Abstract
Acquisition of self-renewal capability by myeloid progenitors to become leukemic stem cells during myeloid leukemia development is poorly understood. Here, we show that Setbp1 overexpression efficiently confers self-renewal capability to myeloid progenitors in vitro, causing their immortalization in the presence of stem cell factor and IL-3. Self-renewal after immortalization requires continuous Setbp1 expression. We also found that Hoxa9 and Hoxa10 mRNA are present at dramatically higher levels in Setbp1-immortalized cells compared with other immortalized cells, and are induced shortly after Setbp1 expression in primary myeloid progenitors. Suppression of either gene in Setbp1-immortalized cells drastically reduces their colony-forming capability. Interestingly, Setbp1 protein associates with Hoxa9 and Hoxa10 promoters in chromatin immunoprecipitation assays in these cells, suggesting that both are direct transcriptional targets of Setbp1. Setbp1 also promotes self-renewal of myeloid progenitors in vivo as its coexpression with BCR/ABL transforms primary mouse myeloid progenitors, generating aggressive leukemias in recipient mice resembling chronic myelogenous leukemia (CML) myeloid blast crisis. Increased SETBP1 mRNA levels were also detected in a subset of CML advanced phase/blast crisis patients with high levels of HOXA9 and HOXA10 expression. Thus, Setbp1 activation represents a novel mechanism conferring self-renewal capability to myeloid progenitors in myeloid leukemia development.
Introduction
Increasing evidence supports the cancer stem cell model for myeloid leukemias that each leukemia is a heterogeneous population and only a fraction of the leukemia cells, known as the leukemic stem cells (LSCs) or leukemia initiating cells, are capable of sustaining and regenerating the disease because of their capability of unlimited self-renewal.1,2 Therefore, targeting against the self-renewal of LSCs represents a promising strategy for their elimination and effective treatment of this disease. However, poor understanding of the molecular mechanism(s) underlying LSC self-renewal has severely hampered efforts in finding ways to inhibit such processes.
LSCs of myeloid leukemias were initially thought to be exclusively derived from hematopoietic stem cells (HSCs) that normally possess extensive self-renewal capability; however, recent studies have suggested that the more mature progenitor cells can also give rise to these malignant cells by acquiring self-renewal capacity through mutations.3,4 Identification and characterization of such mutations will provide crucial insights into the molecular mechanisms controlling LSC self-renewal. For human acute myeloid leukemias (AMLs), a limited number of such mutations including MLL-ENL, MLL-AF9, and MOZ-TIF2 were identified because of their capability to transform mouse granulocyte and macrophage progenitors (GMPs).3–5 In chronic myelogenous leukemia (CML), whereas LSCs in the chronic phase of the disease are probably derived directly from HSCs,6 GMPs have been proposed as the source for LSCs in CML myeloid blast crisis.7,8 BCR/ABL alone is not able to increase self-renewal in GMPs,5,9 suggesting that other mutations must contribute to the gaining of self-renewal capability by these cells. However, relatively few mutations, including NUP98/HOXA9 and overexpression of Hes1, have been functionally confirmed by their ability to cooperate with BCR/ABL to transform mouse GMPs.9,10 Therefore, identification of additional genes/pathways that could confer self-renewal capability to myeloid progenitors during leukemia development will be critical to gain a complete understanding of LSC self-renewal mechanisms.
Mutations known to confer self-renewal capability to LSCs of myeloid leukemias have also been shown to immortalize hematopoietic progenitors in vitro.5,11 We attempted in our previous studies to identify novel LSC self-renewal regulators by screening for genes capable of immortalizing myeloid progenitors in culture through retroviral insertional mutagenesis.12,13 Two immortalized progenitor lines established in these studies contain independent retroviral insertions activating Setbp1 expression. In addition, SETBP1 was frequently activated by vector insertions in expanding myeloid clones in 1 patient of the recent gene therapy trial for X-linked chronic granulomatous disease (CGD), suggesting that it may help confer self-renewal capability to myeloid progenitors in vivo.13 These findings point to a potential role of Setbp1 in conferring self-renewal capability to LSCs in myeloid leukemias.
SETBP1 encodes a predominantly nuclear localized large protein of 1542 amino acids with largely unclear function. SETBP1 was initially found to interact with SET, which is a small protein inhibitor for tumor suppressors PP2A and NM23-H1.14–16 SET was found to fuse with another gene CAN through chromosome translocation in a case of acute undifferentiated leukemia.17,18 Inhibition of PP2A by SET overexpression was also identified as a mechanism for the progression of CML.19 Recent studies have suggested direct involvement of SETBP1 in leukemia development. SETBP1 has been reported to be involved in chromosome translocation in a case of acute T-cell leukemia.20 More recently, SETBP1 was found activated by chromosome translocation in a case of AML.21 The same study also identified SETBP1 overexpression in more than 27% of 192 AML patients and suggested that SETBP1 activation causes PP2A inhibition in AML cells through stabilization of SET.21
In this study, we provide in vitro and in vivo evidence to support a new role for Setbp1 in conferring self-renewal capability to LSCs in myeloid leukemias, and have also uncovered a critical transcriptional mechanism underlying Setbp1-induced self-renewal. Our examination of CML patients also suggests that SETBP1 activation may play a role in human CML progression.
Methods
Mice
The C57BL/6 and C57BL/6-Ly5.2 mice (Charles River) were maintained in the animal facility of Laboratory of Animal Medicine at Uniformed Services University of the Health Sciences (USUHS). All mouse experiments were carried out according to protocols approved by the USUHS Institutional Animal Care and Use Committee.
Patient samples
Cord blood (CB) samples were kindly provided by StemCyte. CML samples were obtained from patients seen at the City of Hope National Medical Center (COHNMC). Mononuclear cells were isolated using Ficoll separation. All subjects signed an informed consent form. Sample acquisition was approved by the Institutional Review Boards at the COHNMC, in accordance with an assurance filed with and approved by the Department of Health and Human Services, and met all requirements of the Declaration of Helsinki.
Retroviral constructs
Setbp1 cDNA was amplified by reverse transcription-polymerase chain reaction (RT-PCR) from BM70 cells, confirmed by sequencing, and cloned into the HpaI and EcoRI site of pMYs-IRES-GFP vector for the generation of pMYs-Setbp1-IRES-GFP. For generating pMYs-3xFLAGSetbp1-IRES-GFP, 3x FLAG DNA sequence was PCR-amplified from pDest-737 and inserted in frame behind the start codon of Setbp1 cDNA. MSCV-BCR/ABL-IRES-GFP virus was previously described.22 Virus was produced by transient transfection of Plat-E cells using Fugene 6 (Roche) and tittered by infecting NIH-3T3 cells (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry
LSK cells (Lin−Sca-1+c-kit+), CMPs (IL-7Rα−Sca-1−c-Kit+FcγR-II/IIILoCD34+), and GMPs (IL-7Rα−Sca-1−c-Kit+FcγR-II/IIIHiCD34+) were purified from C57BL/6 mice (7-12 weeks old) as described using a FACSAria cell sorter.23,24 For flow cytometry analysis of leukemic bone marrow (BM) and spleen, samples were analyzed on a BD LSRII flow cytometer. Dead cells were excluded by staining with Sytox Blue (supplemental Methods).
