Abstract
The grass family includes some 10,000 species, and it encompasses tremendous morphological, physiological, ecological, and genetic diversity. The phylogeny of the family is becoming increasingly well understood. There were two major radiations of grasses, an early diversification leading to the subfamilies Pooideae, Bambusoideae, and Oryzoideae, and a later one leading to Panicoideae, Chloridoideae, Centothecoideae, and Arundinoideae. The phylogeny can be used to determine the direction of changes in genome arrangement and genome size.
The grass family includes all the major cereals, such as wheat, maize, rice, barley, and oats, and most of the minor grains as well, such as rye, common millet, finger millet, teff, and many others that are less familiar. It also includes such economically important species as sugar cane and sorghum. Understanding the grass family is thus central to understanding the crops that feed the world.
The family encompasses far more physiological, morphological, and genetic diversity than just the major cereal crops. It is the fourth largest family of flowering plants, and is divided into 650 (1) to 765 (2, 3) genera. It includes 8,000–10,000 species, which is about double the number of species of mammals (4), and roughly the same number as birds (5). Members of the family occur on all continents, including Antarctica, which means that there is a grass species adapted to virtually every terrestrial habitat on earth.
The genomic similarities now being discovered among all the cereal crops imply that the entire family can be viewed as a single genetic system (6, 7). This means that the diversity in the family can potentially be harnessed for agronomic uses.
Physiological Diversity
The family includes many species adapted to dry and/or saline habitats. Examples include Thinopyrum (= Agropyron) elongatum, native to the Mediterranean region and already used as a source of germ plasm in wheat breeding; Chasmanthium latifolium, native to the Southeastern U.S.; Austrofestuca littoralis, native to Australia; and Dregeochloa pumila, native to southwestern Africa. The species are not related to each other, indicating that their tolerance of hot, sandy, and saline conditions has developed independently. The genetic basis of their drought and salt tolerance is unknown. However, a study that mapped quantitative trait loci (QTL) for salt or drought tolerance for any one of the species could potentially identify whether the genes involved were the same, whether there were novel alleles in the wild species, or whether hitherto unknown genes were controlling the trait.
Grasses also vary for photosynthetic pathway, with many of them—e.g., maize, sorghum, and sugar cane—exhibiting C4 photosynthesis. C4 photosynthesis is an addition to the standard C3 pathway (8–10) and increases the efficiency of CO2 fixation. C4 plants use phosphoenolpyruvate carboxylase (PEPC) in the mesophyll to add atmospheric CO2 to the 3-carbon phosphoenolpyruvate (PEP), creating a 4-carbon compound (oxaloacetate). The 4-carbon compound is then transported to the cells surrounding the vascular bundle, where the newly fixed carbon is removed and attached instead to ribulose 1,5-bisphosphate (RuBP) by RuBP carboxylase/oxygenase (RuBisCO). The latter reaction is the first of the conventional C3 pathway. C4 plants thus sequester their entire C3 pathway in bundle sheath cells and create a novel carbon fixation pathway in the mesophyll. Although the photosynthetic genes involved in the C4 pathway are familiar, the signal that causes them to be deployed in the C4 manner is unknown.
In addition to physiological diversity, there is also morphological diversity, full description of which can fill a book. Many species are dwarfs—e.g., Phleum alpinum, the alpine timothy, closely related to, but about half the size of, the familiar Phleum pratense. Others are enormous, such as the many genera of woody bamboos, which are a familiar part of the tropical landscape in Asia (e.g., Dendrocalamus) and South America (e.g., Chusquea). Still others, such as Cladoraphis, are highly branched and rigid, and can best be described as shrubby. Study of any of these would certainly illuminate mechanisms of cell wall formation, lignification, and distribution of sclerenchyma and silica in the plant, all characters that are involved in crop architecture, and possibly resistance to pests.
Grass Classification and Phylogeny
With a group this large, it is no surprise that there have been many attempts to produce a useful classification. Early in the 19th century, Robert Brown (11, 12) delineated a group similar to what we now call the Panicoideae and then combined everything else into a large and heterogeneous Pooideae. This would be only of historical interest, but for the fact that A. S. Hitchcock in his Manual of the Grasses of the United States (13), followed Brown’s classification, and Chase, in her second edition of the manual, retained it (14). Thus anyone who has studied grass taxonomy in the United States has learned, and still works with, Brown’s 1814 classification.
