Abstract
The protozoan parasite Giardia intestinalis (also known as Giardia lamblia) is a major waterborne pathogen. During its life cycle, Giardia alternates between the actively growing trophozoite, which has two diploid nuclei with low levels of allelic heterozygosity, and the infectious cyst, which has four nuclei and a tough outer wall. Although the formation of the cyst wall has been studied extensively, we still lack basic knowledge about many fundamental aspects of the cyst, including the sources of the four nuclei and their distribution during the transformation from cyst into trophozoite. In this study, we tracked the identities of the nuclei in the trophozoite and cyst using integrated nuclear markers and immunofluorescence staining. We demonstrate that the cyst is formed from a single trophozoite by a mitotic division without cytokinesis and not by the fusion of two trophozoites. During excystation, the cell completes cytokinesis to form two daughter trophozoites. The non-identical nuclear pairs derived from the parent trophozoite remain associated in the cyst and are distributed to daughter cells during excystation as pairs. Thus, nuclear sorting (such that each daughter cell receives a pair of identical nuclei) does not appear to be a mechanism by which Giardia reduces heterozygosity between its nuclei. Rather, we show that the cyst nuclei exchange chromosomal genetic material, perhaps as a way to reduce heterozygosity in the absence of meiosis and sex, which have not been described in Giardia. These results shed light on fundamental aspects of the Giardia life cycle and have implications for our understanding of the population genetics and cell biology of this binucleate parasite.
Key words: Cytoskeleton, Giardia, Encystation, Excystation, Meiosis, Homologous recombination
Introduction
The divergent eukaryotic parasite Giardia intestinalis (syn. Giardia lamblia, Giardia duodenalis) is a major cause of diarrheal disease throughout the world. Additionally, as a diplomonad, it is a member of one of the earliest-diverging eukaryotic lineages based on single- and multi-gene phylogenies (Baldauf et al., 2000; Baldauf, 2003; Best et al., 2004; Arisue et al., 2005; Ciccarelli et al., 2006; Morrison et al., 2007). The Giardia life cycle consists of two stages: a flagellated trophozoite with two nuclei and an infectious cyst with four nuclei. The two diploid (2N=10) nuclei in the trophozoite contain complete copies of the genome and are both transcriptionally active (Kabnick and Peattie, 1990; Bernander et al., 2001; Yu et al., 2002). They also remain independent in the trophozoite, dividing with separate spindles by a semi-open mitosis, and one copy of each parental nucleus is inherited by each daughter cell (Yu et al., 2002; Sagolla et al., 2006).
The differentiation from trophozoite into cyst occurs when a trophozoite, which is usually found attached to the wall of the small intestine, is swept towards the large intestine. Changes in pH and cholesterol availability prompt the trophozoite to encyst, forming a cyst with four nuclei and a thick outer wall (Gillin et al., 1989; Luján et al., 1996; Luján et al., 1997). The formation of the cyst wall, which involves the regulated secretion of cyst wall protein components, has been a topic of intense study over the past few decades (see Lauwaet et al., 2007a; Faso and Hehl, 2011). The cyst is water resistant and can persist for weeks in the environment until it is ingested by a new host. After passing through the stomach, the cyst undergoes excystation, releasing an ‘excyzoite’ that rapidly divides to produce two daughter trophozoites (Buchel et al., 1987; Bernander et al., 2001).
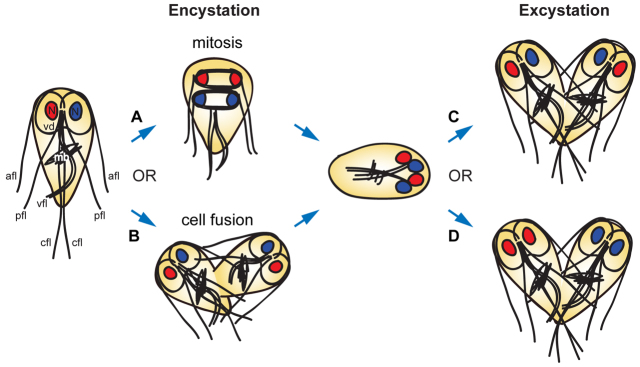
Although many advances have been made in our understanding of cyst wall formation, particularly with regard to ultrastructural aspects of the transition and to cyst wall morphology (Sheffield and Bjorvat, 1977; Luchtel et al., 1980; Buchel et al., 1987; Erlandsen et al., 1989; Hetsko et al., 1998; Lanfredi-Rangel et al., 2003; Palm et al., 2005; Chávez-Munguía et al., 2007; Midlej and Benchimol, 2009; Bittencourt-Silvestre et al., 2010; Faso and Hehl, 2011), the cytoskeletal changes underlying both encystation and excystation have received less attention. The trophozoite microtubule cytoskeleton comprises four pairs of flagella, with eight basal bodies located between the two nuclei, as well as a ventral adhesive disc (used to attach to the intestinal epithelium) and a prominent bundle of microtubules of unknown function called the median body (Fig. 1) (Elmendorf et al., 2003; Dawson and House, 2010). These structures are reorganized and/or disassembled during cyst formation, with only internalized flagella and disc fragments thought to remain in the mature cyst (Elmendorf et al., 2003).
Fig. 1.
Overview of alternative hypotheses for cyst formation and nuclear distribution during excystation. Shown on the left is a diagram of a Giardia trophozoite, with the major components of the microtubule cytoskeleton labeled: the ventral adhesive disc (vd), used for attachment to the intestinal epithelium; four sets of flagella (afl, anterolateral; pfl, posterolateral; vfl, ventral; cfl, caudal); the median body (mb), a microtubule array of unknown function; and the two nuclei (N). Although it has often been assumed that the Giardia cyst is formed after an incomplete mitotic division (A), the alternative route of cell fusion between two trophozoites (B) has never been excluded. In addition, during excystation, two possible routes for the partitioning of nuclei to daughter cells would have dramatically different consequences for Giardia populations. If each daughter cell receives one copy of each parental nucleus (C), any nuclear heterozygosity is maintained throughout the life cycle. By contrast, if the nuclei were distributed such that each daughter cell received two identical nuclei (D), heterozygosity would be eliminated.
It has been assumed that the cyst is formed after an incomplete mitotic division (as opposed to the fusion of two trophozoites) on the basis of morphological observations (reviewed by Adam, 2001; Lauwaet et al., 2007a), flow cytometric analysis of ploidy (Bernander et al., 2001) and the finding that an encystation restriction point exists at the G2 phase of the cell cycle (Reiner et al., 2008). However, to our knowledge, the absence of cell fusion has never been directly tested (Fig. 1A,B), and mitotic spindles in encysting cells have never been observed. Addressing this question using solely morphological observations is difficult because an encysting cell resulting from cell fusion would probably resemble one resulting from incomplete mitosis [i.e. it would contain four nuclei, two discs (or disc fragments) and up to 16 flagella (fewer if some were disassembled upon fusion)]. Similarly, flow cytometry analysis alone cannot conclusively distinguish between mitosis and fusion because both would produce cysts with the same final DNA content.
Related to the question of how the quadrinucleate cyst is formed are the identities of the nuclei in the cyst and during excystation (Fig. 1C,D). The genomes of most Giardia isolates contain very low levels of heterozygosity (Morrison et al., 2007; Teodorovic et al., 2007; Lasek-Nesselquist et al., 2009; Jerlström-Hultqvist et al., 2010), suggesting that the two nuclear genomes are able to exchange genetic information during the Giardia life cycle. The Giardia genome also contains seven homologs of meiosis-specific genes: Spo11, Hop1, Dmc1a, Dmc1b, Hop2, Mnd1 and Mer3 (Ramesh et al., 2005; Malik et al., 2008), which in most organisms are expressed only during meiosis and are specifically involved in meiotic homologous recombination. However, neither meiosis nor a sexual stage has been identified in the Giardia life cycle. We have previously shown that nuclear fusion and the exchange of episomal plasmids can occur between nuclei in the Giardia cyst, a process we have termed ‘diplomixis’ (Poxleitner et al., 2008); we have also shown that Spo11, Hop1 and Dmc1a are expressed only during encystation. Thus, we hypothesized that diplomixis could involve homologous recombination between the nuclear genomes. If accompanied by recombination and/or gene conversion within nuclei in the trophozoite, this process could help reduce heterozygosity between the two nuclei.
However, another way to maintain low heterozygosity could be the sorting of nuclei during excystation such that each daughter cell receives two identical nuclei (rather than one of each original nucleus from the parent trophozoite) (Fig. 1C,D). Therefore, the goals of the present study were twofold: (i) To track the identities of the nuclei during encystation and excystation, and (ii) to determine whether diplomixis involves the exchange of chromosomal genetic material between nuclei. The results of these studies have implications for our understanding of both the basic biology and the population genetics of this parasite and could also shed light on the evolution of meiosis.
