Abstract
The transcription factors Foxa1 and Foxa2 promote the specification of midbrain dopaminergic (mDA) neurons and the floor plate. Whether their role is direct has remained unclear as they also regulate the expression of Shh, which has similar roles. We characterized the Foxa2 cis-regulatory network by chromatin immunoprecipitation followed by high-throughput sequencing of mDA progenitors. This identified 9160 high-quality Foxa2 binding sites associated with 5409 genes, providing mechanistic insights into Foxa2-mediated positive and negative regulatory events. Foxa2 regulates directly and positively key determinants of mDA neurons, including Lmx1a, Lmx1b, Msx1 and Ferd3l, while negatively inhibiting transcription factors expressed in ventrolateral midbrain such as Helt, Tle4, Otx1, Sox1 and Tal2. Furthermore, Foxa2 negatively regulates extrinsic and intrinsic components of the Shh signaling pathway, possibly by binding to the same enhancer regions of co-regulated genes as Gli1. Foxa2 also regulates the expression of floor plate factors that control axon trajectories around the midline of the embryo, thereby contributing to the axon guidance function of the floor plate. Finally, this study identified multiple Foxa2-regulated enhancers that are active in the floor plate of the midbrain or along the length of the embryo in mouse and chick. This work represents the first comprehensive characterization of Foxa2 targets in mDA progenitors and provides a framework for elaborating gene regulatory networks in a functionally important progenitor population.
Keywords: Dopaminergic neuron specification, Foxa2, Forkhead, Chromatin immunoprecipitation, Mouse, Floor plate
INTRODUCTION
Foxa1 and Foxa2 (Foxa1/2) are members of the Foxa family of forkhead/winged helix transcription factors, which have diverse roles in regulating development, tissue homeostasis and tumorigenesis (reviewed by Hannenhalli and Kaestner, 2009). Foxa1/2 regulate the development of endoderm tissues, including liver (Lee et al., 2005), lung (Wan et al., 2005) and pancreas (Gao et al., 2008). Within the central nervous system (CNS), Foxa1/2 regulate the specification and differentiation of midbrain dopaminergic (mDA) neurons (Lin et al., 2009) and hindbrain serotonergic neurons (Jacob et al., 2007). Foxa2 also regulates adaptive behavior during fasting in the hypothalamus (Silva et al., 2009).
mDA progenitors arise from the floor plate (FP) region of the midbrain and caudal diencephalon (Andersson et al., 2006b; Ono et al., 2007). We have previously established by analyzing mouse mutants that Foxa1 and Foxa2 function cooperatively to regulate mDA progenitor specification and differentiation in a dose-dependent manner by regulating the expression of key regulatory genes such as Lmx1a, Lmx1b and Neurog2 (Ferri et al., 2007; Lin et al., 2009). However, it was not possible to establish intrinsic roles for Foxa1/2 in regulating the expression of these genes as Foxa1/2 directly regulate sonic hedgehog (Shh) expression, and Shh also functions as an upstream regulator of the expression of these genes (Andersson et al., 2006b). Moreover, Foxa1/2 cooperatively regulate distinct sets of target genes in mDA progenitors, immature neurons and mature neurons (Ang, 2009). How Foxa1/2 regulate distinct target genes at different phases of mDA neuron lineage development is not known.
We set out to identify Foxa2 target genes on a genome-wide scale using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). Our studies led to the identification of 9160 Foxa2-bound regions (FBRs); 178 and 108 genes annotated to Foxa2-bound regions were specifically expressed in the FP and ventrolateral regions of the midbrain, respectively. Interestingly, Foxa2 positively regulates the expression of FP-specific genes while negatively regulating the expression of ventrolateral-specific genes in mDA progenitors to specify mDA and FP identity. These results are consistent with studies in mouse mutants, which have demonstrated a function of Foxa genes in promoting ventral FP and mDA identity at the expense of lateral midbrain GABAergic neuron identity (Lin et al., 2009). Foxa2 also regulates the expression of factors involved in guiding axons around the FP in both rostral and caudal regions of the CNS. We also show that Foxa2 negatively regulates multiple components of the Shh signaling pathway by inhibiting Gli1 and Gli2 expression as well as the expression of some common Gli1-upregulated target genes, possibly by binding to the same enhancer regions. In addition, our data will also serve as an important resource for identifying novel genes that regulate mDA progenitor function.
MATERIALS AND METHODS
Differentiation of embryonic stem (ES) cells
E14.1 (NesE-Lmx1a) ES cells were cultured and differentiated as described (Andersson et al., 2006b).
ChIP-qPCR reactions
Foxa2 ChIP experiments were performed on chromatin prepared from dissected E12.5 ventral midbrain tissue using Foxa2-specific antiserum as described (Lin et al., 2009). Primers of genomic regions are provided in supplementary material Table S4. The Foxa2 open reading frame was used as a negative control for non-specific enrichment. The fold enrichment of each target site was calculated as 2 to the power of the cycle. The enrichment of target sequences in ChIP material was calculated relative to the input.
ChIP-seq and data analysis
The ChIP sample was generated from NestinLmx1a stably transfected ES cells that had been differentiated into mDA progenitors (day 5 in vitro) as described (Andersson et al., 2006b). Cells were cross-linked as described (Lin et al., 2009). After washing in PBS to remove excess formaldehyde and glycine, 2×107 fixed cells were homogenized in 300 μl cold whole-cell lysis buffer (10 mM Tris-HCl pH 8.0, 10 mM NaCl, 3 mM MgCl2, 1% NP40, 1% SDS) and protease inhibitors. After incubating on ice for 10 minutes, lysates were sonicated using a Diagenode Bioruptor (30-second on/off pulses for 10 minutes, on the high setting). Debris was removed by centrifugation at 13,000 g for 10 minutes, and the supernatant was collected and snap frozen in liquid nitrogen. As input, 10 μl of sonicated chromatin was incubated in PBS with 200 mM NaCl overnight at 65°C, treated with proteinase K and purified using the QIAquick PCR Purification Kit (Qiagen). Immunoprecipitations were performed as described (Lin et al., 2009).
For ChIP-seq experiments, the immunoprecipitated DNA was modified for sequencing following the manufacturer's protocol (Illumina). ChIP-seq libraries were sequenced on the Illumina GAIIx using two lanes per sample for increased coverage at the EMBL Genomics Core Facility, Heidelberg, Germany. Sequencing reads were aligned to the mouse genome (mm9) using MAQ v0.6.8 (Li et al., 2008), where reads with low mapping quality (<13), greater than three mismatches or that mapped to more than a single genomic location were excluded from downstream analysis. Foxa2 binding sites were then identified with MACS v1.3.4 (Zhang et al., 2008). Peak annotation was performed using PeakAnalyzer (Salmon-Divon et al., 2010) and CEAS, where peak summits (as defined by MACS) and genes from the UCSC Refseq track were used as input (Shin et al., 2009) and MEME was used for de novo motif analysis (Bailey and Elkan, 1994). RegionMiner release 4.0 (Genomatix, Munich, Germany) was used to determine over-representation relative to the genome of the Foxa2 motif identified by MEME. Genomic coordinates defined by MACS with less than 5% false discovery rate (FDR) are provided in supplementary material Table S1. ChIP-seq data are available at ArrayExpress under accession E-MTAB-1137.
