorelli V. The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18(21):2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rampalli S, Li L, Mak E, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14(12):1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acharyya S, Sharma SM, Cheng AS, et al. TNF inhibits Notch-1 in skeletal muscle cells by Ezh2 and DNA methylation mediated repression: implications in duchenne muscular dystrophy. PLoS ONE. 2010;5(8):e12479. doi: 10.1371/journal.pone.0012479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H, Gu X, Su IH, et al. Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic β-cell proliferation. Genes Dev. 2009;23(8):906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung C, Lingbeek M, Shakhova O, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428(6980):337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- 59.Roman-Trufero M, Mendez-Gomez HR, Perez C, et al. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells. 2009;27(7):1559–1570. doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- 60.Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26(11):2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- 61.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27(10):3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirabayashi Y, Suzki N, Tsuboi M, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63(5):600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 63.Luna-Fineman S, Shannon KM, Lange BJ. Childhood monosomy 7: epidemiology, biology, and mechanistic implications. Blood. 1995;85(8):1985–1999. [PubMed] [Google Scholar]
- 64.Dohner K, Brown J, Hehmann U, et al. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92(11):4031–4035. [PubMed] [Google Scholar]
- 65.Iwase S, Lan F, Bayliss P, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128(6):1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 66.Kleine-Kohlbrecher D, Christensen J, Vandamme J, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38(2):165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Bokhoven H, Kramer JM. Disruption of the epigenetic code. an emerging mechanism in mental retardation. Neurobiol Dis. 2010;39(1):3–12. doi: 10.1016/j.nbd.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Shima Y, Kitabayashi I. Deregulated transcription factors in leukemia. Int J Hematol. 2011;94(2):134–141. doi: 10.1007/s12185-011-0905-9. [DOI] [PubMed] [Google Scholar]
- 69.Kobelka CE. Exome sequencing: expanding the genetic testing toolbox. Clin Genet. 2010;78(2):132–134. doi: 10.1111/j.1399-0004.2010.01452_1.x. [DOI] [PubMed] [Google Scholar]
- 70.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12(9):628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 71▪.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. Discusses the advantages and limitations of using exome sequencing to define causative mutations underlying genetic diseases. [DOI] [PubMed] [Google Scholar]
- 72.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 73.Liu H, Cheng EH, Hsieh JJ. MLL fusions: pathways to leukemia. Cancer Biol Ther. 2009;8(13):1204–1211. doi: 10.4161/cbt.8.13.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2010;152(2):141–154. doi: 10.1111/j.1365-2141.2010.08459.x. [DOI] [PubMed] [Google Scholar]
- 75.Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22(24):3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90(5):1799–1806. [PubMed] [Google Scholar]
- 79.Heuser M, Yap DB, Leung M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113(7):1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 80.Velichutina I, Shaknovich R, Geng H, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116(24):5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17(9):2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 82.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yap DD, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Makishima H, Jankowska AM, Tiu RV, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24(10):1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 85.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 86.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 87.Jankowska AM, Makishima H, Tiu RV, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2 and DNMT3A. Blood. 2011;118(14):3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grossmann V, Kohlmann A, Eder C, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25(5):877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 89.Van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stec I, Wright TJ, Van Ommen GJ, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7(7):1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 92.Martinez-Garcia E, Popovic R, Min DJ, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117(1):211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94▪.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. Reports inactivating mutations in the histone acetyltransferases CREBBP and EP300, and loss of CBP acetyltransferase function toward its nonhistone substrates, p53 and BCL6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oike Y, Takakura N, Hata A, et al. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood. 1999;93(9):2771–2779. [PubMed] [Google Scholar]
- 96▪▪.Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. First report implicating MLL2 and MLL3 as tumor suppressors in pediatric medulloblastoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 102.Roelfsema JH, Peters DJ. Rubinstein–Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med. 2007;9(23):1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- 103.De Sario A. Clinical and molecular overview of inherited disorders resulting from epigenomic dysregulation. Eur J Med Genet. 2009;52(6):363–372. doi: 10.1016/j.ejmg.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 104.Alarcon JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein–Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 105.Bartholdi D, Roelfsema JH, Papadia F, et al. Genetic heterogeneity in Rubinstein–Taybi syndrome: delineation of the phenotype of the first patients carrying mutations in EP300. J Med Genet. 2007;44(5):327–333. doi: 10.1136/jmg.2006.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller RW, Rubinstein JH. Tumors in Rubinstein–Taybi syndrome. Am J Med Genet. 1995;56(1):112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- 107.Kleefstra T, Smidt M, Banning MJ, et al. Disruption of the gene Euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet. 2005;42(4):299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Verhoeven WM, Egger JI, Vermeulen K, Van De Warrenburg BP, Kleefstra T. Kleefstra syndrome in three adult patients: further delineation of the behavioral and neurological phenotype shows aspects of a neurodegenerative course. Am J Med Genet A. 2011;155(10):2409–2415. doi: 10.1002/ajmg.a.34186. [DOI] [PubMed] [Google Scholar]
- 109.Cheung HC, Yatsenko SA, Kadapakkam M, et al. Constitutional tandem duplication of 9q34 that truncates EHMT1 in a child with ganglioglioma. Pediatr Blood Cancer. 2012;58(5):801–805. doi: 10.1002/pbc.23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jensen LR, Amende M, Gurok U, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005;76(2):227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rujirabanjerd S, Nelson J, Tarpey PS, et al. Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur J Hum Genet. 2010;18(3):330–335. doi: 10.1038/ejhg.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tahiliani M, Mei P, Fang R, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447(7144):601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 113▪▪.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. Reports that mutations in MLL2 cause Kabuki syndrome. This is the first example in which mutations within an MLL family member play a causal role in a multiple malformation disorder. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paulussen AD, Stegmann AP, Blok MJ, et al. MLL2 mutation spectrum in 45 patients with Kabuki syndrome. Hum Mutat. 2010;32(2):E2018–E2025. doi: 10.1002/humu.21416. [DOI] [PubMed] [Google Scholar]
- 115.Hannibal MC, Buckingham KJ, Ng SB, et al. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am J Med Genet A. 2011;155A(7):1511–1516. doi: 10.1002/ajmg.a.34074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y, Bogershausen N, Alanay Y, et al. A mutation screen in patients with Kabuki syndrome. Hum Genet. 2011;130(6):715–724. doi: 10.1007/s00439-011-1004-y. [DOI] [PubMed] [Google Scholar]
- 117.Niikawa N, Matsuura N, Fukushima Y, Ohsawa T, Kajii T. Kabuki make-up syndrome: a syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J Pediatr. 1981;99(4):565–569. doi: 10.1016/s0022-3476(81)80255-7. [DOI] [PubMed] [Google Scholar]
- 118.Kuroki Y, Suzuki Y, Chyo H, Hata A, Matsui I. A new malformation syndrome of long palpebral fissures, large ears, depressed nasal tip, and skeletal anomalies associated with postnatal dwarfism and mental retardation. J Pediatr. 1981;99(4):570–573. doi: 10.1016/s0022-3476(81)80256-9. [DOI] [PubMed] [Google Scholar]
- 119.Pless B, Oehm C, Knauer S, Stauber RH, Dingermann T, Marschalek R. The heterodimerization domains of MLL-FYRN and FYRC – are potential target structures in t(4;11) leukemia. Leukemia. 2011;25(4):663–670. doi: 10.1038/leu.2010.308. [DOI] [PubMed] [Google Scholar]