Results
Cysts are formed by an incomplete mitotic division
In vitro, encystation can be prompted by exposing cells to basic medium containing high levels of bile (which prevents cholesterol absorption) for 24–48 hours (Gillin et al., 1988; Gillin et al., 1989; Kane et al., 1991; Luján et al., 1996). Similarly, excystation can be triggered by a two-stage procedure in which cells are first exposed to acidic conditions for 30 minutes, mimicking passage through the stomach, followed by a higher-pH solution containing proteases for an additional 30 minutes, which helps break open the cyst wall (Bingham and Meyer, 1979; Rice and Schaefer, 1981; Schaefer et al., 1984; Buchel et al., 1987; Hautus et al., 1988; Boucher and Gillin, 1990).
To determine whether cysts can be derived from the fusion of two trophozoites, we first mixed two strains with different epitope-tagged markers [histone 2A tagged with three hemagglutinin molecules (3HA) and median body protein tagged with three Myc molecules (3myc)] and encysted them in vitro. None of the resulting cysts contained both markers (supplementary material Table S1). This result indicates that, at least in vitro, cysts are generally derived from a single trophozoite. Following water treatment (to destroy any unencysted cells), excystation and growth to confluency of these mixed cultures, we found that trophozoites always had only one of the two markers (supplementary material Table S1), suggesting that cell fusion also does not occur during excystation. Additionally, in a larger-scale experiment performed with 5×107 live cysts (500-ml cultures), encystation and excystation of a mixture of two strains with different integrated drug resistance markers (to neomycin and puromycin) failed to produce any trophozoites with resistance to both drugs (data not shown). Because we typically observe an in vitro excystation efficiency on the order of ~1%, the production of recombinant cells (which would require cell fusion followed by chromosomal recombination or nuclear sorting), if it occurs, would have to happen in fewer than 1 in 500,000 cells.
Next, we sought to characterize the incomplete mitotic division during encystation by staining cells with antibodies against tubulin, centrin (a basal body and spindle pole marker) and cyst wall protein (CWP) after 24 hours of encystation (Fig. 2). Before the division, the trophozoites produce encystation-specific vesicles (ESVs) full of CWP, and two clusters of four centrin spots (marking the basal bodies) are visible between the nuclei (Fig. 2A). Anaphase (Fig. 2B) and telophase (Fig. 2C) generally resemble the mitotic division in trophozoites, with two parallel spindles and the basal bodies (centrioles) at the spindle poles. However, unlike in the trophozoite, the encysting cells are concurrently in the process of exporting ESVs to the periphery of the cell for deposition. At the end of telophase of the encystation mitotic division (Fig. 2D), the process diverges further from its vegetative counterpart: cytokinesis does not occur, the cell does not grow new flagella, and its existing flagella appear to shorten, perhaps as a result of internalization.
Fig. 2.
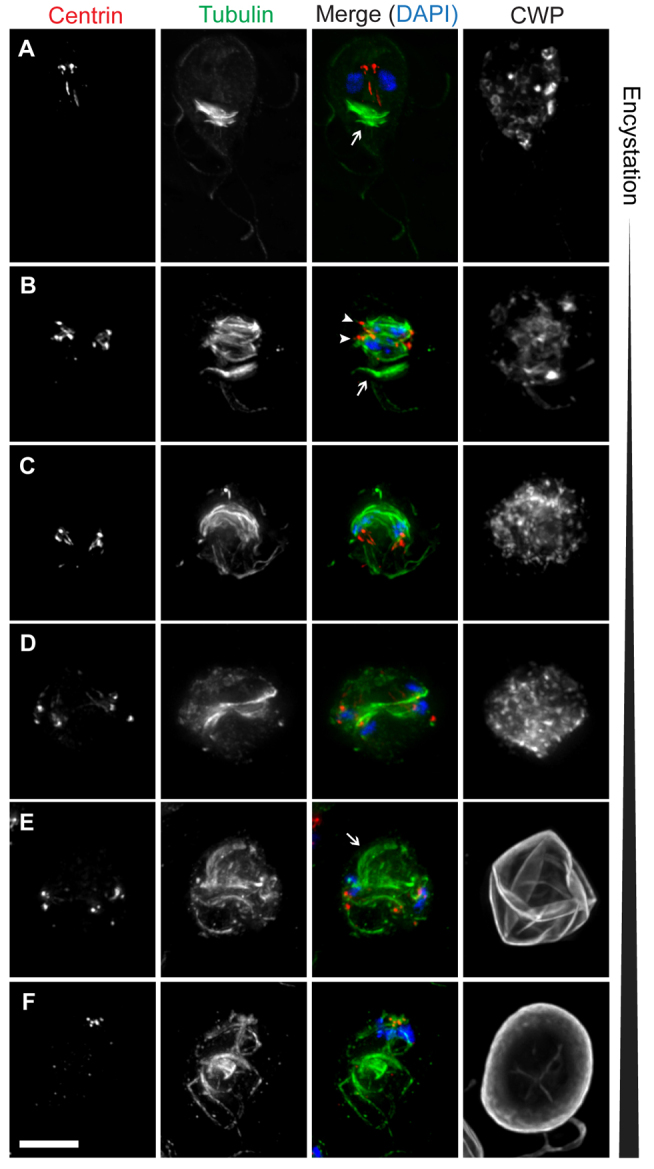
The Giardia cyst is formed by an incomplete mitotic division. Encysting cells were stained with DAPI to label DNA (blue) and antibodies to label tubulin (green), centrin (red), and cyst wall protein (CWP) (gray). (A) An encysting trophozoite, demonstrating typical centrin and tubulin staining and the presence of two nuclei. The median body, a cluster of microtubules of unknown function, is indicated (arrow). (B) An encysting cell in anaphase. The two spindles (arrowheads) and median body (arrow) are indicated. (C,D) Encysting cells in early and late stages of telophase. (E) Early cyst in which the nuclear pairs are still at opposite ends of the cell. The arrow indicates what appears to be a nascent disc fragment associated with one pair of nuclei. (F) Mature cyst in which the nuclear pairs have migrated to one pole of the cell. Scale bar: 5 μm.
At this stage, the two pairs of nuclei are found at opposite poles of the cell (approximately at the previous location of the spindle poles) and each pair remains associated with a bundle of internal axonemes, which appear to be derived from the axonemes that remain between the nuclei during mitosis (Nohynková et al., 2006) (Fig. 2E). It is at this time that the cyst wall is rapidly deposited; by the time the pairs of nuclei have migrated to the same pole of the cyst, the cyst wall has completely formed. The two nuclei within each pair are closely associated and the pairs remain motile for some time in the cyst (Poxleitner et al., 2008), possibly owing to their association with the internal axonemes; this is also presumably the mechanism by which they migrate together. The mature cyst contains four nuclei, two bundles of internal axonemes with eight centrin loci and several ventral disc fragments (Fig. 2F).
It has been reported that the mature cyst contains four fragments of the parental disc that are reassembled during excystation (Elmendorf et al., 2003; Palm et al., 2005). However, on the basis of staining of cells with antibodies to label the tubulin cytoskeleton and the disc (antibody against delta giardin), we observed what appeared to be the initial stages of two new daughter discs being formed during the later stages of the encystation mitotic division (supplementary material Fig. S1A). At the same time, the parental disc was being disassembled, as has been described during cytokinesis in the trophozoite (Tůmová et al., 2007). We most often saw two disc fragments in the mature cyst and early excyzoites; however, in some cases, up to four fragments were visible, although they were still usually found in two clusters (supplementary material Fig. S1B,C). In the excyzoite, the nascent disc fragments appeared to move to the correct position and orientation to complete their development into two new discs (supplementary material Fig. S1D).
Reconstruction of the cytoskeleton during excystation
Next, we stained the cytoskeleton of excysting cells to monitor the transition from excyzoite into trophozoite (Fig. 3). As previously reported, the excyzoite emerges from a single pole of the cyst, which is weakened by the exposure to acidic conditions in the stomach and intestinal proteases (Coggins and Schaefer, 1986; Buchel et al., 1987; Boucher and Gillin, 1990; Hetsko et al., 1998) (Fig. 3A). The flagella emerge first, followed by the body of the cell (Fig. 3B). At this stage, the cytoskeleton of the excyzoite still resembles that of the cyst, with internal bundles of axonemes, two pairs of nuclei and eight centrin spots (Fig. 3A–C). These cells sometimes display one or several structures that resemble ESVs (arrow in Fig. 3C) and contain CWP; these could result from the residual translation of cyst wall protein transcripts, which are extremely abundant during encystation (Hetsko et al., 1998), or could represent leftover vesicles from encystation or excystation that have not yet been eliminated.
Fig. 3.