Functional analysis by Gene Ontology (GO)
Significance of enrichment between any two sets of genes was determined by applying the hypergeometric distribution as described by Gennet et al. (Gennet et al., 2011), where x is the number of genes shared by two gene lists (FP=178, VL=108), n is the number of genes identified by ChIP-seq (5409), M is the number of genes of the second list (FP=444, VL=312), and N is the number of all Ensembl genes (37,361). Note that the number of genes was used rather than the probe number (entities) as provided by Gennet et al. because some genes in the microarray experiment are annotated by multiple probes. An equivalently sized list of random genes was used as a negative control. GO analysis was performed using GOToolBox (http://genome.crg.es/GOToolBox), using the Mouse Genome Informatics identities of the list of genes. Gene lists and GO term results are provided in supplementary material Table S2.
Generation and analysis of transgenic mice
Transgenes for injection were separated from vector sequences by electrophoresis in 1% agarose gels, purified on Qiagen PCR extraction columns, precipitated and resuspended in injection buffer (10 mM Tris-HCl pH 7.5, 0.1 mM EDTA). Transgenic mice were generated by standard procedures (Scardigli et al., 2001) using fertilized eggs from (CBA/CA × C57Bl10) crosses. Transgenic embryos were identified by PCR with lacZ primers: F, 5′-CGAGTGTGATCATCTGGTCG-3′; and R, 5′-TTACCTTGTGGAGCGACATC-3′. E10-10.5 embryos were dissected from the uterus in cold PBS and fixed for 30 minutes at room temperature in 4% formaldehyde in PBS (pH 7.2). Whole-mount β-gal staining and analysis of the embryos were performed as described (Scardigli et al., 2001). Reporter gene expression within the developing neural tube was examined using whole-mount or 7 μm paraffin sections.
Generation of reporter constructs
Regulatory sequences beneath Foxa2 peak regions called by MACS were selected according to the limits defined by the highest sequence conservation among multiple vertebrate species from the UCSC genome browser (http://genome.ucsc.edu/). All regulatory sequences assayed were cloned into the XbaI site of a reporter vector comprising the β-globin promoter, lacZ cDNA, SV40 large T antigen poly(A) site, and sequences mediating Lmx1a E1, E2, E3, Lmx1b E1, and Corin E1 activity in an arrangement that retains the native orientation of these genes. All DNA sequences tested were generated by PCR amplification, the primer sequences for which are listed in supplementary material Table S4. Point mutations designed to disrupt DNA binding at the recognition sequences for Foxa2 were performed using the QuikChange Mutagenesis Kit (Agilent Technologies) with 5′-GGGCAGTGCTACCCACTCAGCTCCTGC-3′ (mutated residues underlined). The integrity and orientation of all constructs containing PCR-generated fragments were confirmed by DNA sequencing.
Generation and genotyping of mutant embryos and animals
All mouse strains were maintained in a mixed MF1-129/SV background. En1KICre/+, Foxa2flox/flox and Foxa1loxp/loxp mouse strains were generated as described (Sapir et al., 2004; Hallonet et al., 2002; Gao et al., 2008). Here, we refer to the Foxa1loxp allele as Foxa1flox. Foxa1flox/flox;Foxa2flox/flox mice were generated by crossing Foxa1flox/flox with Foxa2flox/flox animals. To obtain conditional Foxa1 and Foxa2 double mutants, we first crossed En1KICre/+ mice with Foxa1flox/flox;Foxa2flox/flox animals. Subsequently, En1KICre/+;Foxa1flox/+;Foxa2flox/+ F1 male animals were mated to Foxa1flox/flox;Foxa2flox/flox females to generate En1KIcre/+;Foxa1flox/flox;Foxa1flox/flox double mutants. The Foxa2flox and Foxa1flox alleles were detected by PCR (Hallonet et al., 2002; Gao et al., 2008); the Cre transgene was detected using the pair of primers and PCR conditions described previously (Indra et al., 1999). NestinCre/+;Foxa1−/−;Foxa2flox/flox animals were generated as described (Ferri et al., 2007). Animals were handled according to the Society of Neuroscience Policy on the Use of Animals in Neuroscience Research, as well as the European Communities Council Directive.
RNA extraction and cDNA synthesis
Ventral midbrain tissue was dissected from three Foxa1flox/flox;Foxa2flox/flox control embryos and three En1KICre/+;Foxa1flox/flox;Foxa2flox/flox embryos. Total RNA isolation was carried out using the Pico Pure RNA Extraction Kit (Arcturus) according to the manufacturer's instructions. RNA was reverse transcribed using Superscript III (Invitrogen) to obtain cDNA for use in quantitative (q) PCR reactions. For quantitative analysis of the expression level of mRNAs, real-time PCR analyses using Platinum SYBR Green Super Mix (Invitrogen) were performed in triplicate using the 7900 PCR System (ABI). Oligonucleotides amplifying small amplicons were designed using Primer3 (BioTools, http://biotools.umassmed.edu). Amplifications were performed in 20 μl containing 0.5 μM each primer. 0.5× SYBR Green (Invitrogen) and 2 μl 50-fold diluted cDNA, with 45 cycles at 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and 79°C for 5 seconds. The dissociation curve of each PCR product was determined to ensure that the observed fluorescent signals were only from specific PCR products. After each PCR cycle, the fluorescent signals were detected at 79°C. The fluorescent signals from specific PCR products of cDNA prepared from mutant or control (wild-type littermate) ventral midbrain tissue were normalized to that of Gapdh (2Ct Gapdh–Ct gene tested), and then relative values were calculated by setting the normalized value of the control as 1. All reactions were repeated using at least three independent samples (biological replicates), and one-way ANOVA and Dunnett's test were used to calculate the significance of the fold change in the expression of genes in the mutant compared with the wild-type control. Primers are provided in supplementary material Table S5.
Chick embryo electroporation
The lacZ reporter constructs containing the selected genomic regions bound by Foxa2 were electroporated into naïve chick midbrain neuroepithelium at Hamilton and Hamburger stage 10. The embryos were allowed to develop for 48 hours and then analyzed by whole-mount β-gal staining as described above. The empty vector was used as a negative control.
Immunohistochemistry
Cells were fixed in 4% paraformaldehyde (PFA) for 10 minutes at room temperature. Chick embryos were fixed in 4% PFA for 1 hour on ice. Cells or cryosectioned chick tissue were then washed with 1× PBS and incubated for 10 minutes in blocking solution (5% fetal bovine serum and 0.1% Triton X-100 in PBS). Primary antibody was added and cells were incubated at 4°C overnight. Cells were then washed three times with PBS containing 0.1% Triton X-100. Secondary antibody was added and cells were incubated at room temperature for 1 hour. Antibodies used were rabbit anti-Nurr1 (Nr4a2 – Mouse Genome Informatics) (Santa Cruz), mouse anti-tyrosine hydroxylase (Th; Chemicon), rabbit anti-Lmx1a (Andersson et al., 2006b), mouse anti-Tuj1 (Tubb3 – Mouse Genome Informatics; Chemicon), mouse anti-nestin (Chemicon), rabbit anti-Foxa2 (Wan et al., 2005), rabbit anti-β-gal (Biogenesis) and sheep anti-GFP (Biogenesis).