Real-time RT-PCR
For mouse cells or tissues, total RNA was extracted using RNAeasy Plus mini kit (QIAGEN). Oligo-dT-primed cDNA samples were prepared using Superscript III (Invitrogen), and real-time PCR analysis was performed in triplicates using SYBR green detection reagents (Invitrogen) on a 7500 real time PCR system (Applied Biosystems). Quantitative detection of SETBP1, HOXA9, HOXA10, and BCR transcripts in human samples was performed by TaqMan assay using the ABI Prism 7900 sequence detector (Applied Biosystems; supplemental Methods)
Retroviral transduction and BM transplantation
For testing leukemia induction using progenitors generated in vitro, infections were performed on retronectin-coated plates in the presence of mouse stem cell factor (SCF; 100 ng/mL) and IL-3 (10 ng/mL). GMP infections were performed in the presence of mouse SCF (100 ng/mL) and IL-11(10 ng/mL). C57BL/6-Ly5.2 recipient mice (7- to 12-week-old females) were irradiated twice at a total dose of 1000 R using a GammaCell 40 irradiator. Transduced cells in indicated numbers, along with supporting BM cells (7 × 105 cells/recipient), from unirradiated C57BL/6-Ly5.2 mice were subsequently transplanted into recipient mice via tail-vein injection. For secondary, tertiary, and quaternary transplantations, 1 × 106 spleen cells from preceding leukemic recipients were injected (supplemental Methods).
ChIP
Chromatin immunoprecipitations (ChIPs) were performed using ChIP-IT Express kit (Active Motif). Hoxa9 locus primers were previously described.25 See supplemental Methods for Hoxa10 locus-specific primer sequences and PCR conditions.
Results
Ectopic expression of Setbp1 efficiently immortalizes primary myeloid progenitors in culture
To test whether overexpression of Setbp1 alone is capable of immortalizing myeloid progenitors in culture, we cloned murine Setbp1 cDNA into the pMYs-GFP retroviral vector and compared the ability of the resulting Setbp1 virus (pMYs-Setbp1-IRES-GFP) to control virus (pMYs-IRES-GFP) to immortalize BM progenitors in the presence of SCF plus IL-3. To enrich myeloid progenitors for infection, whole BM cells from C57BL/6 mice were first cultured in serum-free medium in the presence of SCF, TPO, IGF-2, and FGF-1 for 5 days to expand HSCs.26 These expanded HSCs were then induced to differentiate primarily into myeloid progenitors in the presence of mouse SCF plus IL-3 for 4 days which were subsequently transduced and passaged for 4 weeks to assess for immortalization (Figure 1A). At a low titer of 1 × 105 cfu, infection by Setbp1 virus caused immortalization of myeloid progenitors in each of 6 independent experiments (Figure 1B top panel). These cells are truly immortalized as they could be continuously passaged for 6 months until the experiments were terminated. In contrast, infection with empty virus at the same titer was unable to produce any immortalized myeloid progenitors and only mature macrophages were observed in the culture after 4 weeks (Figure 1B bottom panel). Myeloid progenitors immortalized by Setbp1 virus are dependent on IL-3 for proliferation, and differentiation into mature neutrophils can be induced by treatment with mouse granulocyte colony-stimulating factor (G-CSF; Figure 1C). We also examined the clonality of these immortalized cells by Southern blotting analysis of their viral integrations using a GFP-specific probe. These cells contain many integrations with various intensities (Figure 1D), suggesting that they are polyclonal. Therefore, ectopic Setbp1 expression alone is probably sufficient to cause immortalization of these myeloid progenitors. We also found that Setbp1 mRNA is expressed at significantly higher levels in purified mouse LSK cells enriched for HSCs compared with more mature myeloid progenitors including common myeloid progenitors (CMPs), and GMPs and that infection of purified mouse GMPs with Setbp1 virus, but not empty virus, efficiently generated immortalized myeloid progenitors in the presence of SCF and IL-3 (Figure 1E and data not shown), further suggesting that Setbp1 may have specific functions in normal HSCs or multipotent progenitors and its abnormal reactivation in committed myeloid progenitors causes immortalization of these cells.
Figure 1.
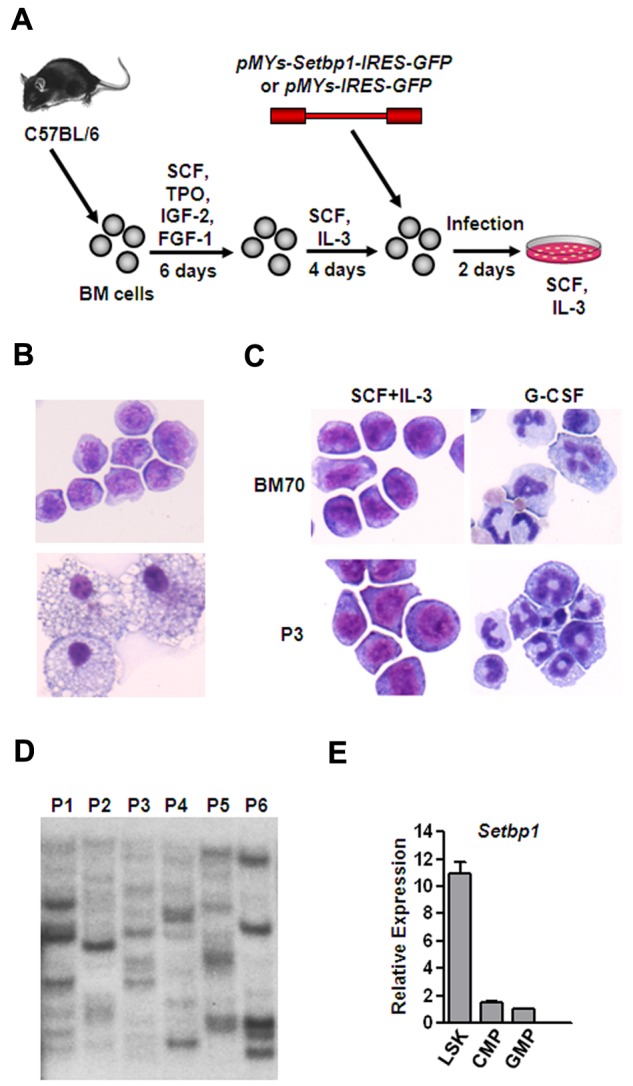
Efficient immortalization of myeloid progenitors by Setbp1 expression. (A) Schematic diagram of the immortalization procedure. (B) Representative cytospin preparation and Wright-Giemsa staining of cells infected with Setbp1 (top panel) or empty retrovirus (bottom panel) at 1 month after infection. Original magnification ×400. Images were obtained using a Nikon Eclipse E800 microscope and a Qimaging Micropublisher 5.0 digital camera. (C) Cytospin preparation of Setbp1-immortalized cells (BM70 and P3 population) before and after treatment with G-CSF for 2 days. BM70 cells are immortalized by insertional activation of endogenous Setbp1.12,13 Original magnification ×400. (D) Southern blotting analysis of viral integrations present in 6 Setbp1-immortalized myeloid progenitor populations (P1-P6) using a GFP-specific probe. Seven ug of genomic DNA from each population was digested with EcoRI, resulting the generation of a single GFP-containing DNA fragment from each provirus. Each band represents an independent integration. (E) Real-time RT-PCR analysis of total RNA isolated from purified LSK, CMP, and GMP populations of C57BL/6 mice using Setbp1-specific primers. Relative expression levels were calculated by normalizing to β-actin mRNA levels in the same sample and also in LSK cells. The mean and SD of each relative expression level is shown.