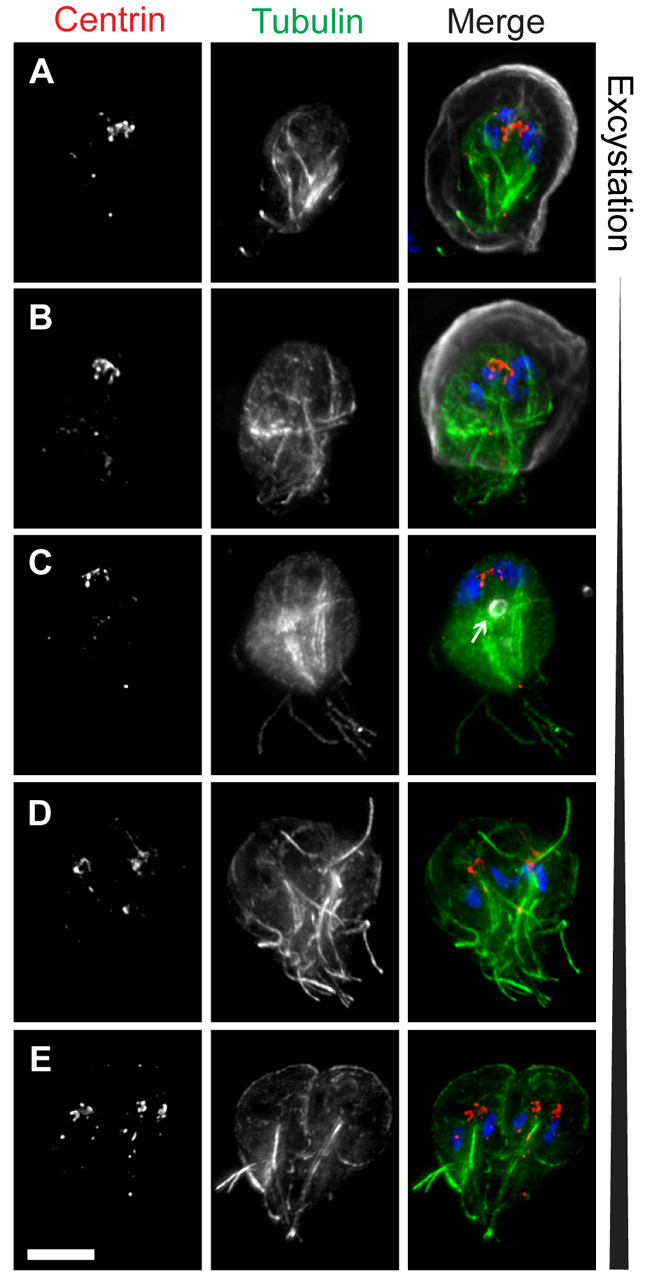
Reassembly of the microtubule cytoskeleton during excystation. Excysting cells were stained with DAPI to label DNA (blue) and antibodies to label tubulin (green), centrin (red), and cyst wall protein (CWP) (gray). (A) Cyst in the early stages of excystation (note the rupture in the cyst wall at the bottom of the image). (B) An excyzoite emerging from a cyst. (C) Completely excysted cell. A residual encystation-specific vesicle is indicated (arrow). (D,E) Later-stage excyzoites in which the typical heart-shaped cytokinetic morphology is restored, including two complete discs and 16 flagella. Scale bar: 5 μm.
The axonemes in the excyzoite are clustered into two prominent bundles, each associated with a nuclear pair (Fig. 3C). Although this configuration of axonemes is different from that of a trophozoite, the cell is still capable of swimming (Buchel et al., 1987; Hautus et al., 1988). After additional rearrangements, the excyzoite begins to resemble a heart-shaped (pre-cytokinesis) cell that is essentially indistinguishable from that of a dividing trophozoite, with the exception that each nucleus is thought to contain a 4C (replicated diploid) DNA content (Fig. 3D–E) (Bernander et al., 2001). By this stage, the number of basal bodies and flagella has increased from 8 to 16, and the cell rapidly completes the construction of two ventral discs (Fig. 3E). Following cytokinesis, the resulting 8C trophozoites presumably divide immediately to produce two 4C trophozoites (Bernander et al., 2001).
Nuclear identity and genetic exchange during differentiation
The association of pairs of nuclei in the cyst and in the excyzoite suggests that the nuclear pairs from the parent trophozoite are preserved throughout differentiation, meaning that a mechanism other than nuclear sorting must exist to reduce allelic heterozygosity in Giardia populations (compare Fig. 1C and Fig. 1D). To test this hypothesis, we integrated a plasmid containing a 10-kb array of LacO repeats into the chromosomes of a population of trophozoites. To ensure the presence of only a single integrated construct, we then made clonal populations and tested for the absence of episomes by PCR (supplementary material Fig. S2). We then performed fluorescence in situ hybridization (FISH) using a fluorescent probe to label the LacO repeat. As for episomes (Yu et al., 2002), integrated plasmids were always found in only one of the two nuclei in the trophozoite (Fig. 4A). Most cells contained a single spot, but occasionally we observed two spots very close together in a single nucleus; we interpret these to represent either replicated chromatids that became separated (most trophozoites rest in G2, but few cells contained two spots) or cases in which the homolog gained the marker through mitotic recombination or gene conversion. In the pre-encystation population, 76% of cells (n=100) contained one spot (or two very close together) in a single nucleus, whereas 24% of cells contained no spot (Fig. 4A). Because this population was clonal and the marker was integrated, and therefore stable even in the absence of drug selection, we interpret this result as a common artifact of FISH; however, it also possible that at least some cells lose the repeat cassette through recombination. After encystation, excystation, and growth of excysted trophozoites to confluency, all in the absence of drug selection, we observed that 49±2% of cells (±s.d.; average of three biological replicates, each n=100) had one spot, 31±2% had no spots and 20±2% had two spots (one in each nucleus) (Fig. 4A). Thus, the production of non-parental patterns is common and consistent.
Fig. 4.
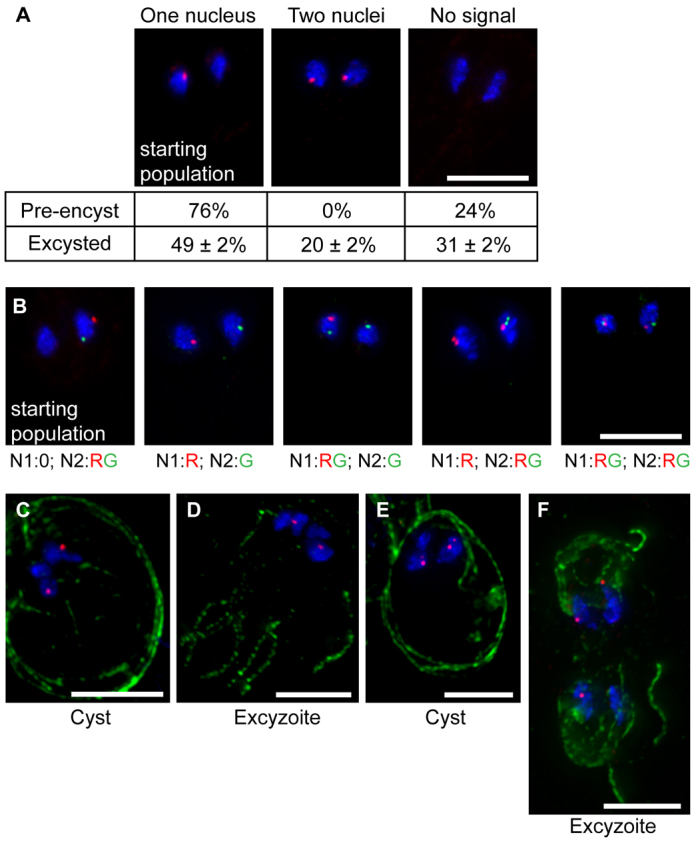
Chromosomal exchange occurs between nuclei during differentiation. (A) A clonal population of cells containing an integrated LacO array in one nucleus was subjected to FISH to track the marker in the starting population (pre-encystation trophozoites) and in excysted trophozoites. DAPI-stained DNA is shown in blue and the LacO probe is shown in red. Percentages of cells with 0, 1 and 2 spots are shown below each image. (B) Patterns observed after excystation of a clonal population containing two markers, LacO (red) and TetO (green), integrated in the same nucleus (first panel). The number of spots in each nucleus is indicated below each image (e.g. N1:0; N2:RG=0 spots in nucleus 1; one red spot and one green spot in nucleus 2). (C–F) Cysts and excyzoites from the experiment described in A. DAPI-stained DNA is shown in blue, and the LacO probe is shown in red. Cyst wall protein (CWP) staining is shown in green in C and E, and tubulin staining is shown in green in D and F. Panels C and D show a cyst and an excyzoite, respectively, with one spot in each nuclear pair; panels E and F show a cyst and an excyzoite, respectively, in which one nuclear pair contains two spots. Scale bars: 5 μm.