In situ hybridization of brain sections
Section in situ hybridization of E10.5 mouse brains was performed as described (Lin et al., 2009). The following probes were used: Tal2 [a PCR fragment corresponding to the first 600 bp of the Tal2 cDNA clone (NCBI gene ID: 21350)], Sox1 (Collignon et al., 1996), Tle4, Otx1 and Pax3 (Puelles et al., 2003).
RESULTS
Genome-wide characterization of Foxa2 binding sites in mDA progenitors
To identify direct target genes of Foxa2, we chose a genome-wide approach using ChIP-seq (Barski et al., 2007; Johnson et al., 2007; Robertson et al., 2007). As sufficient numbers of mDA progenitors could not be obtained from mouse embryos at E10.5, we obtained mDA progenitors by in vitro differentiation of NestinLmx1a-transfected ES cells (Andersson et al., 2006b). We efficiently generated 60-80% pure mDA progenitors after 5 days of differentiation of NestinLmx1a-transfected ES cells in the presence of Fgf8 and Shh (supplementary material Fig. S1A,B) (Andersson et al., 2006b). These in vitro generated mDA progenitors robustly expressed Foxa2 at day 5 and differentiated further into Nurr1+ Th+ mature mDA neurons by day 8, as previously described (supplementary material Fig. S1A,B) (Andersson et al., 2006b). In addition, in vitro differentiated mDA progenitors have been shown to develop into bona fide mDA neurons that integrate and innervate the striatum of 6-hydroxydopamine lesioned neonatal rats when transplanted into the striatum (Friling et al., 2009), indicating that they have a similar functional potential as embryonic mDA progenitors.
Ten million NestinLmx1a-transfected ES cells were used to prepare chromatin and a Foxa2-specific antibody was used for ChIP (Mavromatakis et al., 2011). Following sequencing, peak calling was performed using MACS with a threshold of FDR<5%, resulting in a dataset of 9160 high confidence peaks. All FBRs were annotated to genes that have their transcription start site (TSS) closest to the peak. Several known and validated FBRs in the CNS were included in this study, such as the Shh brain enhancer (Epstein et al., 1999), Foxa2 enhancer (Sasaki and Hogan, 1996), Foxa1 promoter (Peterson et al., 1997) and Ferd3l (Nato3) promoter (Mansour et al., 2011), but not the Nkx2.2 and Th promoters (Lin et al., 2009). As the latter two genes are not expressed in our in vitro ES cell differentiation system, these gene loci might be in a closed chromatin configuration and inaccessible for Foxa2 binding.
Validation of this dataset was performed by ChIP-qPCR on a selected subset of FBRs located in genes known to be expressed in the midbrain FP, including Lmx1a, Lmx1b (Andersson et al., 2006b), Msx1 (Andersson et al., 2006b), Ferd3l (Ono et al., 2010; Segev et al., 2001), Corin (Ono et al., 2007), Slit2 (Wang et al., 1999), Cobl (Gasca et al., 1995), Rfx3 (Baas et al., 2006), Bmp6 and Bmp7 (Kim et al., 2001), using chromatin from mouse E12.5 ventral midbrain that contains mostly FP tissue. Foxa2-dependent enrichment was confirmed in 17 out of 19 of these FBRs (Fig. 1A,B). We also confirmed Foxa2 binding in 16 out of 20 additional FBRs located in selected transcriptionally active genes (supplementary material Fig. S2). Together, these results indicate a high-quality dataset.
Fig. 1.
Chromatin immunoprecipitation of in vitro-differentiated murine mDA progenitors. (A) Peaks of Foxa2 enrichment identified by ChIP-seq are shown for selected mDA progenitor-expressed genes. The plots show the peak heights as numbers of sequenced reads. Arrows indicate peak locations called by MACS. (B) Independent ChIP-qPCR experiments were performed with chromatin prepared from E12.5 ventral midbrain tissue for validation of selected candidate targets, where enrichment is shown as percentage of input (*P<0.05). Multiple peaks within a gene region, corresponding to the list in supplementary material Table S1, are numbered E1 (for Element 1), and so on, from left to right. -ve1, Foxa2 open reading frame (Mavromatakis et al., 2011). Error bars indicate s.e.m.
Binding site analysis
The peaks in FBRs had an average width of 290 bp and an average height of 16 tags (Fig. 1A). De novo motif analysis of their sequences using MEME (Zhang et al., 2008) revealed, as expected, a substantial enrichment in Foxa2 motif sites (Fig. 2A). Of the peaks, 7298 (80%) contained at least one Foxa2 motif within 60 bp of the peak summit (Fig. 2A). To further characterize the distribution of the peaks, we compared their locations with UCSC RefSeq genes using CEAS (Shin et al., 2009) and found that 46.7% of the high confidence peaks were located within a gene region extending from the TSS to the 3′UTR downstream (Fig. 2B). In total, 44.2% of all peaks were located within an intron, 0.7% within an exon, 0.8% within the 5′UTR and 1% within the 3′UTR. The remaining Foxa2 peaks were in intergenic regions located further than 10 kb upstream or downstream of the annotated genes. Interestingly, Foxa2 binding was enriched in promoter regions, with 3.7% of peaks located within 1 kb and 11% within 10 kb upstream of the TSS when compared with the genome background (Fig. 2B). Foxa2 binding was not enriched in regions directly downstream of genes (Fig. 2B).
Fig. 2.
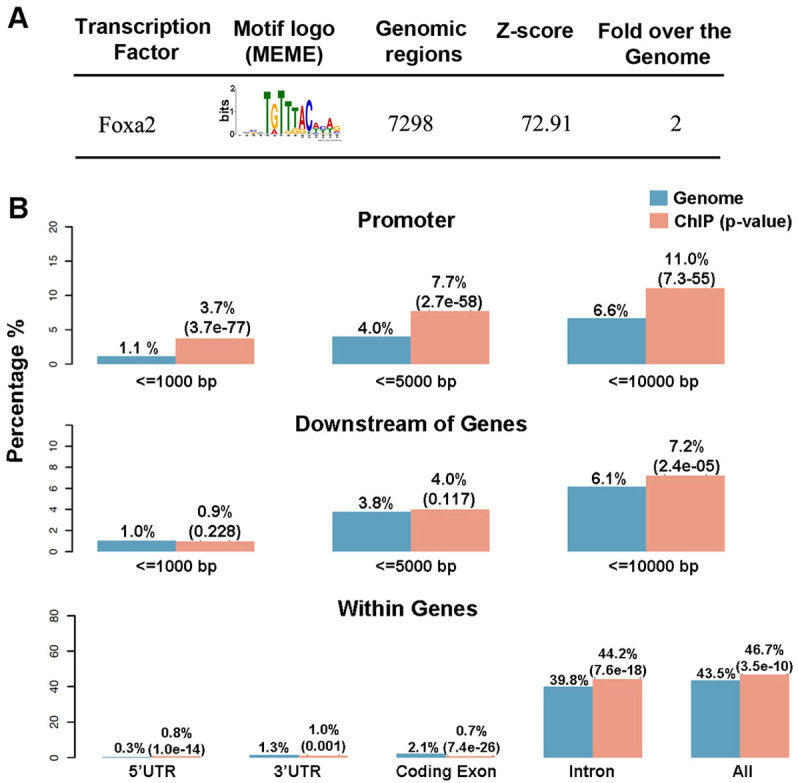
Analyses of motifs and peak distribution in Foxa2-bound regions. (A) MEME identified de novo transcription factor binding motifs within 60 bp of the center of Foxa2 peaks. Genomatix results include: (1) the number of peaks that contained the transcription factor binding motif; (2) the Z-score of over-representation against the selected background (randomized sequences); and (3) the over-representation against the mouse genome, which is the fold factor of the number of Foxa2 motifs within the ChIP-seq dataset compared with an equally sized sample of the genome. (B) The distribution and enrichment relative to the genome background of Foxa2 peaks with respect to different gene features. The P-value is given in parenthesis.