Work in the 1930s and 1940s revealed how much variability was encompassed in Brown’s Pooideae, which led to a revised classification by Stebbins and Crampton (15). They maintained Brown’s subfamily Panicoideae, and they identified several more groups of similar genera, which they called subfamilies Oryzoideae, Bambusoideae, Pooideae, and Chloridoideae. All the leftovers were classified under the name “Arundinoideae.” Most recent taxonomic studies, notably those of Clayton and Renvoize (1) and Watson and Dallwitz (3), are variations on the Stebbins and Crampton (15) classification. In general, the groups they name are the same, but the names given may differ slightly. With respect to crop plants, the major inconsistencies are (i) whether Oryzoideae is a separate subfamily or is a tribe (which then must be called Oryzeae) within Bambusoideae, and (ii) whether Maydeae is a separate tribe within the Panicoideae or whether its members (Zea, Tripsacum, and Coix) can be placed conveniently within the Andropogoneae.
This is not the place to discuss theory of classification. In general, though, the difficulty with a classification is that it has no inherent directionality. The Bambusoideae clearly differ from the Pooideae, but it is impossible to say whether the characteristics of the bamboos are ancestral or derived relative to the Pooideae, or indeed whether one can tell. It is analogous to a mutagenized batch of seed, but with no indication of which were wild-type and which were mutant. This lack of directionality obviously affects the interpretation of whether the mutations represented gain-of-function or loss-of-function, and it affects the nature of the mechanisms proposed.
A phylogeny can provide the directionality lacking in a classification. The grasses are part of a larger group of monocots, the Poales. Anatomical and morphological features (16, 17), as well as chloroplast genome arrangements (18), sequences of the large subunit of ribulose-bisphosphate carboxylase/oxygenase [rbcL (19)] and of phytochrome [phy (20)] all indicate that the closest relative of the grasses is the genus Joinvillea, the only member of the family Joinvilleaceae. Joinvillea is a large plant of forest margins and disturbed sites in Southeast Asia and western parts of the South Pacific. It serves as a point of comparison (outgroup) for phylogenetic studies in the grasses.
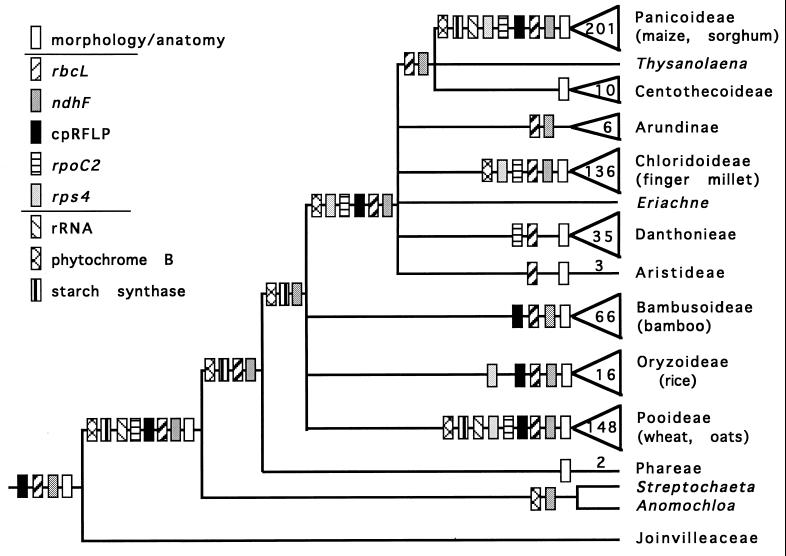
An early attempt to work out the phylogeny of the grasses was that of Kellogg and Campbell (21), using morphological and anatomical characteristics. Since then, phylogenies have been produced by using chloroplast restriction site variation (22) and sequences of four different chloroplast genes—rbcL (19, 23, 24), NADH dehydrogenase [ndhF; (25)], the β" subunit of RNA polymerase II [rpoC2 (26, 27)], and ribosomal protein 4 [rps4 (28)]. In addition there are data on three nuclear loci, the ribosomal RNA [rRNA (29)], granule-bound starch synthase I (GBSSI or waxy; R. J. Mason-Gamer and E.A.K., unpublished data), and phytochrome (ref. 20; S. Y. Mathews, R. C. Tsai, and E.A.K., unpublished data). Although these studies differ widely in the species they include, they have many results in common (Fig. 1). They all indicate that the family is a single lineage (is monophyletic), except for rps4, which did not include any outgroups and thus assumed rather than tested the unity of the family. The earliest diverging branch in the family leads to two neotropical genera, Streptochaeta and Anomochloa, and the next to the tribe Phareae, which includes only the two genera Pharus and Leptaspis. The position of these genera was indicated by the ndhF sequences of Clark et al. (25) and has since been confirmed by rbcL (19), phytochrome B (S. Y. Mathews, R. C. Tsai, and E.A.K., unpublished data) and GBSSI (R. J. Mason-Gamer and E.A.K., unpublished data). The overwhelming majority of the family diverged well after the appearance of Streptochaeta, Anomochloa, and the Phareae. An early radiation led to three distinct lineages—the Pooideae, the Bambusoideae, and the Oryzoideae. (Note that the latter two are clearly separate lines, so should be treated as separate subfamilies.) Somewhat later, there was a second major radiation, which led to the Panicoideae, Chloridoideae, the tiny subfamily Centothecoideae, and the multiple lineages making up the miscellaneous nongroup, the Arundinoideae. This second radiation [sometimes given the acronym the PACC clade (20)] contains all the C4 lineages, as well as several groups of C3 plants.