The observed pattern could be generated in two ways: (i) genetic exchange between nuclei, or (ii) nuclear sorting such that during excystation each daughter cell received two identical nuclei. However, because only 20% of cells contained two spots, nuclear sorting would have had to occur in a minority of excysting cells (excyzoites). To begin to distinguish between these possibilities, we examined a strain that contained the same LacO repeat construct, but also contained a TetO repeat construct integrated into a different chromosome in the same nucleus (Fig. 4B). As with the LacO construct, the absence of episomal plasmid in this clonal line was confirmed by PCR (data not shown). In this strain, 71% of pre-encysting trophozoites (n=100) contained both markers in a single nucleus; the remaining 29% were missing one or both markers, probably owing to artifacts and/or recombination, as noted above. However, in 25% of excysted trophozoites (n=100), we observed cells with all possible non-parental arrangements of markers (i.e. 14% with one in each nucleus, 10% with two in one nucleus and one in the other, and 1% with two in both nuclei), further suggesting that chromosomal genetic exchange occurs between nuclei in the cyst and/or early excyzoite (Fig. 4B). The remaining 75% of excysted trophozoites contained either the parental arrangement or were missing one or both markers. In theory, all of the observed non-parental arrangements could also be compatible with nuclear sorting combined with signal loss; however, our level of signal loss is about 30% overall (on the basis of our observations of pre-encysting cells), so if nuclear sorting were solely responsible for these patterns, we would expect to see higher levels of cells with both markers in both nuclei. Thus, although we cannot completely exclude rare nuclear sorting on the basis of these data, we can conclude that it is not responsible for all of the observed non-parental arrangements of markers.
To rule further out nuclear sorting, we also performed FISH on cysts and excyzoites (Fig. 4C–F). As expected, the nuclei of most cysts contained two clustered pairs of nuclei, each with one labeled nucleus (Fig. 4C). This pattern was recapitulated in most excyzoites (Fig. 4D), again supporting the idea that the non-identical nuclear pairs remain associated throughout encystation and excystation. Although we were able to observe an occasional cyst with three labeled nuclei (Fig. 4E), as would be expected on the basis of our observation of two spots in the nuclei of excysted trophozoites, this was rare; however, we did notice that the spots in the cyst were often positioned between the masses of DAPI-stained DNA (Fig. 4C), suggesting that the nuclei might remain fused in the cyst and only separate during excystation. Consistently, we were able to identify an early excyzoite with three labeled nuclei (Fig. 4F) (i.e. where one nuclear pair underwent genetic exchange involving movement of the integrated marker, but the other did not). Therefore, our results indicate that the predominant mode of nuclear inheritance for excysting trophozoites involves the inheritance of one copy of each parental nucleus, and chromosomal genetic exchange occurs between nuclear pairs in the cyst.
On the basis of our previous observations of the expression patterns of the Giardia meiotic gene homologs, we predict that this genetic exchange involves homologous recombination. However, because the previous studies were performed using tagged genes on multi-copy episomes (albeit under the native promoter), it is possible that the observed patterns might have been caused by overexpression. Thus, in the present study, we used plasmid integration (Gourguechon and Cande, 2011) to endogenously tag the seven meiotic gene homologs with 3HA epitope tags. Because the genes were tagged at their endogenous locus, the constructs were under the control of the native promoter. We then examined the expression and localization of these proteins in both trophozoites and cysts. Mer3, which in yeast is a meiosis-specific DNA helicase (Nakagawa and Kolodner, 2002), did not localize to the nuclei of trophozoites or cysts (data not shown), suggesting that it might not, in fact, be a Mer3 ortholog; consistently, it is annotated as an RNA helicase in the Giardia genome database (ORF GL50803_87022 at giardiadb.org). As shown in supplementary material Fig. S3, and consistent with our previous findings, only Dmc1b was expressed at high levels in the trophozoite; Mnd1 and Hop2 (not previously studied) were present at low levels, and Spo11, Dmc1a and Hop1 were absent. By contrast, Dmc1b was absent in cysts, whereas Spo11, Dmc1a and Hop1 were upregulated, appearing in encysting trophozoites and remaining in fully encysted cells (supplementary material Fig. S3) (Poxleitner et al., 2008). Mnd1 and Hop2 remained at low (but detectable) levels in cysts. In all cases, and as expected, the proteins were localized to the nuclei in a punctate pattern. We also attempted to knockdown Dmc1a and Spo11 by electroporation with morpholinos (Carpenter and Cande, 2009) immediately before encystation to assess their effects on exchange of the LacO marker, but we were unable to attain measurable levels of knockdown, possibly owing to the morpholinos being diluted during the encystation process (data not shown).
Finally, because we never observe spindles in fully formed cysts (i.e. there is no organized separation of chromosomes such as that seen in a meiotic division), the cell must have some way in which to anchor its chromosomes to prevent widespread aneuploidy following diplomixis. On the basis of the telomere bouquets observed in meiosis in many organisms, we predicted that telomere anchoring might be involved in Giardia as well. Thus, we performed FISH on trophozoites and cysts using a probe to detect the Giardia telomeric repeat sequence TAGGG (Fig. 5). Interestingly, the telomeres were clustered, usually in two spots per nucleus, in both trophozoites (Fig. 5A) and cysts (Fig. 5B). A preliminary BLAST search for telomere-binding proteins in the Giardia genome revealed only telomerase (GiardiaDB accession GL50803_16225), suggesting that the proteins involved in tethering the chromosomes to the nuclear envelope in Giardia are either highly divergent from those in yeast and humans and/or are completely novel. This clustering might provide a mechanism by which the nuclei are able to undergo recombination without drastically changing the ploidy of each nucleus. However, it should be noted that some aneuploidy between nuclei, usually involving the addition or subtraction of one chromosome, has been observed in Giardia trophozoites (Tůmová et al., 2006); diplomixis could be the mechanism by which this aneuploidy was initially created.
Fig. 5.
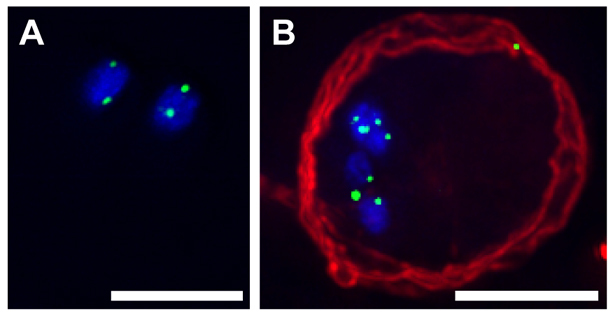
Telomere clustering in trophozoites and cysts. FISH was performed on trophozoites (A) and cysts (B), using a probe to label the telomeric repeat sequence. DAPI-stained DNA is shown in blue, the telomere probe is shown in green and antibody staining against cyst wall protein is shown in red. Scale bars: 5 μm.
Discussion
As an organism with two nuclei, Giardia faces unique challenges in coordinating nuclear inheritance during its life cycle. Although the functional equivalence of the nuclei in Giardia and their independent distribution during mitosis in the trophozoite are relatively well established (Kabnick and Peattie, 1990; Yu et al., 2002), their fates during encystation and excystation have only been studied with respect to genome ploidy (Bernander et al., 2001), despite the significance of this knowledge to our understanding of patterns of allelic heterozygosity and genetic diversity in Giardia populations. Therefore, the goal of the present study was to determine how the four nuclei are derived in the cyst and how these nuclei are distributed during excystation.
Cysts are produced by mitosis and not cell fusion
On the basis of strain mixing experiments (supplementary material Table S1) and immunofluorescence staining of encysting cells (Fig. 2), we have demonstrated that the quadrinucleate cyst is produced through an incomplete mitotic division that occurs rapidly during the final stages of cyst wall deposition. In addition, we have provided a detailed description of the temporal organization of the microtubule cytoskeleton during encystation and excystation. Although the fusion of two trophozoites to form a cyst – which, if followed by nuclear fusion and genetic exchange, would resemble a parasexual cycle with outcrossing – is still theoretically possible, this event would have to be rare and/or happen only under certain conditions, such as in the host. It is also possible that fusion only happens between different strains or between cells with different variant surface proteins (Lasek-Nesselquist et al., 2009). However, given that encystation takes place during transit through the small and large intestine, we speculate that a strict requirement for two cells to find each other and fuse in order to form a cyst might be disadvantageous; thus, encystation from a single parent cell appears to be the predominant (if not sole) mode of cyst formation.
Nuclei are inherited in non-identical pairs
Our observations of the cytoskeletal components of encysting and excysting cells both synthesize and build upon the results of previous ultrastructural and transmission electron microscopy (TEM) studies (Sheffield and Bjorvat, 1977; Luchtel et al., 1980; Buchel et al., 1987; Hetsko et al., 1998; Bernander et al., 2001; Palm et al., 2005; Midlej and Benchimol, 2009; Bittencourt-Silvestre et al., 2010), which have primarily focused on mature cysts. We found that the encystation division resembles the vegetative trophozoite mitotic division until approximately telophase. At that point, instead of continuing on into cytokinesis, the cell internalizes its flagella as the cyst wall rapidly forms. At the same time, cyst wall components are being produced and transported to the periphery of the cell, a process that appears to be independent from, albeit simultaneous with, the nuclear division (Stefanic et al., 2009). In addition, although trophozoites in the early stages of encystation display larger rounded ESVs, by the time the nuclear division takes place, the cyst wall components have been transported to the periphery of the cell and exist in a more diffuse network, as previously reported (Konrad et al., 2010).