Foxa2 and Shh signaling have similar roles in ventral midbrain patterning (Blaess et al., 2006; Lin et al., 2009); however, we recently found that Foxa2 also attenuates Shh signaling by repressing the expression of its intracellular transducers Gli1, Gli2 and Gli3 (Mavromatakis et al., 2011). We therefore examined whether targets of Gli1 that were previously identified by ChIP-chip in neural progenitors (Vokes et al., 2007) are also bound by Foxa2. Foxa2 binding overlapped with ten out of 25 Gli1 binding regions, including those located in Ptch1, Nkx2.9, Gli1, Hhip, Shh and Prdx2, many of which are known to function as Gli-dependent enhancers (Sasaki et al., 1997; Dai et al., 1999; Santagati et al., 2003; Agren et al., 2004; Hallikas et al., 2006) (Fig. 3 and supplementary material Table S3). These results therefore suggest that Foxa2 and Gli1 co-regulate a number of common targets through nearby binding sites in the genome. In addition, Foxa2 also bound sites in Gli2 and Gli3 (Mavromatakis et al., 2011) that are not known to be bound by Gli1 (Fig. 3), suggesting the possibility that Foxa2 directly regulates all three Gli genes.
Fig. 3.
Foxa2 peaks in genes of the Shh pathway. (A) Foxa2 binding sites were observed in genomic regions previously identified as bound by Gli1 (Vokes et al., 2007), including in the genes Ptch1, Nkx2-9, Gli1, Hhip and Foxa2. Peaks found in the Shh and Gli3 genes are also included. (B) ChIP-qPCR experiments performed with chromatin from mouse E12.5 ventral midbrain tissue were used to validate the data (*P<0.05). Error bars indicate s.e.m. -ve1, Foxa2 open reading frame; -ve2, Gli-ve (an upstream region of Gli2) (Mavromatakis et al., 2011).
Foxa2 directly and positively regulates the expression of key determinants of the mDA lineage
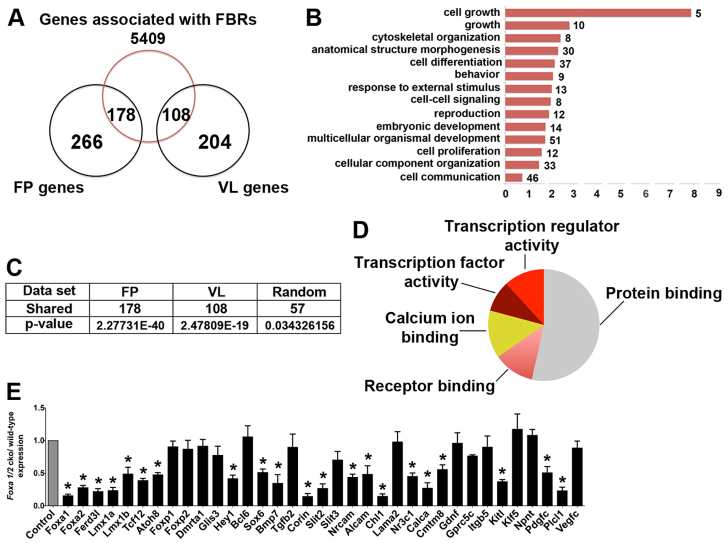
In order to identify a core set of Foxa2 target genes that are functionally relevant for mDA neuronal progenitor and FP specification, we mapped FBRs to the nearest TSS of Ensembl genes. We then compared this set of Foxa2 candidate target genes with a list of 444 genes that are specifically expressed in the midbrain FP as generated by expression profiling of E10.5 mouse embryos (Gennet et al., 2011). Interestingly, 40% (178) of these FP genes were bound by Foxa2 (hypergeometric analysis, P=2.27×10−40; Fig. 4C), suggesting that Foxa2 directly regulates many of the genes expressed in this tissue. We then surveyed the functional annotations of these putative Foxa2 targets using Gene Ontology (GO). A wide spectrum of biological processes was significantly enriched, including cell growth, embryonic development, cell differentiation and cell communication (Fig. 4B). Foxa2 targets also display a diverse range of molecular functions, including transcriptional regulator activity, receptor binding and calcium ion binding (Fig. 4D).
Fig. 4.
Foxa2 directly regulates genes with multiple developmental functions. (A) The overlap between genes associated with Foxa2 binding events and genes expressed predominantly in the floor plate (FP) or the ventrolateral region (VL) of the midbrain of E10.5 mouse embryos. FP and VL data were obtained from Gennet et al. (Gennet et al., 2011). (B) Enrichment of Gene Ontology (GO) terms from GO Slim for biological processes associated with FP-specific Foxa2-bound genes. The number of target genes in each category is shown. (C) The number of Foxa2 target genes in each region with statistical analysis. (D) Enrichment of GO terms for molecular functions (GO Slim) associated with FP-specific Foxa2-bound genes. (E) qRT-PCR analysis of selected FP-specific Foxa2-bound genes using ventral midbrain tissue from E10.5 wild-type and Foxa1/2 cko embryos (*P<0.05). Error bars indicate s.e.m.
To further address the functional significance of Foxa2 binding, we analyzed by qRT-PCR the expression of some of these targets in the midbrain FP of wild-type and Foxa1/2 conditional mutant mouse embryos at E10.5. We selected targets involved in transcriptional regulation and receptor binding, two classes of genes with potentially important roles in regulating cell identity. The expression of nine out of 17 Foxa2 targets with transcriptional activity and nine out of 18 targets with receptor binding activity was severely reduced in En1Cre;Foxa1flox/flox;Foxa2flox/flox (referred to as Foxa1/2 cko) mutants compared with control embryos (Fig. 4E). Analysis of this sample therefore suggests that ~50% of the candidate FP targets identified in Fig. 4A are regulated in Foxa1/2 cko embryos. Interestingly, five of the 17 bound and regulated targets identified in Fig. 4E, namely Lmx1a (Andersson et al., 2006b), Msx1 (Andersson et al., 2006b), Ferdl3 (Ono et al., 2010), Foxa1 and Foxa2 (Andersson et al., 2006b), are known to have essential roles in regulating mDA progenitor specification, suggesting that Foxa2 directly controls the expression of essential determinants of the lineage. Lmx1b also has a role in regulating the specification and differentiation of mDA neurons (Andersson et al., 2006b) and is bound (supplementary material Table S1) and regulated (Lin et al., 2009) by Foxa2. Its omission from the FP-specific gene list is presumably because its expression is not restricted to the FP but is also present in lateral midbrain progenitors. Taken together, these results suggest that Foxa2 has multiple direct inputs into the molecular pathways regulating the specification and differentiation of the mDA progenitor lineage.