Continuous Setbp1 expression is required for the self-renewal of immortalized myeloid progenitors
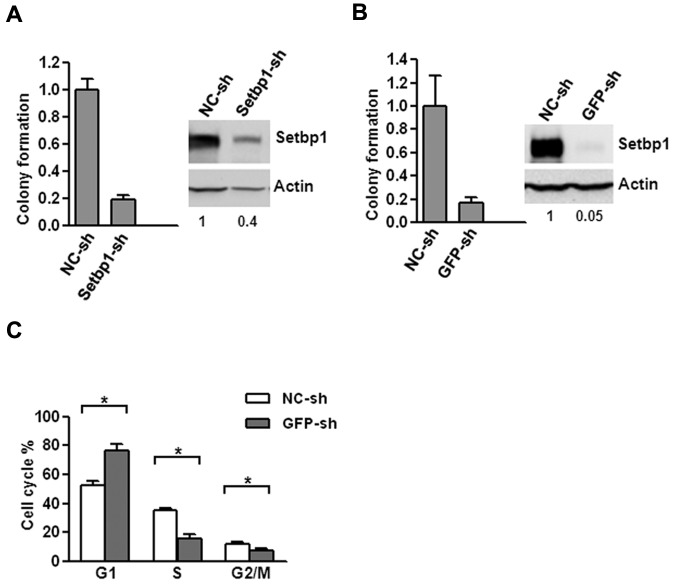
To further confirm that Setbp1 expression is responsible for immortalization, we examined the effects of Setbp1 knockdown on Setbp1-immortalized myeloid progenitors. Lentiviral delivery of a Setbp1-specific shRNA (Setbp1-sh) versus a nontargeting shRNA (NC-sh) caused approximately 60% knockdown of Setbp1 protein levels in BM70 cells immortalized by insertional activation of endogenous Setbp1(Figure 2A).12,13 This knockdown coincided with poor expansion of BM70 cells in liquid medium as well as approximately 80% reduction in their colony-forming potential on methylcellulose (Figure 2A), suggesting that Setbp1 expression is essential for the continuous proliferation/self-renewal of immortalized myeloid progenitors.
Figure 2.
Setbp1 knockdown inhibits the proliferation of Setbp1-immortalized myeloid progenitors. (A) Left panel, mean and SD of colony formation potential of BM70 cells in the presence of SCF and IL-3 at 48 hours after infection with a Setbp1-specific shRNA (Setbp1-sh) and control shRNA (NC-sh); right panel, representative Western blot analysis of Setbp1 and actin protein in the same infected cells at 72 hours after infection. Relative Setbp1 protein levels after normalization to actin levels in the same sample are indicated. (B) Mean and SD of colony formation potential in the presence of SCF and IL-3 (left panel) and Western blotting analyses (right panel) of S3 cells at 48 and 72 hours, respectively, after infection with GFP-specific shRNA (GFP-sh) and control shRNA (NC-sh). (C) Cell cycle distribution of S3 cells infected with GFP-specific and control shRNA determined by PI-staining at 96 hours after infection. The mean and SD of each phase is shown (*P < .05).
To exclude possible off-target effects of Setbp1-sh, we also tested a different strategy of reducing Setbp1 expression. Because a single mRNA is synthesized from our pMYs-Setbp1-IRES-GFP virus for the production of both Setbp1 and GFP protein, we tested suppressing Setbp1 expression by a shRNA targeting GFP portion of the mRNA in S3 myeloid progenitor cells which had been immortalized by this virus using the same procedure described in Figure 1A. Indeed, a 95% knockdown of Setbp1 protein levels was achieved in these cells using a lentivirus expressing a GFP-specific shRNA (Figure 2B, GFP-sh). Similar to BM70 cells infected with Setbp1-sh, poor expansion in liquid culture and dramatically reduced colony-forming potential were also observed for S3 cells after infection with GFP-sh compared with the control virus (Figure 2B), confirming that specific knockdown of Setbp1 expression in Setbp1-immortalized myeloid progenitors inhibits their continuous expansion. To determine the nature of this inhibition, we further analyzed apoptosis, differentiation, and cell cycle status of S3 cells after infection with GFP-sh virus, which results in more efficient Setbp1 knockdown (Figure 2). We could not detect any significant increases in annexin V staining at 72 and 96 hours after lentiviral infection of S3 cells (data not shown), suggesting that apoptosis did not contribute significantly to inhibition of cell expansion. No significant increase in morphologic differentiation was observed by cytospin analysis of the infected cells of these time points (data not shown). In contrast, cell cycle analysis of the infected S3 cells by propidium iodide (PI) staining at the same time points revealed a gradual and significant increase of cells in G1 phase and concomitant reductions of cells in G2/M and S phase (Figure 2C and data not shown), suggesting that Setbp1 reduction blocks cell cycle progression and subsequently expansion of these immortalized cells. In combination, these data demonstrate that Setbp1 expression remains essential for the continuous proliferation/self-renewal of myeloid progenitors after immortalization and suggests that Setbp1 plays a direct role in promoting self-renewal of these immortalized cells.
Setbp1 activates Hoxa9 and Hoxa10 expression in myeloid progenitors
To identify potential mechanisms responsible for Setbp1-induced self-renewal of immortalized cells, we examined the expression of a number of known regulators of hematopoietic self-renewal by real-time RT-PCR in Setbp1-immortalized cells in comparison with myeloid progenitors immortalized by other mechanisms. We found that Hoxa9 and Hoxa10 mRNAs are expressed at dramatically higher levels in Setbp1-immortalized cells including BM70, SF1 (a mouse HSC-like line immortalized by retroviral insertional activation of endogenous Setbp113), S1 and S2 (2 myeloid progenitor lines immortalized by Setbp1 virus using the same protocol outlined in Figure 1A), than myeloid progenitors immortalized by other mechanisms (Figure 3A). We also analyzed Hoxa9 and Hoxa10 mRNA expression in primary myeloid progenitor cells generated, as described in Figure 1A, 48 hours after their infection by Setbp1 or empty virus (Figure 3B). Significant increases in Hoxa9 and Hoxa10 mRNA levels were detected in cells transduced by the Setbp1 virus versus empty virus (Figure 3B), suggesting that both are up-regulated soon after Setbp1 expression. Therefore, both Hoxa9 and Hoxa10 are probably downstream targets of Setbp1.
Figure 3.