On the basis of the patterns of centrin and tubulin staining in our study, the bundles of axonemes associated with each nuclear pair in the cyst appear to be derived from the axonemes found associated with the nuclei during mitosis (Nohynková et al., 2006). According to that study, each daughter cell (or pair of nuclei, in this case) receives one anterolateral flagellum, one caudal flagellum, and either the posterolateral or ventral flagellar pair. These results are consistent with those of a TEM-based study that reported two bundles of three internal axonemes associated with each nuclear pair in the cyst, as well as two additional flagella found in the periphery of the cell (Sheffield and Bjorvat, 1977). It remains unclear whether the flagella are actively depolymerized during encystation or whether they are simply internalized.
On the basis of our observations of the growth of two nascent daughter discs during the encystation mitotic division and the overall morphological similarity of these nascent discs to the fragments in the cyst, we hypothesize that disc growth is paused during encystation and then resumed during excystation. This scenario would be more consistent with a what is observed during cytokinesis in the trophozoite, in which two new ventral discs grow on the opposite side of the cell from the parental ventral disc (Tůmová et al., 2007). However, because we cannot distinguish between old and new discs, it remains possible that the disc fragments in the cyst are derived from the parental disc and are retained as raw material for the construction of new discs during excystation.
This is the first study, to our knowledge, to use immunofluorescence staining to examine the Giardia cytoskeleton in depth during excystation, although previous studies have looked at the localizaton of two proteins, PP2A and beta-giardin, in excyzoites (Palm et al., 2005; Lauwaet et al., 2007b). As reported previously (Coggins and Schaefer, 1984; Coggins and Schaefer, 1986; Buchel et al., 1987; Erlandsen et al., 1989), we observed that the excyzoite exits the cyst at one pole, starting with its flagella. The early excyzoite has a similar cytoskeletal organization to the cyst, with each pair of nuclei remaining associated with a bundle of internal axonemes; however, it rapidly re-molds its cytoskeleton to resemble a cytokinetic trophozoite, which is the stage where it paused after the mitotic division during encystation. Its nuclei become decondensed and the distance between nuclei in each pair increases, presumably bringing an end to any nuclear fusion that has occurred. At this stage, each nascent trophozoite is thought to have an 8C genetic content (based on a 16C total in the cyst) (Bernander et al., 2001); thus, after cytokinesis is completed, the cell is primed to complete another round of mitosis without an additional S phase, as the chromosomes were already duplicated in the S phase during encystation. Because the number of centrin spots increases from eight in the excyzoite to 16 in the heart-shaped cells, we conclude that basal body duplication is also occurring during this time. Taken together, our results suggest that the cytoskeletal changes that happen during encystation, including disc assembly, basal body duplication and flagellar development, are very similar on the molecular level to their counterparts in the trophozoite (Nohynková et al., 2006; Tůmová et al., 2007; Dawson and House, 2010); the primary difference is the timing of these events in relation to spindle formation.
Importantly, we have determined that during excystation, the daughter trophozoites receive one copy of each parental nucleus. To date, the genomes of three Giardia assemblages (genotypes) have been sequenced. Two of these, assemblages A and E, contain extremely low levels of allelic heterozygosity – less than 0.01% (Morrison et al., 2007; Jerlström-Hultqvist et al., 2010). These results are surprising because, in a presumed asexual organism with two independent nuclei, the nuclear genomes – and perhaps even homologous chromosomes – would be expected to diverge over time owing to genetic drift (Birky, 2010). Fusion and genetic exchange (diplomixis) between the nuclei in the cyst, if accompanied by recombination and gene conversion within nuclei in the trophozoite, could potentially explain these low levels of heterozygosity (Poxleitner et al., 2008; Andersson, 2011). However, the distribution of identical nuclei to daughter cells during excystation would also explain, independently of diplomixis, the low levels of heterozygosity observed in Giardia populations (Fig. 1). Thus, our observation that nuclei are distributed in non-identical pairs during excystation is significant and implies that there are other mechanisms in place, including but perhaps not limited to diplomixis, to reduce genomic heterozygosity in Giardia. One of these could be highly efficient DNA damage repair in the trophozoite, which rests in the G2 phase of the cell cycle (Bernander et al., 2001); indeed, it is tempting to speculate that this unusual characteristic (most eukaryotic cells rest in G1) arose to allow ample time for repair using the sister chromatid as a template. The expression of Dmc1b in the trophozoite suggests that this Dmc1 homolog could mediate homologous recombination at this stage, and its similarity to Dmc1 (Ramesh et al., 2005) could point to an ability to mediate repair using the homolog as a template. This ability would also be required to maintain the low levels of heterozygosity observed in laboratory strains that have been passaged for long periods of time (Morrison et al., 2007; Teodorovic et al., 2007), as well as for heterozygosity to be reduced within nuclei following diplomixis (Andersson, 2011).
Chromosomal genetic exchange and meiotic gene expression patterns are consistent with homologous recombination
We were surprised to find consistently high rates of chromosomal genetic exchange (~20%) between nuclei in the cyst. This is probably an underestimate of the amount of exchange occurring, as our assay can only detect events involving transfer and preservation of the marker. This finding is consistent with diplomixis being used as a mechanism to reduce nuclear heterozygosity: to be effective, genetic exchange would be expected to be relatively common and involve large portions of the genome. We currently cannot distinguish between homologous recombination and whole-chromosome exchange. One way to directly detect recombination would be to place two integrated markers far apart on the same chromosome and observe their separation; however, this would be technically difficult (if not impossible) in Giardia, both because it is tetraploid and because there would be no clear way to confirm that the markers are in the same chromosome. Nevertheless, we predict that diplomixis involves homologous recombination for two reasons: first, three of the meiotic gene homologs (Dmc1a, Spo11 and Hop1) are expressed specifically at this stage in the life cycle, and, second, it is difficult to envision a mechanism for whole-chromosome exchange that would not result in widespread aneuploidy. Therefore, we predict that the meiotic gene homologs are involved in mediating homologous recombination between the nuclei during diplomixis. If this turns out to be the case, it would be one of the first examples of these proteins functioning in a non-meiotic context. Spo11 has been found to mediate homologous recombination during the parasexual cycle of the fungus Candida albicans (Forche et al., 2008). However, the lack of a sexual cycle in this organism is clearly a case of loss, whereas it is unclear whether Giardia or its relatives ever had a true meiotic cycle. Future studies of the functions of the meiotic gene homologs during diplomixis in Giardia could shed light on the evolution of the meiotic homologous recombination machinery.
On the basis of the observations from the present study, we envision a model whereby tethered chromosomes are able to interact and recombine with chromosomes from a neighboring nucleus (Fig. 6A–D) at some point either in the cyst or early excyzoite. In this way, widespread crossing over (facilitated by the meiotic gene homologs) between chromosomes from different nuclei could produce the observed patterns, and subsequent recombination and gene conversion could then act to homogenize the homologous chromosomes within each nucleus. Consistent with this hypothesis, we occasionally observed two paired spots rather than one in some cells (see fourth panel in Fig. 4B). These spots could represent replicated chromosomes that became separated; however, these patterns, as well as the increase in unlabeled nuclei following excystation (Fig. 4A), could also result from recombination and gene conversion between homologous chromosomes in the same nucleus.
Fig. 6.