Figure 1.
Summary (semistrict consensus) of phylogenetic data on the grass family. An appropriately shaded rectangle marks any clade supported by a particular set of data and not strongly contradicted by any other set of data. Joinvilleaceae is the sister group of the grasses; all other taxa are grasses. Triangles indicate a large clade. Numbers refer to number of genera. Sources of data and references are listed in text. Some evidence indicates that Bambusoideae, Oryzoideae, and Pooideae actually form a single clade (20, 25), but this grouping is not yet well supported, so the relationship is shown as ambiguous.
The early fossil record of the grasses is scanty. The oldest reliable fossils are pollen grains, Monoporites annulatus, dated as Maastrictian, the top of the Cretaceous, ≈65–70 million years ago (Mya) (30). Grass flowers have been recovered from the Paleocene/Eocene boundary (56 Mya) (31).
Genome Rearrangements
Maps of the nuclear genomes of wheat, barley, rye, oats, maize, sugar cane, sorghum, rice, pearl millet, and foxtail millet are all available [see paper by M. Gale, this colloquium (32)], with others such as ryegrass and finger millet well underway. These data, combined with the phylogeny, allow some generalizations about genome structure, and also inferences about the direction of some changes. For example, the linkage group represented by rice chromosome 9 is inserted between rice 7a and 7b in all Panicoideae studied so far, and rice 10 into rice 3. Because of the similarity among all members of the Panicoideae, I infer that these arrangements will be present throughout the subfamily. Similarly, rice 10 should be inserted into rice 5 and rice 8 into rice 6 in all Pooideae, as in oats and the Triticeae.
Phylogenetic studies commonly assume that ancestral species look much like their descendants. Thus the ancestral panicoid probably had linkage groups corresponding to 7a-9-7b and 3c-10-3b-3a, linkages shared by all known descendants. Such inferences are obvious and noncontroversial when the descendants all share the same trait.
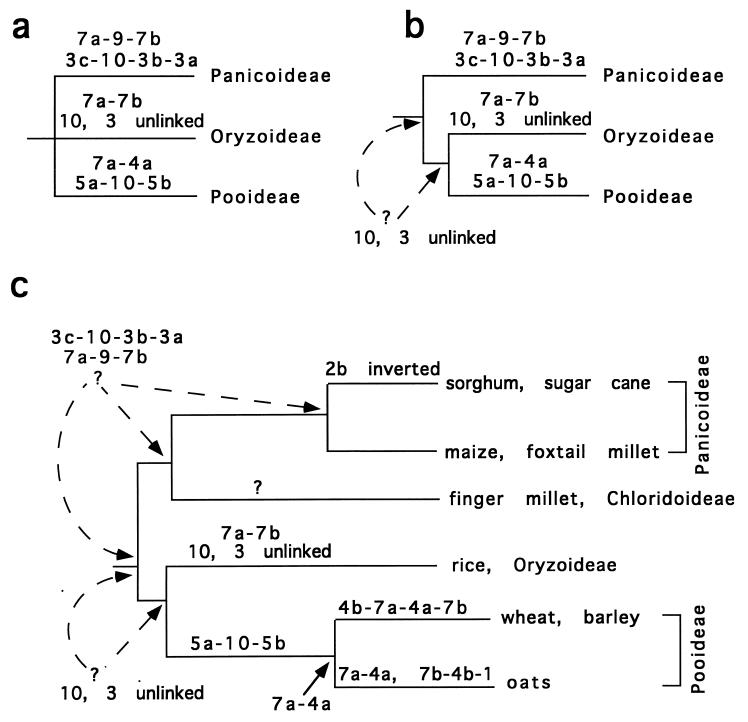
When descendants have different traits (e.g., different linkages of 7a and 7b), the ability to extrapolate to an ancestor becomes weaker. On the basis of available genome data and the very conservative phylogeny shown in Fig. 1, it is not possible to infer that any particular genome arrangement is ancestral. This difficulty is more obvious if branches of the tree for which no genome data are available are removed and the tree is redrawn to show only the relationship among the Panicoideae, Pooideae, and Oryzoideae (Fig. 2a). The linkage of 7a-9-7b that occurs in the Panicoids does not occur in either of the other two subfamilies, and it could be either the primitive or the derived arrangement. Similarly, only in rice are 7a and 7b joined into a single chromosome; the 7a-7b linkage could be ancestral for Oryza, for Oryzoideae, or for the grasses as a whole. In Triticeae, 7a and 7b are separated in the linkage group 4b-7a-4a-7b, and in oats they are on separate chromosomes (7a-4a and 7b-4b-1, except a piece of 4b is with 7a-4a). Because oats and Triticeae are sister taxa, and because they share the 7a-4a linkage, I infer that the ancestral pooid also had this linkage; but with available data, no inference is possible about ancestral grass arrangement.