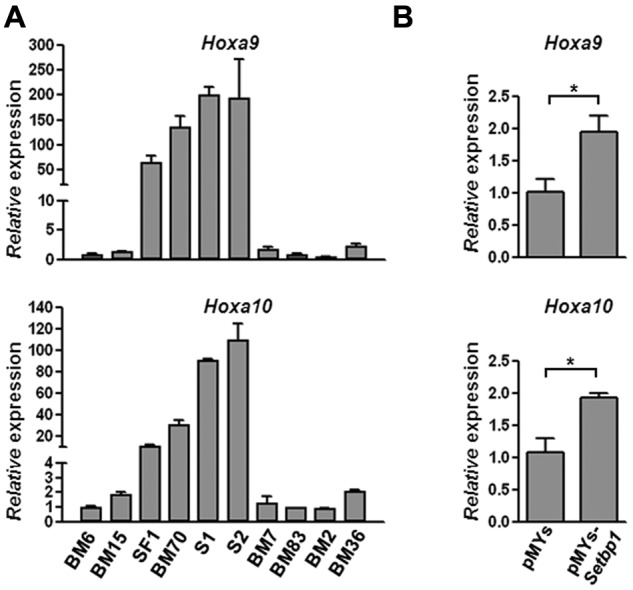
Setbp1 expression increases Hoxa9 and Hoxa10 mRNA levels. (A) Real-time RT-PCR analysis of total RNA from indicated immortalized progenitor lines using primers specific for Hoxa9 (top panel) and Hoxa10 (bottom panel). Lines immortalized by activation of endogenous Setbp1: SF1 and BM70; lines immortalized by retroviral Setbp1 expression: S1 and S2; Evi1-immortalized lines: BM7 and BM83; Prdm16-immortalized lines: BM2 and BM36; lines immortalized by unknown mechanisms: BM6 and BM15. Relative expression levels were calculated by normalizing to β-actin mRNA levels in the same sample and also in BM6 cells. The mean and SD of each relative expression level is shown. (B) Real-time RT-PCR analysis of Hoxa9 (top panel) and Hoxa10 (bottom panel) mRNA levels using total RNA from BM progenitors 48 hours after transduction by Setbp1 virus or empty virus. Relative expression levels were calculated by normalizing to β-actin mRNA levels in the same sample and also in cells infected with empty virus (*P < .05).
Hoxa9 and Hoxa10 are essential for Setbp1-induced immortalization
To test whether Hoxa9 and Hoxa10 are important downstream mediators of Setbp1-induced self-renewal, we examined effects of their knockdown on Setbp1-immortalized cells. Transduction by lentiviruses expressing 2 different Hoxa9-targeting shRNAs caused approximately 95% and 89% knockdown in Hoxa9 protein levels, respectively in BM70 cells (Figure 4A). Hoxa9 knockdowns were accompanied by dramatic reductions in colony-forming potential of BM70 cells (Figure 4A). Similar reductions after Hoxa9 knockdown were also observed for S3 cells (supplemental Figure 2). We did not observe any significant increases of annexin V staining in both cell lines 72 hours after infection with Hoxa9 shRNAs (data not shown), suggesting that reductions in colony-forming potential were not because of increased apoptosis. Cytospin analysis at 72 and 96 hour after infection also showed no significant increase in neutrophil or macrophage production, indicating that Hoxa9 knockdown may not increase differentiation. In contrast, cell cycle analysis of the infected BM70 cells at 48 and 72 hours after infection revealed that there was a gradual increase of cells in G1 phase and concomitant reduction of cells in G2/M and S phase (Figure 4C and data not shown). Taken together, these results suggest that the reduction in colony-forming potential of Setbp1-immortalized cells after Hoxa9 knockdown is primarily because of blocking cell cycle progression and Hoxa9 is essential for Setbp1-induced self-renewal.
Figure 4.
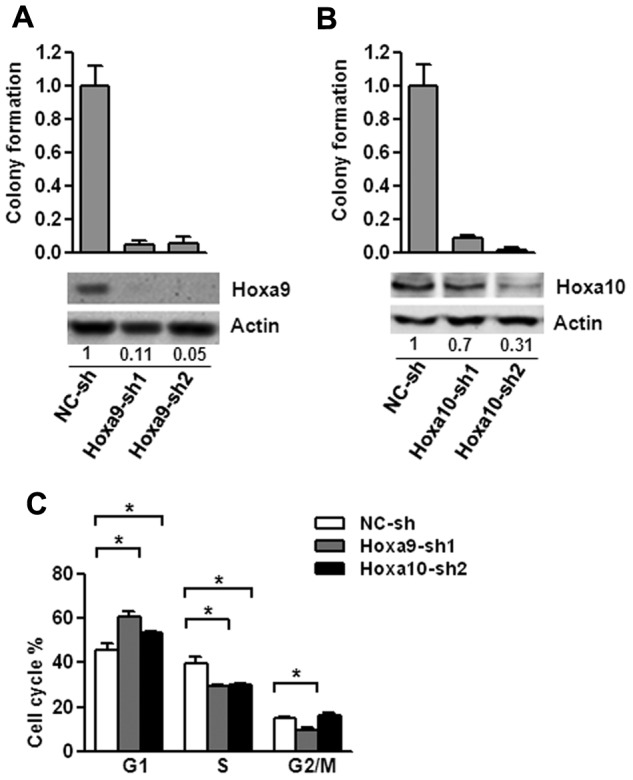
Hoxa9 and Hoxa10 are essential for the proliferation of Setbp1-immortalized myeloid progenitors. (A) Top panel, mean and SD of colony formation potential of BM70 cells in the presence of SCF and IL-3 at 48 hours after infection with Hoxa9-specific shRNAs (Hoxa9-sh1 and Hoxa9-sh2) and control shRNA (NC-sh); bottom panel, representative Western blotting analysis of Hoxa9 and actin protein in the infected cells of the top panel at 72 hours after infection. Relative Hoxa9 protein levels after normalization to actin levels in the same sample are indicated. (B) Mean and SD of colony formation potential in the presence of SCF and IL-3 (top panel) and Western blotting analyses (bottom panel) of BM70 cells at 48 and 72 hours, respectively, after infection with Hoxa10-specific shRNAs (Hoxa10-sh1 and Hoxa10-sh2) and control shRNA (NC-sh). Relative Hoxa10 protein levels after normalization to actin levels are indicated. (C) Cell cycle distribution of BM70 cells infected with Hoxa9-sh1 and Hoxa10-sh2 and control shRNA determined by PI-staining at 72 hours after infection. The mean and SD of each phase is shown (*P < .05).
Significant negative effects on colony-forming potential were also observed on Hoxa10 knockdown in BM70 and S3 cells using 2 individual Hoxa10-specific shRNAs with different knockdown efficiencies (Figure 4B, supplemental Figure 2). Similar to Hoxa9 knockdown, this effect is probably because of blocking cell cycle progression, as there is a significant increase in the percentage of G1 phase cells and a decrease in cells of S phase after infection with Hoxa10-sh2 virus, whereas no significant changes in differentiation or apoptosis were observed (Figure 4C and data not shown). Therefore, Hoxa10 activation is also essential for the proliferation/self-renewal of Setbp1-immortalized myeloid progenitors.