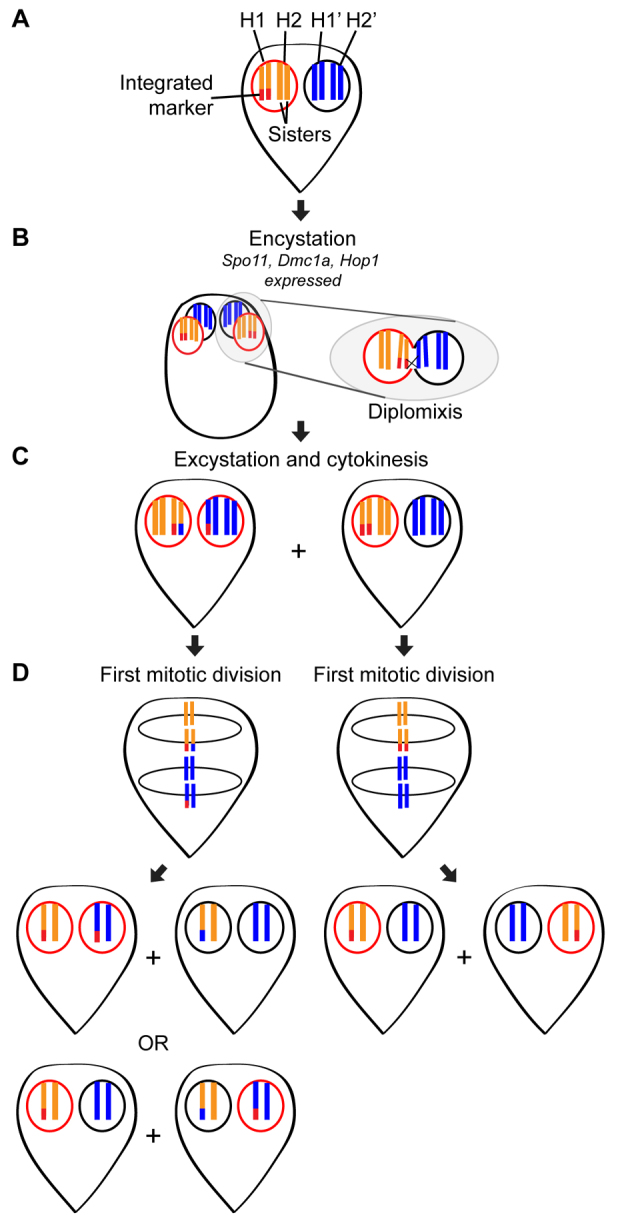
Hypothetical model of homologous recombination during diplomixis. (A) Shown is a simplified cell containing an integrated marker (red) in one of its four homologous chromosomes (H1 and H2, shown in orange, in the left nucleus, and H1′ and H2′, shown in blue, in the right nucleus). Throughout the figure, the presence of the integrated marker is indicated by a red outline around the nucleus in which it is found. Because trophozoites typically rest in G2 of the cell cycle, this trophozoite is shown with its chromosomes already duplicated (8C genome content; 4C in each nucleus). (B) During encystation, the meiotic gene homologs Spo11, Dmc1a and Hop1 are induced. Both nuclei divide and then go through an S phase, producing a 16C cyst. At some point, the nuclei fuse and exchange genetic material, presumably through homologous recombination. The chromosomes remain tethered to the nuclear envelope by their telomeres, which might prevent the development of aneuploidy following diplomixis. (C) When the cell excysts and completes cytokinesis, two trophozoites are formed, one from each nuclear pair found in the cyst. (D) If diplomixis has occurred (left trophozoite in C), the first mitotic division produces two possible combinations of daughter trophozoites, depending on the orientation of the chromosomes on the spindle. One of these combinations would produce one cell with markers in both nuclei and one cell with no marker, whereas the other would produce the parental arrangement (marker in one nucleus). If diplomixis has not occurred (right trophozoite in C), only the parental arrangement (marker in one nucleus) is produced.
In conclusion, we have demonstrated that, at least in vitro, Giardia cysts are derived from a single cell through an incomplete mitotic division and that daughter trophozoites inherit one copy of each parental nucleus during excystation. Furthermore, as hypothesized on the basis of the results of our previous study (Poxleitner et al., 2008), we have directly demonstrated that chromosomal genetic exchange occurs during diplomixis. This exchange happens at the same time that several homologs of meiosis-specific genes are induced, suggesting that homologous recombination in occurring, presumably as a way to reduce allelic heterozygosity between nuclei. However, although diplomixis could explain the maintenance of low heterozygosity between nuclei, it is also possible that rare meiotic events occur in the wild, as suggested by several recent population genetics studies (Cooper et al., 2007; Lasek-Nesselquist et al., 2009). These studies found evidence for recombination within and between assemblages, which cannot be explained by diplomixis within a single cell, but could potentially be explained by a combination of rare cell fusion followed by diplomixis. Furthermore, in contrast to the low heterozygosity in assemblages A and E, the genome of assemblage B strain GS has 0.5% allelic heterozygosity (Franzén et al., 2009), which has been suggested to be evidence for recent genetic exchange in the lineage (Andersson, 2011).
Future studies should aim to test for fusion and diplomixis or a true meiotic event in vivo, similar to what has been performed to detect sexual stages in parasites like Trypanosomes and Leishmania (Akopyants et al., 2009; Peacock et al., 2011). Identifying this process might be difficult, particularly if it is rare or occurs only under specific conditions. Nevertheless, a more complete understanding of the Giardia life cycle will not only give us insight into the basic biology of this important parasite but might also shed light on the spread of drug resistance in Giardia populations.
Please note that during the final preparation of this article, a study describing the mitotic division during encystation and subsequent nuclear interconnection in the Giardia cyst was published (Jirakova et al., 2012).
Materials and Methods
Culture conditions
Giardia intestinalis trophozoites, WB clone C6 (ATCC 50803), were grown in modified TYI-S-33 medium (Keister, 1983) with bovine bile (Sigma-Aldrich). Cultures were incubated in 16-ml plastic screw top tubes (Fisher Scientific) at 37°C and passaged every three days.
In vitro encystation
Encystation was performed essentially as described previously (Kane et al., 1991). Briefly, trophozoites were seeded into 30- or 50-ml tissue culture flasks (BD Biosciences) and grown to 80–100% confluency. The medium was then decanted and replaced with prewarmed encystation medium (10 mg/ml bovine-ovine bile, pH 7.8). After 24 hours at 37°C, cells were either fixed for observation or incubated on ice for 15 minutes, pelleted at 500 g, and resuspended in regular growth medium (0.5 mg/ml bile, pH 7.1). After an additional 24 hours of incubation at 37°C, the cells were again chilled on ice, pelleted, and resuspended in 50 ml ice-cold, sterile deionized water to destroy all unencysted cells. Cysts were stored at 4°C for 1–3 days, then excysted as described below.
In vitro excystation
Excystation was performed essentially as described previously (Boucher and Gillin, 1990). Briefly, water-treated cysts were pelleted in a 50-ml conical tube, then resuspended in 10 ml prewarmed stage I solution (1× Hank's balanced salt solution (Invitrogen), 57 mM L-cysteine HCl, 32.5 mM reduced glutathione and 25 mM NaHCO3; the pH was adjusted to 4.0 with 0.01 M HCl). The tube was vortexed briefly, then incubated at 37°C for 30 minutes. After another brief vortexing, cells were pelleted at 500 g for 5 minutes. The supernatant was then decanted and replaced with 10 ml prewarmed stage II solution [Tyrode's buffer (Sigma-Aldrich) containing 1 mg/ml Trypsin II-S (Sigma); the pH was adjusted to 8.0 with 1 M NaHCO3]. The tube was vortexed briefly, incubated at 37°C for 30 minutes, then vortexed again and pelleted. The supernatant was decanted and replaced with prewarmed growth medium (see the Culture conditions section), and cells were moved into 16-ml culture tubes and incubated at 37°C. Cells were then either fixed 1–3 hours later for immunofluorescence staining or grown to confluency (~5–7 days).
Creation of constructs for endogenous tagging
For creating integration constructs for endogenous tagging, the appropriate ORF was amplified from WBC6 genomic DNA and inserted into either the vector pKS-3HA-PAC or pKS-3myc-NEO (Gourguechon and Cande, 2011). The amplified ORFs, primers, and restriction sites used for digestion and linearization are shown in supplementary material Table S2. Transformation and selection of Giardia trophozoites was performed as previously described (Gourguechon and Cande, 2011).
Creation of clonal strains containing integrated LacO and TetO constructs
For the LacO array integration construct (supplementary material Fig. S2), a SacI-PvuII fragment containing part of the Spo11 coding sequence [GiardiaDB accession GL50803_15279; located on chromosome 4 (Upcroft et al., 2010)] and a puromycin resistance cassette (from the vector pKS-Spo11-3HA-pac) was inserted into the LacO-array-containing plasmid SR7 (Rohner et al., 2008), a gift from Susan M. Gasser (University of Basel, Basel, Switzerland); cut with restriction enzymes SacI and EcoRV. For integration, the plasmid was linearized with MfeI, which cuts within the sequence for Spo11. After the selection of stable transformants, single-cell clones were obtained by limiting dilution as described previously (Baum et al., 1988). These clones were screened for the presence of the integrated plasmid and absence of episomal plasmid by PCR as shown in supplementary material Fig. S2, using the primers P1 (5′-CATGATTTTCTTGTTTTAGGG-3′) and P2 (5′-TAATACGACTCACTATAGGG-3′) or P1 and P3 (5′-AGACTCTACAGGATACTTTCAG-3′). PCRs were performed by boiling a concentrated culture (200 μl) of approximately 1.3×107 cells for 10 minutes at 95°C. One microliter (~65,000 cells) was then used for PCR amplification.
For the TetO array construct, an EcoRI-XbaI fragment containing the median body protein [MBP; GiardiaDB accession GL50803_16343, located on chromosome 3 (Upcroft et al., 2010)] coding sequence and a neomycin resistance cassette (from the vector pKS-MBP-neo) was inserted into the TetO-array-containing plasmid SR8 (Rohner et al., 2008), a gift from Susan M. Gasser; cut with the restriction enzymes EcoRI and NheI. For integration, the plasmid was linearized with MfeI, which cuts within the sequence for MBP. After the selection of stable transformants, clonal lines were created and confirmed as described above, except using primer P4 (5′-TTCCGAATTTTGAGACACCTC-3′) instead of primer P3 listed above.