Figure 2.
Possible evolutionary patterns of genome rearrangements. (a) Tree congruent with that in Fig. 1, pruned to include only those branches with genome maps. Note that no inference about the ancestral arrangement is possible. (b) Same tree, but with Oryzoideae and Pooideae as sister taxa. Note that now it is possible to infer that linkage groups 10 and 3 were unlinked in the common ancestor of rice and the pooids, but it is still impossible to determine the ancestral state for all grasses. (c) More detailed tree. Broken lines indicate ambiguity in the time of origin of linkage groups 3c-10-3b-3a and 7a-9-7b. Note that some of this ambiguity will be resolved with a map for finger millet.
Data are accumulating, however, to suggest that Oryzoideae and Pooideae are actually sister taxa. If this is true, then the phylogeny is as shown in Fig. 2b, and direction can be determined for some of the changes. The linkage groups 6a-6b-8-6c-6d and 5a-10-5b now appear to be derived within the Pooideae (i.e., arising in the ancestral pooid, well after grass diversification), and the linkage 7a-7b is derived within the Oryzoideae. This derivation implies that there must be some mechanism for joining chromosomes, apparently by replacing the centromeric region of one with an entirely different chromosome plus centromere.
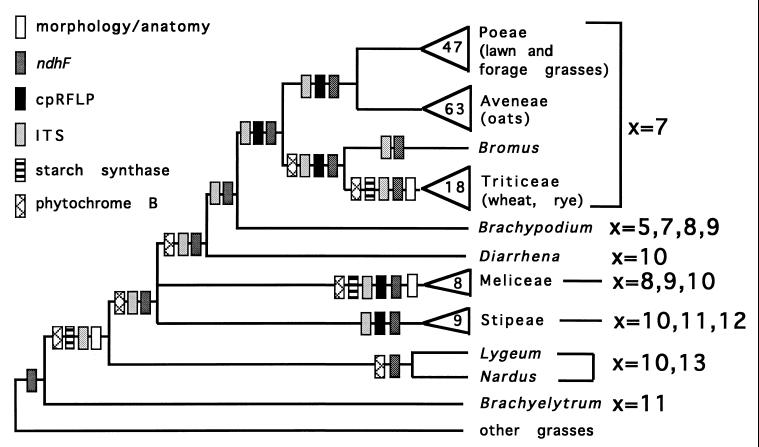
This mechanism is clearly related to changes in chromosome number. The best example of reduction in chromosome number is in the subfamily Pooideae, a phylogeny of which is shown in Fig. 3. This phylogeny is supported by data from morphology and anatomy (21, 33), chloroplast restriction site polymorphisms (22), the internal transcribed spacer of the rRNA (ITS) (34), ndhF (35), and GBSSI (R. J. Mason-Gamer and E.A.K., unpublished data). Chromosome number is variable in the early diverging lineages, as it is in other members of the grass family. The number is smaller, though variable, in the genus Brachypodium. The “core pooids”—Poeae, Aveneae, Triticeae, and Bromeae—all have x = 7. The formation of linkage groups 6a-6b-8-6c-6d and 5a-10-5b (Triticeae chromosomes 7 and 1, Avena chromosomes D and A, respectively) thus may have occurred in the common ancestor of the core pooids, at the same time as the group settled on 7 centromeres. A linkage map of Brachypodium would provide a test of this hypothesis.