Hoxa9 and Hoxa10 represent direct transcriptional targets of Setbp1
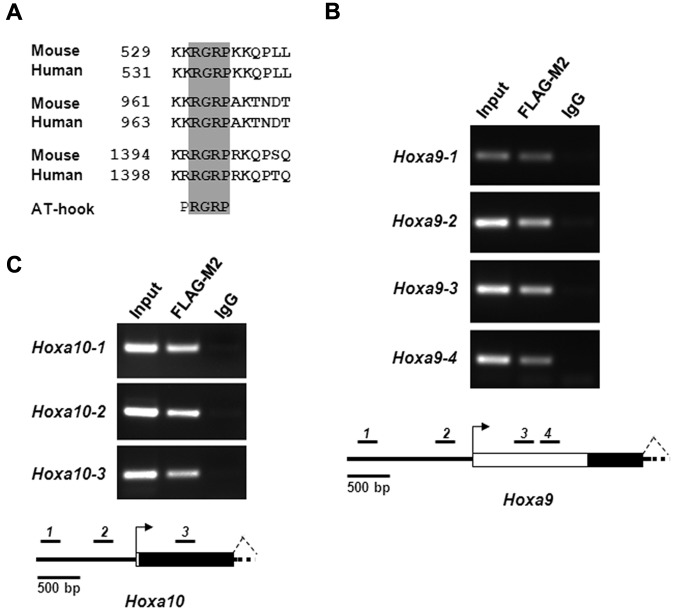
Sequence analysis of both human and mouse Setbp1 proteins reveals that they contain 3 highly conserved AT-hook motifs (Figure 5A), which carry a Pro-Arg-Gly-Arg-Pro consensus sequence (with R-G-R-P being highly conserved). AT-hook motifs are known to bind to the minor groove of stretches of AT-rich DNA in a nonsequence specific manner.27–29 This finding suggests that Setbp1 may function as a transcription factor. The rapid increases in Hoxa9 and Hoxa10 mRNA levels in primary hematopoietic progenitors after transduction with Setbp1-expressing virus further implicate that they may represent direct transcriptional targets of Setbp1. Therefore, we carried out ChIP assays to test whether Setbp1 directly binds to Hoxa9 and Hoxa10 promoters in Setbp1-immortalized myeloid cells. To facilitate the immunoprecipitation of Setbp1 protein, we generated a pMYs retrovirus expressing an N-terminal 3xFLAG-tagged Setbp1 protein (pMYs-3xFLAG-Setbp1-IRES-GFP). Similar to the pMYs-Setbp1-IRES-GFP virus, infection with this virus causes efficient immortalization of mouse myeloid progenitors (data not shown). Myeloid progenitors immortalized by this virus were subsequently used for ChIP assays. Interestingly, multiple regions of Hoxa9 and Hoxa10 promoter and first exon, but not nearby regions at Hoxa6 and Hoxa11, could be efficiently amplified in immunoprecipitates prepared from these myeloid progenitors using the FLAG-M2 antibody and not control IgG (Figure 5B-C, supplemental Figure 3), suggesting that Setbp1 directly and specifically binds to these Hoxa9 and Hoxa10 regions. Same results were also observed using another myeloid progenitor population separately immortalized by the same virus (data not shown). As expected, the immunoprecitation of these Hoxa9 and Hoxa10 regions is specific to 3XFLAG-tagged Setbp1 protein as the same DNA fragments were not amplified from FLAG-M2 immunoprecipitates from S2 cells immortalized by wild-type Setbp1 (data not shown). Moreover, ectopic Setbp1 expression significantly activated Hoxa9 and Hoxa10 promoter activity in luciferase assays in 293T cells (supplemental Figure 4). Collectively, these data suggest that Setbp1 protein may directly activate Hoxa9 and Hoxa10 transcription by binding to their promoters.
Figure 5.
Setbp1 directly activates Hoxa9 and Hoxa10 transcription. (A) Amino acid sequence alignments of AT-hook DNA-binding motifs present in human and mouse Setbp1 protein. Amino acid numbers are indicated. The invariable consensus sequence of AT-hook motifs (RGRP) are highlighted in gray. (B-C) ChIP analysis of myeloid progenitors immortalized by the expression of 3xFLAG-tagged Setbp1 using anti-FLAG M2 antibody and control IgG. PCR products using primers specific to various regions of Hoxa9 (B) or Hoxa10 locus (C) were resolved on ethidium bromide-stained agarose gel (top panel). Results are representative of 3 independent experiments. Diagrams of tested region of Hoxa9 and Hoxa10 locus are also shown (bottom panels). Locations of PCR amplicons are indicated as black bars with corresponding numbers. Transcriptional start sites are indicated as arrows. Exons are indicated as white boxes with coding regions highlighted in black.
Setbp1 cooperates with BCR/ABL (p210) to transform myeloid progenitors
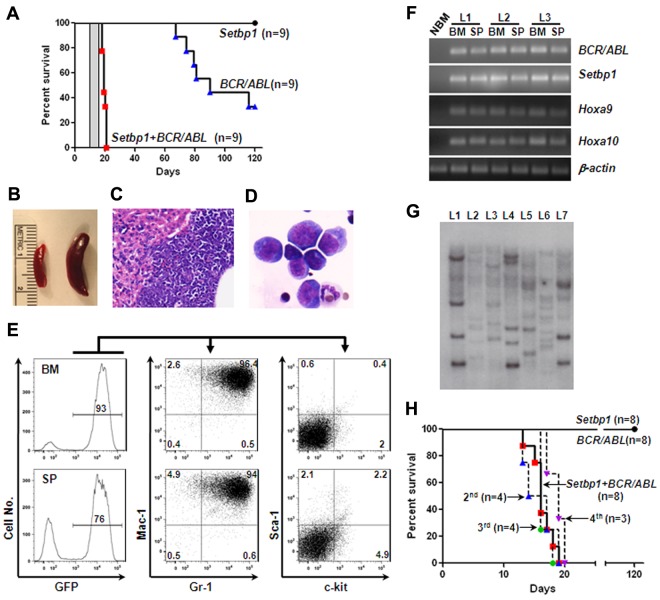
The ability of Setbp1 to immortalize myeloid progenitors in vitro suggests that its activation could play a role in conferring limitless self-renewal capability to LSCs derived from myeloid progenitors. It has been suggested that LSCs of CML myeloid blast crisis originates from committed myeloid progenitors and this conversion requires mutation(s) in addition to BCR/ABL as BCR/ABL alone failed to induce self-renewal of myeloid progenitors.5,7 We therefore reasoned that Setbp1 activation may represent such a self-renewal conferring mutation, which could cooperate with BCR/ABL to transform myeloid progenitors. To test this notion, we generated large number of mouse primary myeloid progenitors as described for testing immortalization of myeloid progenitors, and transduced them with equal titers of MSCV-BCR/ABL-IRES-GFP virus alone, pMYs-Setbp1-IRES-GFP alone, or the combination of both. The transduced cells were subsequently transplanted into lethally irradiated C57BL/6-Ly5.2 recipients (1 × 106 cells/mouse) along with supporting BM cells. Three independent experiments were performed and similar infection efficiencies were achieved among groups as determined by fluorescence-activated cell sorting (FACS) analysis of green fluorescent protein (GFP)–positive cells after infection (supplemental Figure 6). Interestingly, mice receiving cotransduced cells became moribund between 18 to 20 days, whereas recipients of singly infected cells remained healthy during this period (Figure 6A). Pathologic examination of these moribund mice showed that they developed leukemias with enlarged spleens and leukemic infiltrations into the liver (Figure 6B-C). Cytospin preparations of leukemic BM and spleens (not shown) revealed dominance of myeloid blasts, which represent 34% ± 7.8% (mean ± SD) of all nucleated cells in the BM (Figure 6D), suggesting a block in myeloid differentiation. The myeloid classification of the leukemias is further supported by FACS analyses showing that more than 80% of the leukemia cells in the BM or spleen are positive for both Gr-1 and Mac-1 and negative for CD19, CD3, and Ter119 (Figure 6E, supplemental Figure 7). Less than 10% of the leukemia cells were positive for c-kit or Sca-1(Figure 6E). As expected, these leukemias express high levels of BCR/ABL and Setbp1 mRNAs compared with normal BM (Figure 6F), and are transplantable, as secondary recipient mice transplanted with 1 × 106 spleen cells from leukemic primary recipients died of the same disease between 11 to 13 days (Figure 6A area highlighted in gray). Consistent with activation of Hoxa9 and Hoxa10 by Setbp1, dramatically higher levels of both mRNAs and proteins were detected in these leukemias than in control BM (Figure 6F, supplemental Figure 9). These phenotypic characterizations suggest the development of CML myeloid blast crisis in the recipient mice of cotransduced cells. Southern blotting analysis of genomic DNA from leukemic spleens revealed many viral integration bands of various intensities in each of these leukemias (Figure 6G), suggesting that they are highly oligoclonal. Taken together with the uniformly short disease latencies, this result further suggests that additional mutations may not be required for transformation after Setbp1 and BCR/ABL expression.
