Abstract
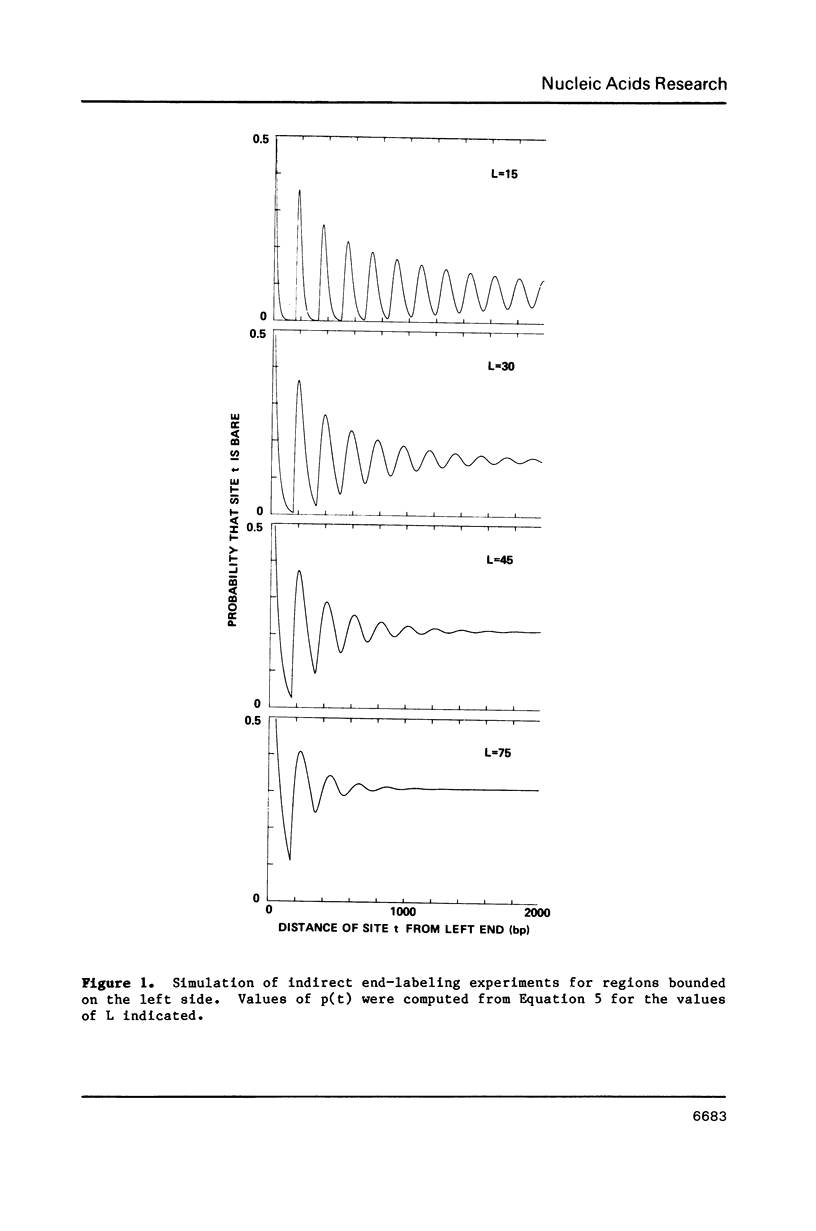
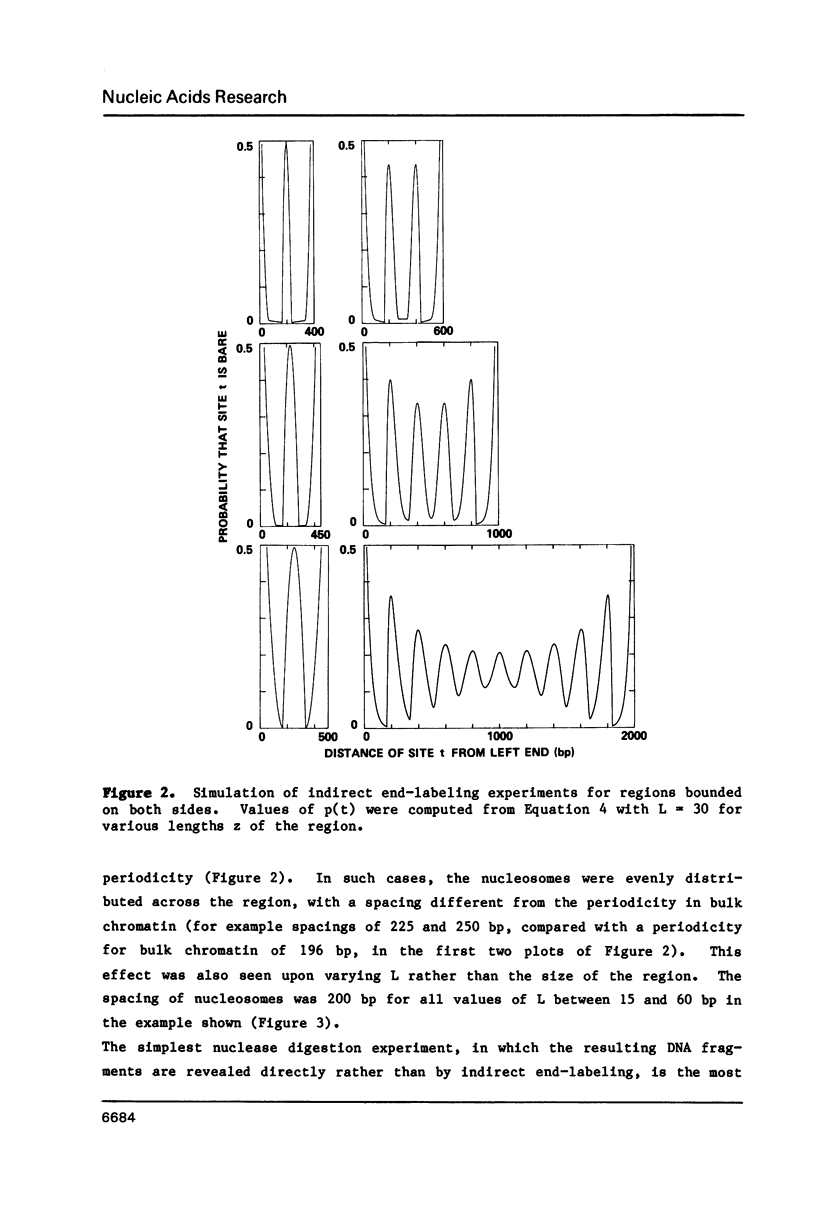
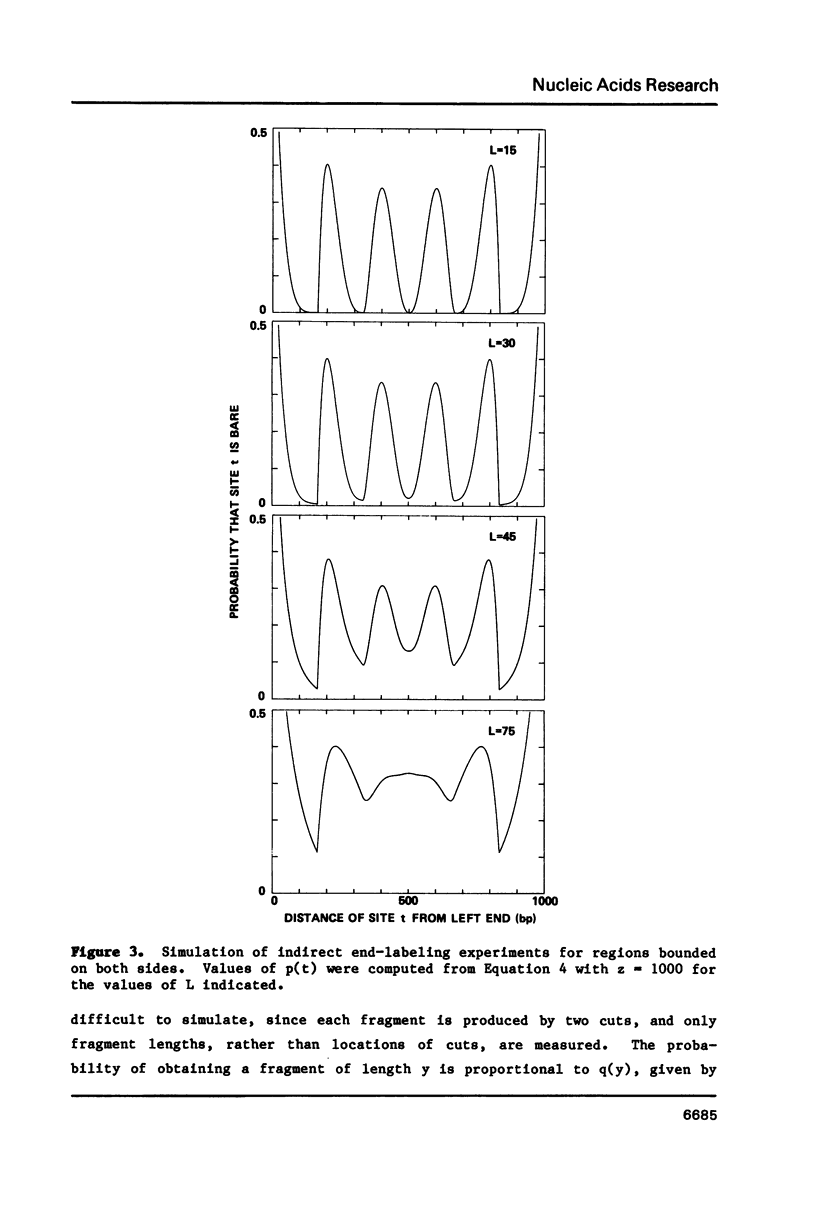
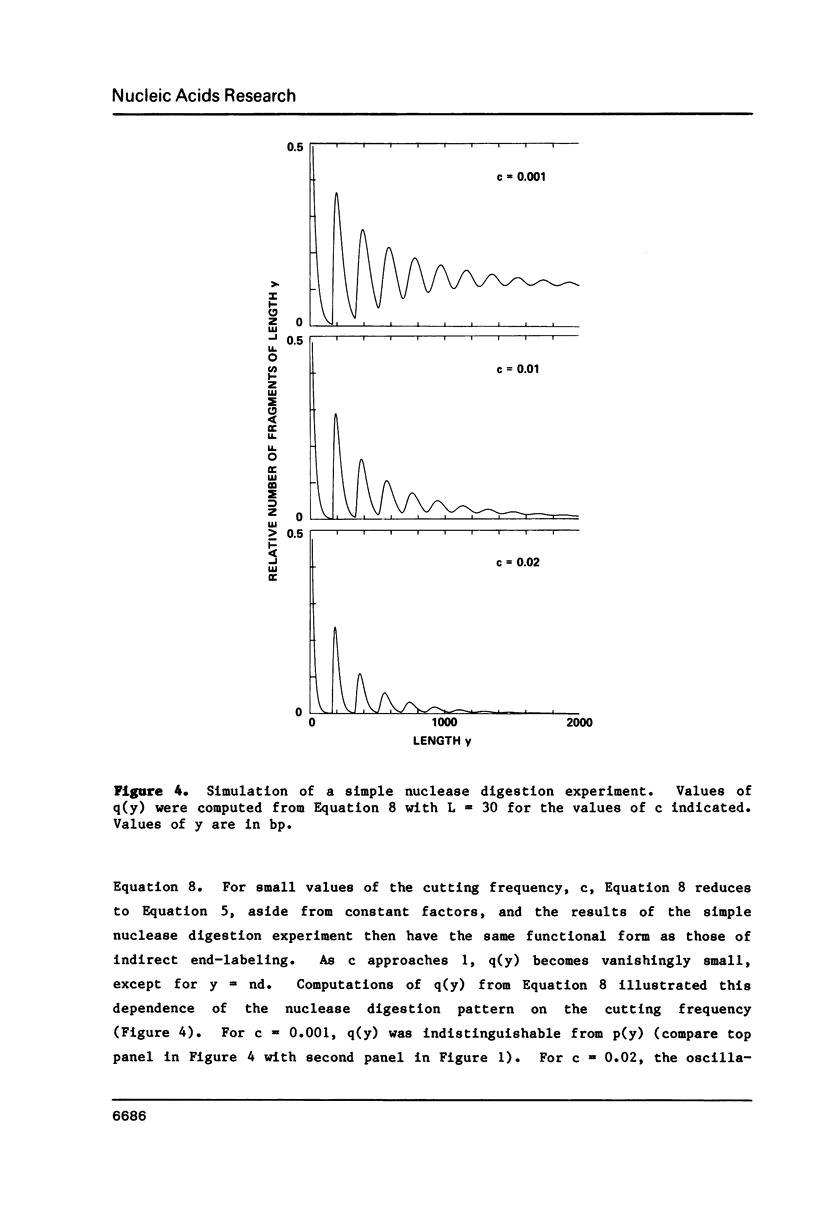
Expressions are derived for distributions of nucleosomes in chromatin. Nucleosomes are placed on DNA at the densities found in bulk chromatin, and their locations are allowed to vary at random. No further assumptions are required to simulate the periodic patterns of digestion obtained with various nucleases. The introduction of a boundary constraint, due for example to sequence-specific protein binding, results in an array of regularly spaced nucleosomes at nonrandom locations, similar to the arrays reported for some genes and other chromosomal regions.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Cantor C. R., Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986 Mar 14;44(5):697–704. doi: 10.1016/0092-8674(86)90835-4. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Hofstetter H., Birnstiel M. L. Nucleosome arrangement on tRNA genes of Xenopus laevis. Cell. 1981 Dec;27(3 Pt 2):459–466. doi: 10.1016/0092-8674(81)90387-1. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budarf M. L., Blackburn E. H. Chromatin structure of the telomeric region and 3'-nontranscribed spacer of Tetrahymena ribosomal RNA genes. J Biol Chem. 1986 Jan 5;261(1):363–369. [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Costlow N. A., Simon J. A., Lis J. T. A hypersensitive site in hsp70 chromatin requires adjacent not internal DNA sequence. Nature. 1985 Jan 10;313(5998):147–149. doi: 10.1038/313147a0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Edwards C. A., Firtel R. A. Site-specific phasing in the chromatin of the rDNA in Dictyostelium discoideum. J Mol Biol. 1984 Nov 25;180(1):73–90. doi: 10.1016/0022-2836(84)90431-5. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. Anatomy of hypersensitive sites. Nature. 1984 May 17;309(5965):213–214. doi: 10.1038/309213a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Cech T. R. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell. 1984 Sep;38(2):501–510. doi: 10.1016/0092-8674(84)90505-1. [DOI] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981 Aug 13;292(5824):579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Levy A., Noll M. Multiple phases of nucleosomes in the hsp 70 genes of Drosophila melanogaster. Nucleic Acids Res. 1980 Dec 20;8(24):6059–6068. doi: 10.1093/nar/8.24.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. The chromatin structure of an actively expressed, single copy yeast gene. Nucleic Acids Res. 1983 Oct 11;11(19):6755–6773. doi: 10.1093/nar/11.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Payvar F., DeFranco D., Firestone G. L., Edgar B., Wrange O., Okret S., Gustafsson J. A., Yamamoto K. R. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983 Dec;35(2 Pt 1):381–392. doi: 10.1016/0092-8674(83)90171-x. [DOI] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Pospelov V. A., Klobeck H. G., Zachau H. G. Correlation between DNase I hypersensitive sites and putative regulatory sequences in human immunoglobulin genes of the kappa light chain type. Nucleic Acids Res. 1984 Sep 25;12(18):7007–7021. doi: 10.1093/nar/12.18.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt J. H. DNase I-hypersensitive sites in the galactose gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Jun;5(6):1522–1524. doi: 10.1128/mcb.5.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N., Felsenfeld G., Rushton B. M., McGhee J. D. A 145-base pair DNA sequence that positions itself precisely and asymmetrically on the nucleosome core. EMBO J. 1984 Nov;3(11):2605–2611. doi: 10.1002/j.1460-2075.1984.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Györgyi C., Finkelstein D. B., Garrard W. T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987 Jan 5;193(1):71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thompson T., Fan H. Mapping of DNase I-hypersensitive sites in the 5' and 3' long terminal repeats of integrated moloney murine leukemia virus proviral DNA. Mol Cell Biol. 1985 Apr;5(4):601–609. doi: 10.1128/mcb.5.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A., Schedl P. Chromatin organization of the 87A7 heat shock locus of Drosophila melanogaster. J Mol Biol. 1984 Feb 5;172(4):385–403. doi: 10.1016/s0022-2836(84)80013-3. [DOI] [PubMed] [Google Scholar]
- Wittig B., Wittig S. A phase relationship associates tRNA structural gene sequences with nucleosome cores. Cell. 1979 Dec;18(4):1173–1183. doi: 10.1016/0092-8674(79)90230-7. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Fittler F., Hörz W. Eight different highly specific nucleosome phases on alpha-satellite DNA in the African green monkey. Nucleic Acids Res. 1983 Jul 11;11(13):4287–4306. doi: 10.1093/nar/11.13.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]