Foxa2 directly and positively regulates the expression of FP axon guidance molecules
In zebrafish embryos, Foxa2 is involved in maintenance of the differentiated character of the FP (Norton et al., 2005). In mouse embryos, Foxa2 is required for the induction of the FP through regulation of the secreted morphogen Shh (Ang and Rossant, 1994; Weinstein et al., 1994), but whether it also has later roles in FP function is largely unknown. The FP expresses secreted and transmembrane proteins that regulate the growth of commissural axons across the midline (reviewed by Brose and Tessier-Lavigne, 2000). FP-specific Foxa2 targets include genes that encode factors with axon guidance activities (reviewed by Kolodkin and Tessier-Lavigne, 2011), such as the Slit ligands Slit2 and Slit3, the TGFβ superfamily members Tgfβ2 and Bmp7, the cell adhesion molecules of the immunoglobulin superfamily Alcam, Chl1 and Nrcam, and Shh (supplementary material Table S2). qRT-PCR experiments indicate that the expression of seven out of nine (78%) of these genes is reduced in Foxa1/2 cko embryos (Fig. 4E), demonstrating a novel role for Foxa2 in promoting the expression of factors that control the guidance of axons across the ventral midline. Since the FP is transformed to more dorsal GABAergic progenitors, we cannot formally rule out the possibility that some of the changes in gene expression (Fig. 4E) in Foxa1/2 cko mutants at E10.5 are a consequence of cell fate transformation (Lin et al., 2009). Therefore, we examined Slit1, Slit2 and Shh expression also in NestinCre/+;Foxa1−/−;Foxa2flox/flox embryos, in which partial specification of the FP still occurs when Foxa1/2 are deleted at E10.5. Shh, Slit1 and Slit2 are not expressed in the FP of these mutant embryos. By contrast, these genes are still expressed in the FP of wild-type embryos at E12.5 (supplementary material Fig. S3).
Analysis of enhancer activity by transient lacZ transgenesis in mouse and chick embryos
To explore the contributions of FBRs to the cis-regulation of gene expression, we selected those associated with Lmx1a, Lmx1b and Corin, three genes expressed in mDA progenitors, for functional analysis by transgenesis. Foxa2 bound to three regions surrounding Lmx1a that are conserved in 15-18 species, referred to as E1, E2 and E3, and two conserved regions around Lmx1b termed E1 and E2 (supplementary material Table S4). These conserved elements were cloned into a minimal β-globin promoter-lacZ reporter plasmid and injected into fertilized mouse zygotes to assess their enhancer activity by X-gal staining at E10.5.
The three Lmx1a genomic regions, 522 bp E1, 881 bp E2 and 446 bp E3, promoted lacZ expression in transgenic embryos (Fig. 5A-C″). Lmx1a E1 transgenic embryos displayed β-galactosidase (β-gal) activity throughout the ventral neural tube, similar to the Foxa2 expression pattern (Fig. 5A-A″), whereas the pattern of β-gal activity in Lmx1a E2 and E3 transgenic embryos was restricted to the rostral part of the endogenous expression domain of Lmx1a (Andersson et al., 2006b), i.e. the caudal forebrain and anterior midbrain (Fig. 5B-C″). The identification of three enhancers for Lmx1a suggests that multiple cis-regulatory elements cooperate to specify the precise spatial expression pattern of this gene.
Fig. 5.
Successful prediction of FP enhancers from Foxa2 binding events in mDA progenitors. (A-E) E10.0-10.5 transgenic mouse embryos expressing lacZ from a minimal promoter and candidate enhancers bound by Foxa2 in the Lmx1a, Lmx1b and Corin genes. (A′-E″) Histological sections through the midbrain (A′-E′) and the spinal cord (A″-E″). (F) Whole-mount lacZ staining of transgenic embryos containing the Lmx1b E1 enhancer with a mutation in the Foxa2 binding site. (G) Multiple species alignment of the Lmx1b E1 enhancer using ClustalW, with the Foxa2 motif indicated by the red box and the nucleotide substitutions in the mutated version of the motif in red. Similar staining in midbrain tissue was observed in at least three transgenic embryos for each of the enhancers illustrated in this figure. Scale bars: 100 μm.
One Lmx1b element, the 206 bp E1 region, governed reporter gene expression in the FP of the midbrain and spinal cord and in future auditory tissue (Fig. 5D-D″), similar to the endogenous domains of Lmx1b expression (Guo et al., 2007). By contrast, none of the five transgenic embryos containing the Lmx1b E2 construct that were analyzed expressed β-gal at E10.5. We generated a mutation in the single Foxa2 binding site in the Lmx1b E1 enhancer to determine the requirement for Foxa2 binding for the activity of this element (Fig. 5G). The Lmx1b E1 Foxa2 mutant construct failed to drive β-gal expression in the FP throughout the anteroposterior axis of the embryo, heart and dorsal part of the otic vessel, whereas ectopic β-gal expression in the somites was retained (Fig. 5F), indicating that expression in the FP is dependent on the Foxa2 site. However, Foxa2 is not expressed in the heart and otic vessel, so the dependence of β-gal expression on the Foxa2 site in these structures suggests that other forkhead proteins with similar binding properties occupy these sites.
Corin is a cell surface protease that is specifically expressed in mDA progenitors in the FP (Ono et al., 2007). Two FBRs were found in the Corin gene, only one of which is conserved. This element, called E1, was able to promote β-gal expression in the FP in the midbrain and caudal regions except in rhombomere 1 (Fig. 5E-E″).
Taken together, these results demonstrate that at least some of the FBRs in the three genes analyzed have enhancer regulatory functions in the FP and that Foxa2 binding is required for enhancer activity in the case of the Lmx1b E1 element.
We also examined whether the FBRs in Lmx1a, Lmx1b and Corin function as enhancers in chick by electroporating the same constructs as above into the midbrain of stage 10 chick embryos and analyzing β-gal activity 48 hours later. In this assay, Lmx1b E1 and Corin E1 promoted β-gal expression in the FP, whereas none of the three FBRs of Lmx1a showed activity (Fig. 6A,B; data not shown). Two additional FBRs, in Bmp7 and Slit2, also promoted β-gal expression in the FP (Fig. 6A). These results indicate that some FBRs have enhancer functions in both mouse and chick.
Fig. 6.