Fluorescence in situ hybridization
FISH was performed essentially as described previously (Poxleitner et al., 2008). However, instead of using nick translation to create the probe, the following probes were used at a concentration of 5 ng/μl: a Cy3-labeled LacO oligonucleotide probe (5′-CCACAAATTGTTATCCGCTCACAATTCCACATGTGG-3′); a FITC-labeled TetO probe (5′-TCGAGTTTACCACTCCCTATCAGTGATAGAGAAAAGTGAAAG-3′); and a FITC-labeled telomere probe (5′-TAGGGTAGGGTAGGGTAGGGTAGGG-3′).
Immunostaining
Fixation and immunostaining were performed essentially as described previously (Carpenter and Cande, 2009). Briefly, cells were fixed in 1×PEM buffer (100 mM PIPES, 1 mM EGTA, 0.1 mM MgSO4) containing 2% paraformaldehyde. After fixation for 30 minutes at 37°C, cells were washed once with PEM buffer, then resuspended in PEM buffer and attached to poly-L-lysine-coated coverslips for 30 minutes. Alternatively, for antibody staining against delta-giardin, cells were fixed for 3 minutes in ice-cold methanol, then washed twice in PBS before being attached to poly-L-lysine coverslips. Fixed cells were permeabilized with 0.1% Triton X-100 for 15 minutes, washed three times with PEM, and blocked in PEMBALG [PEM plus 1% BSA, 0.1% NaN3, 100 mM lysine and 0.5% gelatin from cold water fish skin (Sigma)] for 2 hours. Primary antibody incubations were conducted at room temperature overnight, and antibodies were diluted in PEMBALG as follows: mouse anti-trypanosome alpha-tubulin antibody TAT1, 1:50 (Woods et al., 1989), a gift from Keith Gull (University of Oxford, Oxford, England); 20H5 mouse anti-centrin antibody, 1:100 (Salisbury et al., 1988); rabbit anti-sea urchin tubulin antibody 1:25 (Fujiwara and Pollard, 1978); rat anti-HA antibody 1:100 (Roche); mouse anti-HA antibody 1:100 (Sigma); rabbit anti-Myc antibody 1:100 (Cell Signaling Technology); rabbit anti-delta-giardin antibody 1:100 (Jenkins et al., 2009), a gift from Mark C. Jenkins (US Department of Agriculture, Beltsville, MD, USA). After washing with PEMBALG, cells were incubated with secondary antibodies (goat anti-mouse antibody conjugated to Alexa Fluor 555 and goat anti-rabbit antibody conjugated to Alexa Fluor 488 [Invitrogen]) at room temperature for 2 hours. For labeling with the anti-CWP antibody, cells were then washed three times with PEMBALG, fixed for 10 minutes in 1% paraformaldehyde, washed three times with PEM and blocked for an additional 30 minutes in PEMBALG. The cells were then incubated with a Cy5-labeled mouse anti-CWP antibody (Waterborne, Inc., New Orleans, LA) for 2 hours. After a final three washes each in PEM and PEMBALG, coverslips were mounted onto slides with Prolong Gold Antifade Solution with DAPI (Invitrogen).
Fluorescence deconvolution microscopy
Images were collected with SoftWorX image acquisition software (Applied Precision, Issaquah, WA) on an Olympus IX70 wide-field inverted fluorescence microscope with an Olympus UPlanApo 100×, NA 1.35, oil-immersion objective and Photometrics CCD CH350 camera cooled to −35°C (Roper Scientific, Tuscon, AZ). Serial sections were acquired at 0.2 μm intervals, and data stacks were deconvolved using the SoftWorX deconvolution software. For presentation purposes, two-dimensional maximum intensity projections were created from the three-dimensional data sets using the DeltaVision image analysis software (Applied Precision).
Supplementary Material
Acknowledgements
We thank current and former members of the Cande laboratory, especially L. K. Fritz-Laylin and A. R. Paredez, for helpful discussion. We also thank K. Gull, M. Jenkins and S. Gasser for reagents.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant number A1054693 to W.Z.C.]; and the National Science Foundation (predoctoral fellowship to M.L.C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.103879/-/DC1
References
- Adam R. D. (2001). Biology of Giardia lamblia. Clin. Microbiol. Rev. 14, 447-475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants N. S., Kimblin N., Secundino N., Patrick R., Peters N., Lawyer P., Dobson D. E., Beverley S. M., Sacks D. L. (2009). Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324, 265-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J. O. (2011). Double peaks reveal rare diplomonad sex. Trends Parasitol. 28, 46-52 [DOI] [PubMed] [Google Scholar]
- Arisue N., Hasegawa M., Hashimoto T. (2005). Root of the Eukaryota tree as inferred from combined maximum likelihood analyses of multiple molecular sequence data. Mol. Biol. Evol. 22, 409-420 [DOI] [PubMed] [Google Scholar]
- Baldauf S. L. (2003). The deep roots of eukaryotes. Science 300, 1703-1706 [DOI] [PubMed] [Google Scholar]
- Baldauf S. L., Roger A. J., Wenk-Siefert I., Doolittle W. F. (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290, 972-977 [DOI] [PubMed] [Google Scholar]
- Baum K. F., Berens R. L., Jones R. H., Marr J. J. (1988). A new method for cloning Giardia lamblia, with a discussion of the statistical considerations of limiting dilution. J. Parasitol. 74, 267-269 [PubMed] [Google Scholar]
- Bernander R., Palm J. E., Svärd S. G. (2001). Genome ploidy in different stages of the Giardia lamblia life cycle. Cell. Microbiol. 3, 55-62 [DOI] [PubMed] [Google Scholar]
- Best A. A., Morrison H. G., McArthur A. G., Sogin M. L., Olsen G. J. (2004). Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia. Genome Res. 14, 1537-1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. K., Meyer E. A. (1979). Giardia excystation can be induced in vitro in acidic solutions. Nature 277, 301-302 [DOI] [PubMed] [Google Scholar]
- Birky C. W., Jr (2010). Giardia sex? Yes, but how and how much? Trends Parasitol. 26, 70-74 [DOI] [PubMed] [Google Scholar]
- Bittencourt-Silvestre J., Lemgruber L., de Souza W. (2010). Encystation process of Giardia lamblia: morphological and regulatory aspects. Arch. Microbiol. 192, 259-265 [DOI] [PubMed] [Google Scholar]
- Boucher S. E., Gillin F. D. (1990). Excystation of in vitro-derived Giardia lamblia cysts. Infect. Immun. 58, 3516-3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchel L. A., Gorenflot A., Chochillon C., Savel J., Gobert J. G. (1987). In vitro excystation of Giardia from humans: a scanning electron microscopy study. J. Parasitol. 73, 487-493 [PubMed] [Google Scholar]
- Carpenter M. L., Cande W. Z. (2009). Using morpholinos for gene knockdown in Giardia intestinalis. Eukaryot. Cell 8, 916-919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Munguía B., Omaña-Molina M., González-Lázaro M., González-Robles A., Cedillo-Rivera R., Bonilla P., Martínez-Palomo A. (2007). Ultrastructure of cyst differentiation in parasitic protozoa. Parasitol. Res. 100, 1169-1175 [DOI] [PubMed] [Google Scholar]
- Ciccarelli F. D., Doerks T., von Mering C., Creevey C. J., Snel B., Bork P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283-1287 [DOI] [PubMed] [Google Scholar]
- Coggins J. R., Schaefer F. W., 3rd (1984). Giardia muris: scanning electron microscopy of in vitro excystation. Exp. Parasitol. 57, 62-67 [DOI] [PubMed] [Google Scholar]
- Coggins J. R., Schaefer F. W., 3rd (1986). Giardia muris: ultrastructural analysis of in vitro excystation. Exp. Parasitol. 61, 219-228 [DOI] [PubMed] [Google Scholar]
- Cooper M. A., Adam R. D., Worobey M., Sterling C. R. (2007). Population genetics provides evidence for recombination in Giardia. Curr. Biol. 17, 1984-1988 [DOI] [PubMed] [Google Scholar]
- Dawson S. C., House S. A. (2010). Life with eight flagella: flagellar assembly and division in Giardia. Curr. Opin. Microbiol. 13, 480-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf H. G., Dawson S. C., McCaffery J. M. (2003). The cytoskeleton of Giardia lamblia. Int. J. Parasitol. 33, 3-28 [DOI] [PubMed] [Google Scholar]
- Erlandsen S. L., Bemrick W. J., Pawley J. (1989). High-resolution electron microscopic evidence for the filamentous structure of the cyst wall in Giardia muris and Giardia duodenalis. J. Parasitol. 75, 787-797 [PubMed] [Google Scholar]
- Faso C., Hehl A. B. (2011). Membrane trafficking and organelle biogenesis in Giardia lamblia: use it or lose it. Int. J. Parasitol. 41, 471-480 [DOI] [PubMed] [Google Scholar]
- Forche A., Alby K., Schaefer D., Johnson A. D., Berman J., Bennett R. J. (2008). The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6, e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O., Jerlström-Hultqvist J., Castro E., Sherwood E., Ankarklev J., Reiner D. S., Palm D., Andersson J. O., Andersson B., Svärd S. G. (2009). Draft genome sequencing of giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 5, e1000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K., Pollard T. D. (1978). Simultaneous localization of myosin and tubulin in human tissue culture cells by double antibody staining. J. Cell Biol. 77, 182-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillin F. D., Reiner D. S., Boucher S. E. (1988). Small-intestinal factors promote encystation of Giardia lamblia in vitro. Infect. Immun. 56, 705-707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillin F. D., Boucher S. E., Rossi S. S., Reiner D. S. (1989). Giardia lamblia: the roles of bile, lactic acid, and pH in the completion of the life cycle in vitro. Exp. Parasitol. 69, 164-174 [DOI] [PubMed] [Google Scholar]