Figure 6.
Setbp1 cooperates with BCR/ABL to transform committed myeloid progenitors. (A) Survival curves of lethally irradiated C57BL/6-Ly5.2 mice receiving 1 × 106 in vitro expanded BM progenitors infected with MSCV-BCR/ABL-IRES-GFP virus, pMYs-Setbp1-IRES-GFP virus, or the combination. Results are combined from 3 independent experiments with each containing the 3 indicated groups. Transduction efficiencies: 28%-42% for BCR/ABL groups; 19%-25% for Setbp1 groups; and 21%-31% for cotransduction groups. Secondary recipients receiving 1 × 106 spleen cells from primary leukemic mice died of leukemia between 11 and 13 days as indicated by the gray bar (n = 5). (B) A typical enlarged leukemic spleen (right) compared with a control normal spleen (left). (C) H&E staining of a typical liver section from leukemic mice displaying liver infiltration by the leukemic cells. Original magnification ×100. (D) Wright-Giemsa staining of cytospin preparation from the BM of a leukemic mouse. Original magnification ×400. (E) Staining of GFP positive leukemic cells from the BM and spleen (SP) of a typical moribund mouse with indicated antibodies. Numbers represent the percentages of gated events. (F) Semiquantitative reverse transcription PCR analysis of total RNA extracted from normal mouse bone marrow (NBM), leukemic BM, and spleen (SP) from 3 leukemic mice (L1, 2, and 3) using indicated gene-specific primers. β-actin was included to control for RNA loading. (G) Southern blotting analysis of genomic DNA from leukemic spleens (L1 to L7) using a GFP-specific probe. Samples were digested by EcoRI, and each band represents a separate integration. (H) Survival curves of lethally irradiated C57BL/6-Ly5.2 primary recipient mice receiving purified mouse GMPs (1-1.5 × 105 cells/recipient) infected with MSCV-BCR/ABL-IRES-GFP virus, pMYs-Setbp1-IRES-GFP virus, or the combination as well as secondary (2nd), tertiary (3rd), and quaternary (4th) recipients of the cotransduction group receiving 1 × 106 spleen cells from preceding leukemic mice. Transduction efficiencies: 50%-60% for BCR/ABL groups; 40%-45% for Setbp1 groups; and 34%-50% for cotransduction groups.
Sixty percent of the mice receiving cells infected with BCR/ABL virus alone also developed leukemias in 4 months (Figure 6A). However, close examination of these GFP-positive leukemias by FACS and immunohistochemistry revealed that they are of either B- or T-cell leukemia, and not of myeloid disease (supplemental Figure 8). The absence of any myeloid disease in this group suggests that HSCs were efficiently depleted by induced differentiation in the presence of SCF and IL-3 before transduction and also confirms that BCR/ABL alone is unable to transform committed myeloid progenitors.5,10 Because BCR/ABL has been shown to induce lymphoid leukemias directly from lymphoid progenitors,30,31 the development of lymphoid leukemias in this group is probably because of the infection of B and T precursor cells expanded or generated in our culture conditions.
We also tested whether cotransduction with Setbp1 and BCR/ABL virus can transform GMPs freshly purified from C57BL/6 mice. Similar transduction efficiencies were achieved among all 3 groups, and transduced GMPs were similarly transplanted into lethally irradiated recipient mice (1.5-2 × 105 cells/recipient). Again, all mice receiving the doubly infected GMPs developed myeloid leukemias between 13 to 19 days after transplantation (Figure 6H). Except for higher percentages of myeloid blasts in the BM (44% ± 6.4% versus 34% ± 7.8%, mean ± SD), these GMP-derived leukemias are phenotypically indistinguishable from myeloid leukemias induced using in vitro expanded BM progenitors by FACS analysis (data not shown). In contrast, all mice receiving singly infected GMPs remained healthy when aged for 4 months (Figure 6H). To further establish the self-renewal capability of the transformed GMPs, we also carried out secondary, tertiary, and quaternary transplantations for 2 of the leukemias. All serially transplanted mice died of similar myeloid leukemia between 13 and 20 days after transplantation (Figure 6H), further supporting that LSCs generated by coexpression of Setbp1 and BCR/ABL in GMPs possess unlimited self-renewal capability. To test whether the cooperation between Setbp1 and BCR/ABL requires their expression within the same cell, we examined by PCR the presence of Setbp1 and BCR/ABL proviruses in genomic DNA from single methylcellulose colonies randomly picked from colony assays of 3 GMP-derived leukemias. Both proviruses can be efficiently detected in all 15 tested colonies but not in wild-type C57BL/6 spleen cells (supplemental Figure 10), strongly suggesting that Setbp1 and BCR/ABL cooperate in a cell-autonomous manner. Therefore, these results again support the idea that Setbp1 activation may provide myeloid progenitors with unlimited self-renewal capacity, which renders them competent for transformation by BCR/ABL.
Increased SETBP1 expression is detected in CML myeloid advanced phase or blast crisis patients
The cooperation between Setbp1 and BCR/ABL to transform committed mouse myeloid progenitors suggest that activation of SETBP1 may play a role in the development of human CML myeloid blast crisis. To test this notion, we examined SETBP1 mRNA levels in the BM aspirates of CML myeloid advanced phase/blast crisis (AP/BC) patients versus that of CML chronic phase (CP) patients by real-time RT-PCR (Figure 7). Relatively low levels of SETBP1 mRNA were detected in the BM of normal human volunteers and CP patients. In contrast, we detected significantly elevated levels of SETBP1 mRNA in samples from 2 of thirteen AP/BC patients associated with increased HOXA9 and HOXA10 mRNA expression (Figure 7), suggesting that SETBP1 could be directly involved in CML progression in these patients. Increased SETBP1 expression was also detected in CD34+ cells from several BC patients in a CML microarray analysis (supplemental Figure 11).32 Therefore, SETBP1 activation may contribute to disease progression in a subset of CML patients.
Figure 7.