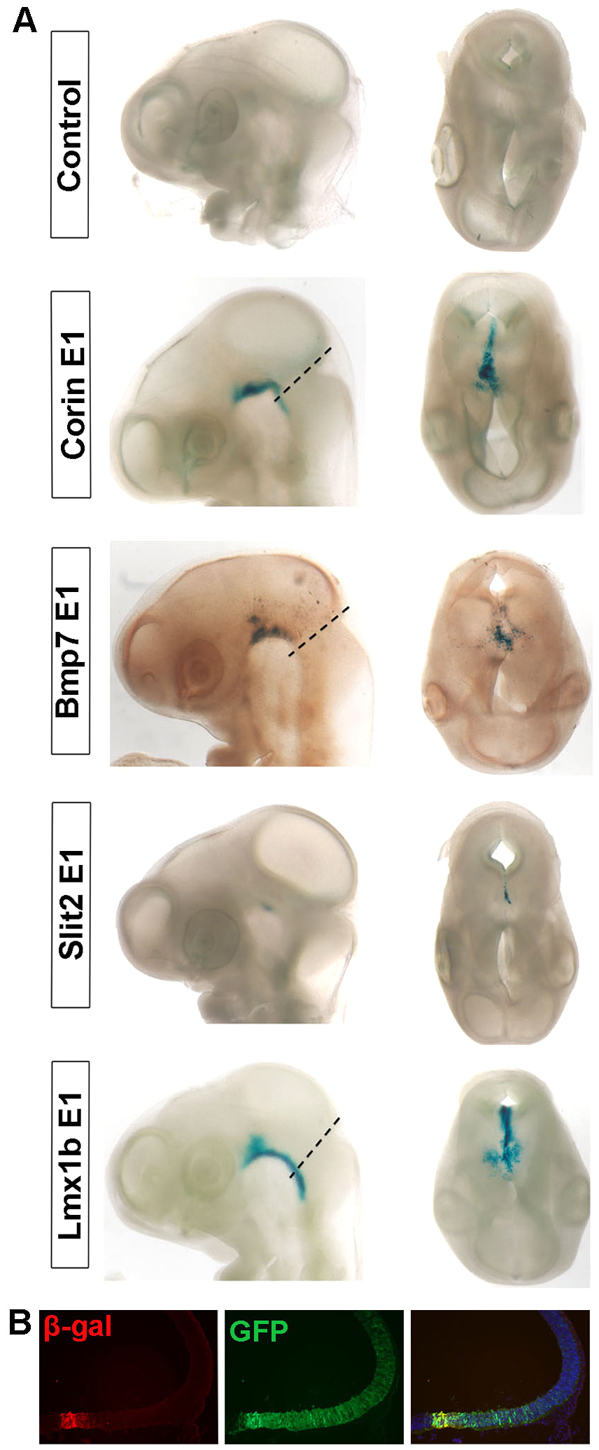
Screening of candidate enhancer elements bound by Foxa2 in chick embryos. (A) The midbrain neuroepithelium of stage 10 chick embryos was electroporated with selected candidate enhancers in the Lmx1a, Lmx1b, Bmp7, Slit2 and Corin genes cloned in a lacZ reporter construct. A GFP construct was co-electroporated with the test constructs to assess electroporation efficiency and the empty lacZ reporter construct was used as control (not shown). Embryos harvested 48 hours post-electroporation are shown in lateral view (left) and dorsal view (right). Dashed lines indicate midbrain-hindbrain boundary. (B) Coronal section of a chick embryo electroporated with the Lmx1b E1 lacZ construct showing GFP+ electroporated cells in most of the midbrain (green), whereas β-gal expression (red) is restricted to the ventral midbrain. The right-hand image is a merge.
Foxa2 inhibits alternative midbrain neuronal fates by direct binding to cell fate determinants
Our earlier functional studies showed that Foxa2 promotes the fate specification of mDA neurons in part by inhibiting the expression of a dorsal and ventrolateral midbrain determinant, Helt, which is essential for the development of most midbrain GABAergic neurons (Miyoshi et al., 2004; Guimera et al., 2006; Nakatani et al., 2007). Since Foxa2 has been shown to act as a transcriptional activator in transfection experiments in vitro, it was unclear whether Foxa2 directly represses Helt to suppress the GABAergic cell fate. Examination of the Foxa2 ChIP-seq data showed that Helt is indeed bound by Foxa2 and is therefore likely to be a direct target of Foxa2 repression (supplementary material Table S1). In addition, Foxa2 also represses a ventrolateral midbrain determinant, Nkx2.2, probably through direct binding (Lin et al., 2009).
Since Foxa2 directly represses ventrolateral determinants such as Helt and Nkx2.2 (Lin et al., 2009), we rationalized that intersecting our list of FBR-associated genes (FBGs) with a list of genes specifically expressed in ventrolateral midbrain might lead to the identification of additional genes that are normally repressed by Foxa2 in the FP. A list of 312 genes specifically expressed in the ventrolateral midbrain was recently published (Gennet et al., 2011). Of the FBGs identified in this study, 108 were present in this list (hypergeometric analysis, P=2.47×10−19; Fig. 4E), including genes that have been shown to be repressed in Foxa1/2 cko mutants, such as Gli1, Gli2 and Gli3 (Mavromatakis et al., 2011), thus providing support for the initial rationale. Examination of the expression of five other transcription factors, namely Tle4, Otx1, Sox1, Tal2 and Pax3, showed that all but Pax3 exhibited expanded ventral midbrain expression in E10.5 Foxa1/2 cko embryos (Fig. 7). These results strongly suggest that Foxa2-bound genes that are expressed in the ventrolateral midbrain represent an enriched list of genes that are repressed by Foxa2 in the FP. Similar to genes that are positively regulated by Foxa2 in the FP, some of the repressed genes might be indirectly regulated by Foxa2, although we have found that Otx1 and Ptch1 are ectopically expressed in the FP of NestinCre;Foxa1−/−;Foxa2flox/flox mutants in the absence of Foxa1 and Foxa2 at E12.5 (supplementary material Fig. S3). These results are consistent with the idea that these genes are directly repressed by Foxa2.
Fig. 7.
Inhibition of ventrolateral determinants in the FP of the midbrain. (A-D′) Tle4, Otx1, Sox1 and Tal2 are ectopically expressed in the FP in addition to their normal expression in the dorsal and/or ventrolateral midbrain regions of mouse E10.5 Foxa1/2 cko mutants, as compared with control littermates. (E,E′) No change was observed for Pax3 expression in the dorsal midbrain between Foxa1/2 cko mutants and control littermates.
DISCUSSION
In this study, we defined by ChIP-seq a comprehensive set of in vivo binding sites for the winged helix transcription factor Foxa2 in mDA progenitors differentiated from NestinLmx1a-transfected ES cells. We detected 9160 FBRs, 80% of which have a Foxa2 binding motif. This result suggests that Foxa2 is recruited to DNA mainly by direct binding. ChIP-qPCR validation of 17 out of 19 selected FBRs using chromatin from mouse E12.5 ventral midbrain tissue suggests a high confidence genome-wide dataset for Foxa2-chromatin interactions.
Intersection of our dataset with a list of genes specifically expressed in FP and ventrolateral midbrain cells produced a statistically significant overlap, suggesting that Foxa2 directly regulates many FP-specific and ventrolateral-specific genes. About 50% of these binding sites appear to be functionally relevant as expression of the associated genes was significantly reduced or lost in Foxa1/2 cko embryos. As detailed above, direct targets of Foxa2 in the FP include genes required in mDA neuron and FP differentiation, indicating a direct role for Foxa2 in promoting these cellular programs. In addition, specification of FP identity also requires a Foxa2-dependent role in the repression of determinants of ventrolateral midbrain fates and attenuation of the Shh signaling pathway (Fig. 8). Finally, the genome-wide analysis of Foxa2 target genes has also revealed a novel role of Foxa2 in the guidance of axons across the FP (Fig. 4). The list of Foxa2 target genes thus represents a valuable resource with which to dissect the role of novel factors in various aspects of FP and mDA neuron biology.
Fig. 8.

Model of the Foxa2-driven regulatory network in the midbrain FP. A model for a Foxa2-driven transcriptional network regulating mDA neuronal and FP specification depicted using standard BioTapestry nomenclature (http://www.biotapestry.org) (Longabaugh et al., 2005). Non-expressed genes are in gray; expressed genes are in black or other colors. Foxa2 targets validated in this study by ChIP-qPCR and expression analysis in mutant embryos are indicated by bold red lines. Other colored broken lines indicate interactions based on other genetic studies in mice or chick (Ono et al., 2010; Andersson et al., 2006a; Yan et al., 2011). Gli-act, Gli activator.