- Gourguechon S., Cande W. Z. (2011). Rapid tagging and integration of genes in Giardia intestinalis. Eukaryot. Cell 10, 142-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautus M. A., Kortbeek L. M., Vetter J. C., Laarman J. J. (1988). In vitro excystation and subsequent axenic growth of Giardia lamblia. Trans. R. Soc. Trop. Med. Hyg. 82, 858-861 [DOI] [PubMed] [Google Scholar]
- Hetsko M. L., McCaffery J. M., Svärd S. G., Meng T.-C., Que X., Gillin F. D. (1998). Cellular and transcriptional changes during excystation of Giardia lamblia in vitro. Exp. Parasitol. 88, 172-183 [DOI] [PubMed] [Google Scholar]
- Jenkins M. C., O'Brien C. N., Murphy C., Schwarz R., Miska K., Rosenthal B., Trout J. M. (2009). Antibodies to the ventral disc protein delta-giardin prevent in vitro binding of Giardia lamblia trophozoites. J. Parasitol. 95, 895-899 [DOI] [PubMed] [Google Scholar]
- Jerlström-Hultqvist J., Franzén O., Ankarklev J., Xu F., Nohýnková E., Andersson J. O., Svärd S. G., Andersson B. (2010). Genome analysis and comparative genomics of a Giardia intestinalis assemblage E isolate. BMC Genomics 11, 543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiráková K., Kulda J., Nohýnková E. (2012) How nuclei of Giardia pass through cell differentiation: semi-open mitosis followed by nuclear interconnection. Protist 163, 465-479 [DOI] [PubMed] [Google Scholar]
- Kabnick K. S., Peattie D. A. (1990). In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J. Cell Sci. 95, 353-360 [DOI] [PubMed] [Google Scholar]
- Kane A. V., Ward H. D., Keusch G. T., Pereira M. E. (1991). In vitro encystation of Giardia lamblia: large-scale production of in vitro cysts and strain and clone differences in encystation efficiency. J. Parasitol. 77, 974-981 [PubMed] [Google Scholar]
- Keister D. B. (1983). Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 77, 487-488 [DOI] [PubMed] [Google Scholar]
- Konrad C., Spycher C., Hehl A. B. (2010). Selective condensation drives partitioning and sequential secretion of cyst wall proteins in differentiating Giardia lamblia. PLoS Pathog. 6, e1000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfredi-Rangel A., Attias M., Reiner D. S., Gillin F. D., De Souza W. (2003). Fine structure of the biogenesis of Giardia lamblia encystation secretory vesicles. J. Struct. Biol. 143, 153-163 [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E., Welch D. M., Thompson R. C., Steuart R. F., Sogin M. L. (2009). Genetic exchange within and between assemblages of Giardia duodenalis. J. Eukaryot. Microbiol. 56, 504-518 [DOI] [PubMed] [Google Scholar]
- Lauwaet T., Davids B. J., Reiner D. S., Gillin F. D. (2007a). Encystation of Giardia lamblia: a model for other parasites. Curr. Opin. Microbiol. 10, 554-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwaet T., Davids B. J., Torres-Escobar A., Birkeland S. R., Cipriano M. J., Preheim S. P., Palm D., Svärd S. G., McArthur A. G., Gillin F. D. (2007b). Protein phosphatase 2A plays a crucial role in Giardia lamblia differentiation. Mol. Biochem. Parasitol. 152, 80-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchtel D. L., Lawrence W. P., DeWalle F. B. (1980). Electron microscopy of Giardia lamblia cysts. Appl. Environ. Microbiol. 40, 821-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luján H. D., Mowatt M. R., Byrd L. G., Nash T. E. (1996). Cholesterol starvation induces differentiation of the intestinal parasite Giardia lamblia. Proc. Natl. Acad. Sci. USA 93, 7628-7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luján H. D., Mowatt M. R., Nash T. E. (1997). Mechanisms of Giardia lamblia differentiation into cysts. Microbiol. Mol. Biol. Rev. 61, 294-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S. B., Pightling A. W., Stefaniak L. M., Schurko A. M., Logsdon J. M., Jr (2008). An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS ONE 3, e2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midlej V., Benchimol M. (2009). Giardia lamblia behavior during encystment: how morphological changes in shape occur. Parasitol. Int. 58, 72-80 [DOI] [PubMed] [Google Scholar]
- Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., et al. (2007). Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317, 1921-1926 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kolodner R. D. (2002). Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol. Cell. Biol. 22, 3281-3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohynková E., Tůmová P., Kulda J. (2006). Cell division of Giardia intestinalis: flagellar developmental cycle involves transformation and exchange of flagella between mastigonts of a diplomonad cell. Eukaryot. Cell 5, 753-761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm D., Weiland M., McArthur A. G., Winiecka-Krusnell J., Cipriano M. J., Birkeland S. R., Pacocha S. E., Davids B., Gillin F., Linder E., et al. (2005). Developmental changes in the adhesive disk during Giardia differentiation. Mol. Biochem. Parasitol. 141, 199-207 [DOI] [PubMed] [Google Scholar]
- Peacock L., Ferris V., Sharma R., Sunter J., Bailey M., Carrington M., Gibson W. (2011). Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proc. Natl. Acad. Sci. USA 108, 3671-3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poxleitner M. K., Carpenter M. L., Mancuso J. J., Wang C. J., Dawson S. C., Cande W. Z. (2008). Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science 319, 1530-1533 [DOI] [PubMed] [Google Scholar]
- Ramesh M. A., Malik S.-B., Logsdon J. M., Jr (2005). A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 15, 185-191 [DOI] [PubMed] [Google Scholar]
- Reiner D. S., Ankarklev J., Troell K., Palm D., Bernander R., Gillin F. D., Andersson J. O., Svärd S. G. (2008). Synchronisation of Giardia lamblia: identification of cell cycle stage-specific genes and a differentiation restriction point. Int. J. Parasitol. 38, 935-944 [DOI] [PubMed] [Google Scholar]
- Rice E. W., Schaefer F. W., 3rd (1981). Improved in vitro excystation procedure for Giardia lamblia cysts. J. Clin. Microbiol. 14, 709-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner S., Gasser S. M., Meister P. (2008). Modules for cloning-free chromatin tagging in Saccharomyces cerevisae. Yeast 25, 235-239 [DOI] [PubMed] [Google Scholar]
- Sagolla M. S., Dawson S. C., Mancuso J. J., Cande W. Z. (2006). Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J. Cell Sci. 119, 4889-4900 [DOI] [PubMed] [Google Scholar]
- Salisbury J. L., Baron A. T., Sanders M. A. (1988). The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 107, 635-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer F. W., 3rd, Rice E. W., Hoff J. C. (1984). Factors promoting in vitro excystation of Giardia muris cysts. Trans. R. Soc. Trop. Med. Hyg. 78, 795-800 [DOI] [PubMed] [Google Scholar]
- Sheffield H. G., Bjorvat B. (1977). Ultrastructure of the cyst of Giardia lamblia. Am. J. Trop. Med. Hyg. 26, 23-30 [DOI] [PubMed] [Google Scholar]
- Stefanic S., Morf L., Kulangara C., Regös A., Sonda S., Schraner E., Spycher C., Wild P., Hehl A. B. (2009). Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. J. Cell Sci. 122, 2846-2856 [DOI] [PubMed] [Google Scholar]
- Teodorovic S., Braverman J. M., Elmendorf H. G. (2007). Unusually low levels of genetic variation among Giardia lamblia isolates. Eukaryot. Cell 6, 1421-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tůmová P., Hofštetrová K., Nohýnková E., Hovorka O., Král J. (2006). Cytogenetic evidence for diversity of two nuclei within a single diplomonad cell of Giardia. Chromosoma 116, 65-78 [DOI] [PubMed] [Google Scholar]
- Tůmová P., Kulda J., Nohýnková E. (2007). Cell division of Giardia intestinalis: assembly and disassembly of the adhesive disc, and the cytokinesis. Cell Motil. Cytoskeleton 64, 288-298 [DOI] [PubMed] [Google Scholar]
- Upcroft J. A., Krauer K. G., Upcroft P. (2010). Chromosome sequence maps of the Giardia lamblia assemblage A isolate WB. Trends Parasitol. 26, 484-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., Gull K. (1989). Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491-500 [DOI] [PubMed] [Google Scholar]
- Yu L. Z., Birky C. W., Jr, Adam R. D. (2002). The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot. Cell 1, 191-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.