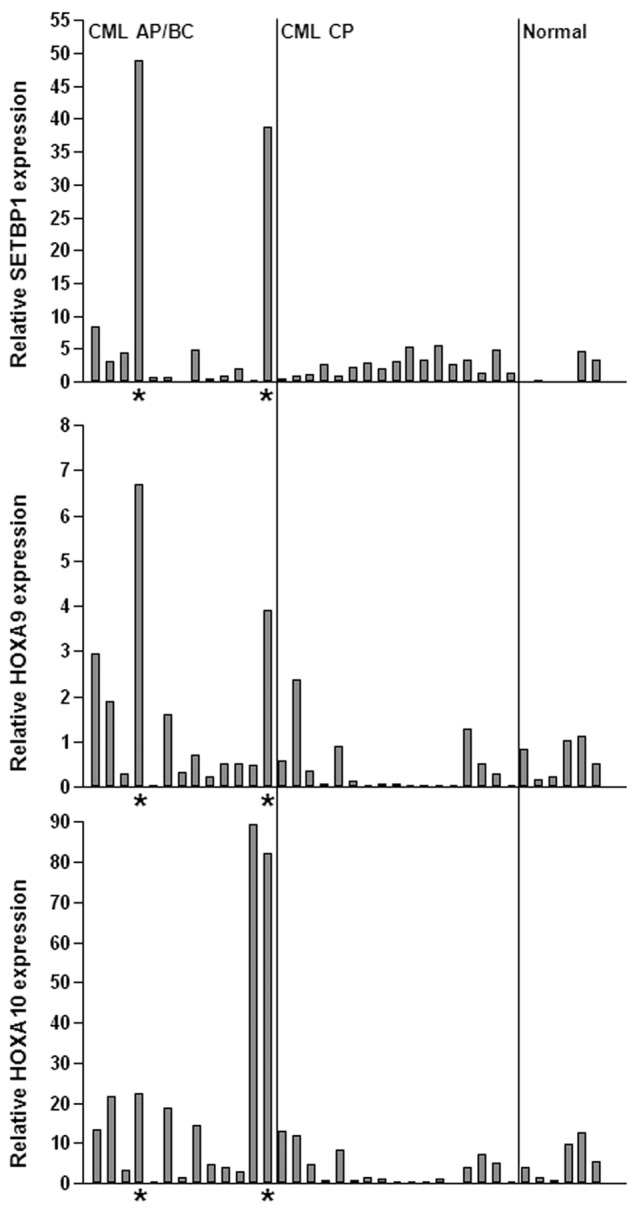
SETBP1 activation is involved in human CML progression. Real-time RT-PCR analysis of SETBP1, HOXA9, and HOXA10 mRNA levels in total RNA isolated from whole BM of healthy volunteers (normal) and CML chronic phase (CML CP) and advanced/blast crisis phase (CML AP/BC) patients (*samples expressing high SETBP1 mRNA levels). Relative expression levels were calculated by normalizing to BCR mRNA levels in the same sample.
Discussion
Here we identify Setbp1 as a novel regulator of LSC self-renewal in myeloid leukemias. We showed that Setbp1 overexpression can efficiently immortalize myeloid progenitors in culture and is also essential to sustain the self-renewal of these immortalized cells. We also provided the first evidence that Setbp1 could function as a transcription factor and identified the transcriptional targets crucial for its self-renewal promoting function in myeloid progenitors. In supporting a role of Setbp1 in conferring self-renewal capability to LSCs in vivo, we demonstrated that Setbp1 can cooperate with BCR/ABL to transform committed myeloid progenitors normally lacking self-renewal capability into LSCs and cause development of myeloid leukemias resembling human CML myeloid blast crisis in mice. Finally, SETBP1 expression increases in a subset of CML myeloid advanced or blast crisis patients, suggesting a conserved self-renewal promoting function of SETBP1 during human CML progression.
It has been proposed recently that SETBP1 expression may contribute to myeloid leukemia development by inhibition of PP2A activity.21 SETBP1 was shown to form a complex with SET and PP2A, enhancing the stability of SET and its inhibition of PP2A. Our results suggest a novel transcriptional mechanism by which SETBP1 contributes to leukemia transformation via activating HOXA9 and HOXA10. Knockdown of either genes caused dramatic reduction of the proliferative potential of Setbp1-immortalized cells. Both genes are also validated oncogenes in myeloid leukemia development. Overexpression of Hoxa9 alone or in cooperation with Meis1 can efficiently induce myeloid leukemia development in BM transduction and transplantation studies in mice.33 Interestingly, Hoxa9 and Hoxa10 overexpression have also been shown to induce immortalization of myeloid progenitors in culture, suggesting that they may promote the self-renewal of myeloid progenitors.4,34 Hoxa10 has also been shown to promote the self-renewal of HSCs.35 Their direct involvement in regulating LSC self-renewal is further supported by their crucial role in mediating transformation by MLL fusion proteins known to be LSC self-renewal regulators for their capability to transform committed myeloid progenitors.4 Increased HOXA9 expression is also associated with development of CML myeloid blast crisis, and it could directly contribute to the disease progression by repressing NUMB expression through MSI2 activation.36 In line with these studies, our results strongly suggest that activation of Hoxa9 and Hoxa10 plays an important role in Setbp1-induced leukemia transformation probably by promoting the self-renewal of LSCs. Although both Set and PP2A can function in the nucleus,37,38 treatment with PP2A activator 1,9-dideoxy-forskolin did not significantly affect Hoxa9 and Hoxa10 mRNA levels in Setbp1-immortalized cells (supplemental Figure 5), suggesting that their activation could be independent of PP2A inhibition induced by Setbp1. However, PP2A inhibition by Setbp1 is probably critical for the final transformation of myeloid progenitors by Setbp1 and BCR/ABL given that PP2A has been shown to strongly antagonize BCR/ABL downstream signaling and induces BCR/ABL degradation.19 It is also probable that additional Setbp1 transcriptional targets may exist and contribute to its transforming capability.
The DNA-binding capability of Setbp1 is probably mediated by its 3 AT-hook motifs. AT-hook motifs were first identified in the high mobility group protein HMGA1, and were shown to bind to the minor groove of stretches of AT-rich DNA in a nonsequence specific manner.27–29 The binding of this motif especially when present at multiple copies can cause DNA bending which may be critical for transcriptional regulation.39 A number of proteins containing this motif are essential components of large chromatin remodeling complexes found in yeast, Drosophila, and mammalian cells.40–42 Therefore, Setbp1 protein may be directly involved in regulating gene transcription as part of a chromatin remodeling complex, which is also consistent with its primary localization in the nucleus.14 Interestingly, Hoxa9 and Hoxa10 promoters are also known targets for the MLL and MLL-fusion proteins that too contain AT-hook motifs and function through formation of large chromatin remodeling complexes. Further studies will be required to clarify whether Setbp1 is a component of the MLL complexes or it may form its own complex independent of MLL proteins. In either scenario, it is probable that a DNA sequence-specific transcription factor is required to recruit Setbp1 to its target promoters as AT-hook motifs do not recognize specific DNA sequence.43
Our results provide additional experimental evidence supporting the notion that LSCs in CML myeloid blast crisis are derived directly from GMPs. BCR/ABL alone is neither sufficient to transform GMPs in vivo nor capable of immortalizing these cells in vitro, suggesting that additional mutations must be acquired by chronic phase GMPs, converting them into self-renewing LSCs in myeloid blast crisis. Aberrant activation of a number of genes/pathways, including Wnt/β-catenin,7 Gata2,44 AML1/MDS1/EVI1,45 and Msi236 have been implicated in the development of CML myeloid blast crisis. However, just 2 mutations, NUP98/HOXA9 and overexpression of Hes1, have been demonstrated so far to cooperate with BCR/ABL to transform GMPs in vivo.9,10 It has been proposed that these genes are responsible for conferring limitless self-renewal capability to GMPs during CML progression because of their capability to immortalize myeloid progenitors in vitro. Our studies have added Setbp1 to this short list of LSC self-renewal promoters involved in blast crisis development. Consistent with a previous study showing that Hoxa9 alone is not sufficient to induce GMP transformation,46 we also found that Setbp1 is not capable of transforming GMPs by itself. It is probable that gaining of self-renewal capability alone may be insufficient to transform committed myeloid progenitors because transplantation of GMPs singly infected with retrovirus expressing NUP98/HOXA9 or Hes1 also did not cause leukemia development in recipient mice. A major cellular effect of BCR/ABL expression in myeloid cells is increase of proliferation; therefore, the cooperation of these self-renewal genes with BCR/ABL suggests that another key mutational event for the transformation of myeloid progenitors may be to increase proliferation. Also in line with this notion, a number of growth-promoting mutations including activation of CSF-2 and Flt3 have been recently identified as cooperating partners for myeloid leukemia development induced by NPM1 mutations which also activate the expression of a number of Hox genes including Hoxa9 and Hoxa10.47
SETBP1 activation was only detected in a subset of CML AP/BC patients in our study; however, this is consistent with the idea that different mechanisms could cause disease progression based on the finding of heterogeneous chromosomal changes commonly associating with BC development.48 It remains unclear how SETBP1 was activated in the 2 SETBP1-positive patients in our sample set. Cytogenetic analysis of their leukemia cells did not reveal any chromosomal rearrangements involving 18q where SETBP1 has been mapped (data not shown), suggesting involvement of other mechanisms, possibly epigenetic regulation or activation of upstream regulator of SETBP1. Given that SETBP1 overexpression was recently found in more than 27% of AML patients,21 our study suggests that SETBP1 activation may also represent a major mechanism regulating the self-renewal of LSCs in human AMLs. Our analysis of a published AML microarray dataset further suggests that activation of HOXA9 and HOXA10 by SETBP1 is probably conserved in AMLs (supplemental Figure 12).49 Therefore, blocking the activities of HOXA9 and HOXA10 may lead to novel therapies to inhibit LSC self-renewal in SETBP1-positive CML blast crisis as well as AML patients.