FBRs identify FP enhancers
Our demonstration that several conserved FBRs have enhancer activity in the FP also suggests that our dataset can be used to identify novel cis-regulatory elements that are active in this region. Three out of six conserved FBRs promoted lacZ reporter expression in the FP along the whole length of the embryo and two out of six specifically in the midbrain FP. Foxa2 binding motifs were found in all five functional enhancers, and mutation of this motif in FBRs in Lmx1b demonstrated that Foxa2 binding is indeed essential for enhancer activity in the FP. Other enhancers that have previously been shown to require Foxa2 for their activity in the FP, including elements in the Shh, Foxa2 (autoregulatory element) and Ferdl3 genes, were also identified as FBRs in our ChIP-seq experiments. Comparison of the enhancers that are only active in the midbrain FP with those that are active in the whole FP might identify other transcription factors that promote or restrict gene expression to the midbrain.
Gene regulatory networks involved in the specification of mDA neuron and FP identity
This study has provided new mechanistic insights into how Foxa1/2 specify mDA progenitors. We demonstrate that Foxa2 directly and positively regulates the expression of essential determinants of this lineage, including Lmx1a/b and Ferd3l (Fig. 8). The latter transcription factors then release the repression of Neurog2 expression by Hes1 (Kele et al., 2006; Andersson et al., 2006a), thereby enabling mDA progenitor cells of the FP to undergo neurogenesis and differentiation into mDA neurons.
We also show that Foxa2 represses in the ventral midbrain multiple components of the Shh signaling pathway, including the Shh receptor Ptch1, the transducers Gli1, Gli2, Gli3, and the transcription factors Nkx2.2 (Mavromatakis et al., 2011) and Nkx2.9 (data not shown). As Foxa2 expression is directly activated by Shh via Gli1 and Gli2, Foxa2 acts as a feedback regulator to modulate the level and duration of Shh signaling. Interestingly, our ChIP-seq data suggest that Foxa2 performs this role by binding the same regulatory regions as Gli factors in a number of Shh pathway genes. The role of Foxa2 in the regulation of these targets is antagonistic to that of Gli, suggesting that Foxa2 might prevent recruitment of Gli to the regulatory elements, thereby blocking its activity. This role of attenuation of Shh signaling is important for the specification of FP identity, as prolonged Shh signaling leads to transformation of the FP into more dorsal midbrain identities (Ribes et al., 2010).
The fact that Foxa2 negatively regulates many Shh target genes as well as transcription factors expressed in ventrolateral midbrain progenitors (Fig. 8) suggests that, in addition to its role as an intrinsic activator, Foxa2 might function as a repressor. Foxa proteins behave as transcriptional activators in cell transfection assays, suggesting that Foxa2 repressor activity might involve the recruitment of a transcriptional repressor. The co-repressors Gro/TLE/Grg have been shown to interact with Foxa proteins (Sekiya and Zaret, 2007) and Tle4 is expressed in the ventral midbrain, suggesting that it might participate in the repressive functions of Foxa1 and Foxa2. Future studies will concentrate on elucidating the identity of co-regulators that interact with Foxa1/2 to positively or negatively regulate distinct cell fates.
Supplementary Material
Acknowledgements
We thank Drs Klaus Kaestner and Paul Robson for initial help with the optimization of chromatin immunoprecipitation experiments and all colleagues who provided in situ hybridization probes.
Footnotes
Funding
This work was supported by the United Kingdom Medical Research Council. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.081034/-/DC1
References
- Agren M., Kogerman P., Kleman M. I., Wessling M., Toftgard R. (2004). Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330, 101-114 [DOI] [PubMed] [Google Scholar]
- Andersson E., Jensen J. B., Parmar M., Guillemot F., Bjorklund A. (2006a). Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development 133, 507-516 [DOI] [PubMed] [Google Scholar]
- Andersson E., Tryggvason U., Deng Q., Friling S., Alekseenko Z., Robert B., Perlmann T., Ericson J. (2006b). Identification of intrinsic determinants of midbrain dopamine neurons. Cell 124, 393-405 [DOI] [PubMed] [Google Scholar]
- Ang S. L. (2009). Foxa1 and Foxa2 transcription factors regulate differentiation of midbrain dopaminergic neurons. Adv. Exp. Med. Biol. 651, 58-65 [DOI] [PubMed] [Google Scholar]
- Ang S. L., Rossant J. (1994). HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561-574 [DOI] [PubMed] [Google Scholar]
- Baas D., Meiniel A., Benadiba C., Bonnafe E., Meiniel O., Reith W., Durand B. (2006). A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur. J. Neurosci. 24, 1020-1030 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28-36 [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823-837 [DOI] [PubMed] [Google Scholar]
- Blaess S., Corrales J. D., Joyner A. L. (2006). Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development 133, 1799-1809 [DOI] [PubMed] [Google Scholar]
- Brose K., Tessier-Lavigne M. (2000). Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr. Opin. Neurobiol. 10, 95-102 [DOI] [PubMed] [Google Scholar]
- Collignon J., Sockanathan S., Hacker A., Cohen-Tannoudji M., Norris D., Rastan S., Stevanovic M., Goodfellow P. N., Lovell-Badge R. (1996). A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development 122, 509-520 [DOI] [PubMed] [Google Scholar]
- Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. (1999). Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274, 8143-8152 [DOI] [PubMed] [Google Scholar]
- Epstein D. J., McMahon A. P., Joyner A. L. (1999). Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development 126, 281-292 [DOI] [PubMed] [Google Scholar]
- Ferri A. L., Lin W., Mavromatakis Y. E., Wang J. C., Sasaki H., Whitsett J. A., Ang S. L. (2007). Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development 134, 2761-2769 [DOI] [PubMed] [Google Scholar]
- Friling S., Andersson E., Thompson L. H., Jonsson M. E., Hebsgaard J. B., Nanou E., Alekseenko Z., Marklund U., Kjellander S., Volakakis N., et al. (2009). Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 106, 7613-7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., LeLay J., Vatamaniuk M. Z., Rieck S., Friedman J. R., Kaestner K. H. (2008). Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 22, 3435-3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasca S., Hill D. P., Klingensmith J., Rossant J. (1995). Characterization of a gene trap insertion into a novel gene, cordon-bleu, expressed in axial structures of the gastrulating mouse embryo. Dev. Genet. 17, 141-154 [DOI] [PubMed] [Google Scholar]
- Gennet N., Gale E., Nan X., Farley E., Takacs K., Oberwallner B., Chambers D., Li M. (2011). Doublesex and mab-3-related transcription factor 5 promotes midbrain dopaminergic identity in pluripotent stem cells by enforcing a ventral-medial progenitor fate. Proc. Natl. Acad. Sci. USA 108, 9131-9136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera J., Weisenhorn D. V., Wurst W. (2006). Megane/Heslike is required for normal GABAergic differentiation in the mouse superior colliculus. Development 133, 3847-3857 [DOI] [PubMed] [Google Scholar]
- Guo C., Qiu H. Y., Huang Y., Chen H., Yang R. Q., Chen S. D., Johnson R. L., Chen Z. F., Ding Y. Q. (2007). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development 134, 317-325 [DOI] [PubMed] [Google Scholar]
- Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J. (2006). Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124, 47-59 [DOI] [PubMed] [Google Scholar]
- Hallonet M., Kaestner K. H., Martin-Parras L., Sasaki H., Betz U. A., Ang S. L. (2002). Maintenance of the specification of the anterior definitive endoderm and forebrain depends on the axial mesendoderm: a study using HNF3beta/Foxa2 conditional mutants. Dev. Biol. 243, 20-33 [DOI] [PubMed] [Google Scholar]
- Hannenhalli S., Kaestner K. H. (2009). The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra A. K., Warot X., Brocard J., Bornert J. M., Xiao J. H., Chambon P., Metzger D. (1999). Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 27, 4324-4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Ferri A. L., Milton C., Prin F., Pla P., Lin W., Gavalas A., Ang S. L., Briscoe J. (2007). Transcriptional repression coordinates the temporal switch from motor to serotonergic neurogenesis. Nat. Neurosci. 10, 1433-1439 [DOI] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497-1502 [DOI] [PubMed] [Google Scholar]
- Kele J., Simplicio N., Ferri A. L., Mira H., Guillemot F., Arenas E., Ang S. L. (2006). Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133, 495-505 [DOI] [PubMed] [Google Scholar]
- Kim R. Y., Robertson E. J., Solloway M. J. (2001). Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev. Biol. 235, 449-466 [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Tessier-Lavigne M. (2011). Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 3, pii: a001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Friedman J. R., Fulmer J. T., Kaestner K. H. (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944-947 [DOI] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851-1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Metzakopian E., Mavromatakis Y. E., Gao N., Balaskas N., Sasaki H., Briscoe J., Whitsett J. A., Goulding M., Kaestner K. H., et al. (2009). Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev. Biol. 333, 386-396 [DOI] [PubMed] [Google Scholar]
- Longabaugh W. J., Davidson E. H., Bolouri H. (2005). Computational representation of developmental genetic regulatory networks. Dev. Biol. 283, 1-16 [DOI] [PubMed] [Google Scholar]
- Mansour A. A., Nissim-Eliraz E., Zisman S., Golan-Lev T., Schatz O., Klar A., Ben-Arie N. (2011). Foxa2 regulates the expression of Nato3 in the floor plate by a novel evolutionarily conserved promoter. Mol. Cell. Neurosci. 46, 187-199 [DOI] [PubMed] [Google Scholar]
- Mavromatakis Y. E., Lin W., Metzakopian E., Ferri A. L., Yan C. H., Sasaki H., Whisett J., Ang S. L. (2011). Foxa1 and Foxa2 positively and negatively regulate Shh signalling to specify ventral midbrain progenitor identity. Mech. Dev. 128, 90-103 [DOI] [PubMed] [Google Scholar]
- Miyoshi G., Bessho Y., Yamada S., Kageyama R. (2004). Identification of a novel basic helix-loop-helix gene, Heslike, and its role in GABAergic neurogenesis. J. Neurosci. 24, 3672-3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani T., Minaki Y., Kumai M., Ono Y. (2007). Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development 134, 2783-2793 [DOI] [PubMed] [Google Scholar]
- Norton W. H., Mangoli M., Lele Z., Pogoda H. M., Diamond B., Mercurio S., Russell C., Teraoka H., Stickney H. L., Rauch G. J., et al. (2005). Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development 132, 645-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Nakatani T., Sakamoto Y., Mizuhara E., Minaki Y., Kumai M., Hamaguchi A., Nishimura M., Inoue Y., Hayashi H., et al. (2007). Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134, 3213-3225 [DOI] [PubMed] [Google Scholar]
- Ono Y., Nakatani T., Minaki Y., Kumai M. (2010). The basic helix-loop-helix transcription factor Nato3 controls neurogenic activity in mesencephalic floor plate cells. Development 137, 1897-1906 [DOI] [PubMed] [Google Scholar]
- Peterson R. S., Clevidence D. E., Ye H., Costa R. H. (1997). Hepatocyte nuclear factor-3 alpha promoter regulation involves recognition by cell-specific factors, thyroid transcription factor-1, and autoactivation. Cell Growth Differ. 8, 869-882 [PubMed] [Google Scholar]
- Puelles E., Acampora D., Lacroix E., Signore M., Annino A., Tuorto F., Filosa S., Corte G., Wurst W., Ang S. L., et al. (2003). Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nat. Neurosci. 6, 453-460 [DOI] [PubMed] [Google Scholar]
- Ribes V., Balaskas N., Sasai N., Cruz C., Dessaud E., Cayuso J., Tozer S., Yang L. L., Novitch B., Marti E., et al. (2010). Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 24, 1186-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., et al. (2007). Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4, 651-657 [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P. (2010). PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11, 415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F., Abe K., Schmidt V., Schmitt-John T., Suzuki M., Yamamura K., Imai K. (2003). Identification of cis-regulatory elements in the mouse Pax9/Nkx2-9 genomic region: implication for evolutionary conserved synteny. Genetics 165, 235-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir T., Geiman E. J., Wang Z., Velasquez T., Mitsui S., Yoshihara Y., Frank E., Alvarez F. J., Goulding M. (2004). Pax6 and engrailed 1 regulate two distinct aspects of renshaw cell development. J. Neurosci. 24, 1255-1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Hogan B. L. (1996). Enhancer analysis of the mouse HNF-3 beta gene: regulatory elements for node/notochord and floor plate are independent and consist of multiple sub-elements. Genes Cells 1, 59-72 [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hui C., Nakafuku M., Kondoh H. (1997). A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124, 1313-1322 [DOI] [PubMed] [Google Scholar]
- Scardigli R., Schuurmans C., Gradwohl G., Guillemot F. (2001). Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31, 203-217 [DOI] [PubMed] [Google Scholar]
- Segev E., Halachmi N., Salzberg A., Ben-Arie N. (2001). Nato3 is an evolutionarily conserved bHLH transcription factor expressed in the CNS of Drosophila and mouse. Mech. Dev. 106, 197-202 [DOI] [PubMed] [Google Scholar]
- Sekiya T., Zaret K. S. (2007). Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell 28, 291-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Liu T., Manrai A. K., Liu X. S. (2009). CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605-2606 [DOI] [PubMed] [Google Scholar]
- Silva J. P., von Meyenn F., Howell J., Thorens B., Wolfrum C., Stoffel M. (2009). Regulation of adaptive behaviour during fasting by hypothalamic Foxa2. Nature 462, 646-650 [DOI] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W. J., Davidson E. H., Wong W. H., McMahon A. P. (2007). Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977-1989 [DOI] [PubMed] [Google Scholar]
- Wan H., Dingle S., Xu Y., Besnard V., Kaestner K. H., Ang S. L., Wert S., Stahlman M. T., Whitsett J. A. (2005). Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J. Biol. Chem. 280, 13809-13816 [DOI] [PubMed] [Google Scholar]
- Wang K. H., Brose K., Arnott D., Kidd T., Goodman C. S., Henzel W., Tessier-Lavigne M. (1999). Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell 96, 771-784 [DOI] [PubMed] [Google Scholar]
- Weinstein D. C., Ruiz i Altaba A., Chen W. S., Hoodless P., Prezioso V. R., Jessell T. M., Darnell J. E., Jr (1994). The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78, 575-588 [DOI] [PubMed] [Google Scholar]
- Yan C. H., Levesque M., Claxton S., Johnson R. L., Ang S. L. (2011). Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J. Neurosci. 31, 12413-12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.