Supplementary Material
Acknowledgments
The authors thank Dr Cynthia Dunbar for providing the MSCV-BCR/ABL-IRES-GFP construct.
This work was supported by National Cancer Institute grant CA143193 (to Y.D.) and the Department of Pediatrics at USUHS.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.O., Y.H., and B.A.V. performed most of the experiments and contributed to data analysis; S.C. and R.B. performed QRT-PCR analyses of patient samples and analyzed results; K.O.G., J.K., N.A.J., and N.G.C. contributed vital reagents; X.C. conducted microarray analysis; V.V. performed immunohistochemistry experiments; and Y.D. designed and performed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yang Du, Dept of Pediatrics, Bldg A, Rm 3042B, USUHS, 4301 Jones Bridge Rd, Bethesda, MD 20814; e-mail: yang.du@usuhs.edu.
References
- 1.Dick JE. Acute myeloid leukemia stem cells. Ann N Y Acad Sci. 2005;1044:1–5. doi: 10.1196/annals.1349.001. [DOI] [PubMed] [Google Scholar]
- 2.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 3.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17(24):3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 5.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Haferlach T, Winkemann M, Nickenig C, et al. Which compartments are involved in Philadelphia-chromosome positive chronic myeloid leukaemia? An answer at the single cell level by combining May-Grunwald-Giemsa staining and fluorescence in situ hybridization techniques. Br J Haematol. 1997;97(1):99–106. doi: 10.1046/j.1365-2141.1997.9662656.x. [DOI] [PubMed] [Google Scholar]
- 7.Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 8.Abrahamsson AE, Geron I, Gotlib J, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A. 2009;106(10):3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neering SJ, Bushnell T, Sozer S, et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110(7):2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakahara F, Sakata-Yanagimoto M, Komeno Y, et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood. 2010;115(14):2872–2881. doi: 10.1182/blood-2009-05-222836. [DOI] [PubMed] [Google Scholar]
- 11.Calvo KR, Sykes DB, Pasillas MP, Kamps MP. Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1. Oncogene. 2002;21(27):4247–4256. doi: 10.1038/sj.onc.1205516. [DOI] [PubMed] [Google Scholar]
- 12.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary BM progenitor cells. Blood. 2005;106(12):3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 14.Minakuchi M, Kakazu N, Gorrin-Rivas MJ, et al. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268(5):1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x. [DOI] [PubMed] [Google Scholar]
- 15.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112(5):659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271(19):11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 17.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12(8):3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozbek U, Kandilci A, van Baal S, et al. SET-CAN, the product of the t(9;9) in acute undifferentiated leukemia, causes expansion of early hematopoietic progenitors and hyperproliferation of stomach mucosa in transgenic mice. Am J Pathol. 2007;171(2):654–666. doi: 10.2353/ajpath.2007.060934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neviani P, Santhanam R, Trotta R, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8(5):355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Panagopoulos I, Kerndrup G, Carlsen N, Strombeck B, Isaksson M, Johansson B. Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12). Br J Haematol. 2007;136(2):294–296. doi: 10.1111/j.1365-2141.2006.06410.x. [DOI] [PubMed] [Google Scholar]
- 21.Cristobal I, Blanco FJ, Garcia-Orti L, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115(3):615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 22.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced BM. Blood. 1998;92(10):3780–3792. [PubMed] [Google Scholar]
- 23.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 24.Ji M, Li H, Suh HC, Klarmann KD, Yokota Y, Keller JR. Id2 intrinsically regulates lymphoid and erythroid development via interaction with different target proteins. Blood. 2008;112(4):1068–1077. doi: 10.1182/blood-2008-01-133504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YX, Yan J, Keeshan K, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci U S A. 2006;103(4):1018–1023. doi: 10.1073/pnas.0510347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang CC, Lodish HF. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood. 2005;105(11):4314–4320. doi: 10.1182/blood-2004-11-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26(19):4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review). Int J Oncol. 2008;32(2):289–305. [PubMed] [Google Scholar]
- 29.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265(15):8573–8582. [PubMed] [Google Scholar]
- 30.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C, Blum J, Chen A, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12(6):528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng C, Li L, Haak M, et al. Gene expression profiling of CD34+ cells identifies a molecular signature of chronic myeloid leukemia blast crisis. Leukemia. 2006;20(6):1028–1034. doi: 10.1038/sj.leu.2404227. [DOI] [PubMed] [Google Scholar]
- 33.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21(1):224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol Cell Biol. 2000;20(9):3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnusson M, Brun AC, Miyake N, et al. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood. 2007;109(9):3687–3696. doi: 10.1182/blood-2006-10-054676. [DOI] [PubMed] [Google Scholar]
- 36.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 466(7307):765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuwaki M, Nagata K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J Biol Chem. 1998;273(51):34511–34518. doi: 10.1074/jbc.273.51.34511. [DOI] [PubMed] [Google Scholar]
- 38.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104(1):119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 39.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277(1-2):63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 40.Bourachot B, Yaniv M, Muchardt C. The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol Cell Biol. 1999;19(6):3931–3939. doi: 10.1128/mcb.19.6.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4(5):715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 42.Xiao H, Sandaltzopoulos R, Wang HM, et al. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8(3):531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 43.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120(2):593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang SJ, Ma LY, Huang QH, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2008;105(6):2076–2081. doi: 10.1073/pnas.0711824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuenco GM, Ren R. Cooperation of BCR-ABL and AML1/MDS1/EVI1 in blocking myeloid differentiation and rapid induction of an acute myelogenous leukemia. Oncogene. 2001;20(57):8236–8248. doi: 10.1038/sj.onc.1205095. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassiliou GS, Cooper JL, Rad R, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 43(5):470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 49.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.