Figure 3.
Summary (semistrict consensus) of phylogenetic data for the subfamily Pooideae. An appropriately shaded rectangle marks any clade supported by a particular set of data and not strongly contradicted by any other set of data. Triangles indicate a large clade. Numbers refer to number of genera. Sources of data and references are listed in the text. Chromosome base numbers are indicated on the right.
The linkage groups 7a-9-7b and 3c-10-3b-3a may be derived in the Panicoideae, or they may be the ancestral arrangement for the family as a whole (Fig. 2a). The critical piece of data will come from the map of finger millet (Eleusine), which is currently being constructed (M. Gale, personal communication).
As additional data become available, it will be possible to infer which arrangements are ancestral and which are derived. This inference is not merely an exercise in history reconstruction, but rather will determine the direction of change, which will in turn demand hypotheses of particular mechanisms and will help estimate the relative frequency of certain sorts of changes.
Genome Size
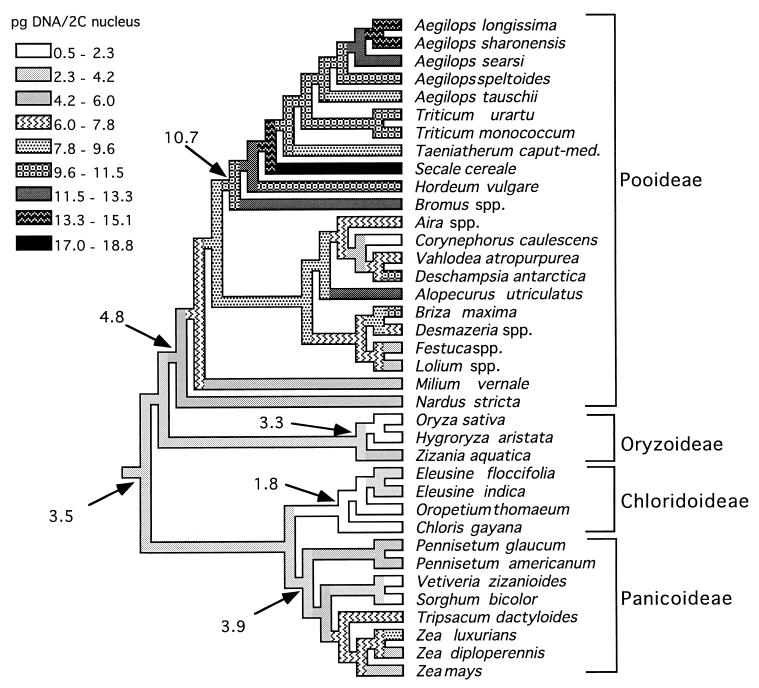
The phylogeny also allows investigation of evolution of genome size, using similar arguments about the similarity of ancestors and descendants. Genome size data are available in papers by Bennett and colleagues (36–39). If data are extracted for 2C values for diploid grasses, these data can be mapped onto the phylogeny (Fig. 4). The ancestral genome size can then be inferred, although making the inference requires certain assumptions. The most critical assumption is the minimum change assumption described above—ancestors looked similar to their descendants. Under this assumption, the ancestral grass genome is calculated to have had about 3.5 pg of DNA per 2C nucleus. There was a steady increase in genome size in the Pooideae, leading to the very large genomes in the Triticeae (the ancestor with 10.7 pg in Fig. 4). Genome size apparently decreased in Corynephorus, in Oryza and Hygroryza, in Oropetium and Chloris, and in Sorghum and Vetiveria. Any general mechanism for increase in genome size should apply to the Triticeae and other members of the Pooideae. Conversely, any general mechanism for decrease should apply in the several independent decreases. The phylogeny, in other words, can direct tests for the generality of mechanisms.
Figure 4.
One possible model for evolution of genome size in the grasses. Data on genome size were extracted from papers by Bennett and colleagues (36–39). The cladogram was based on references cited above; phylogeny in the Triticeae is from Kellogg et al. (40), that in the Poeae/Aveneae is from J. I. Davis (personal communication), that in the Chloridoideae is from Clayton and Renvoize (1), and that in the Panicoideae is from Kellogg and Watson (33) and Doebley (41). Ancestral genome sizes were reconstructed according to squared-change parsimony (42), implemented in MacClade 3.0 (43).
Note that in the case of genome size, inferences about ancestors are sensitive to which taxa are studied, and, more importantly, to the assumption of likelihood of change. Fig. 4 assumes only that ancestors should look as much as possible like their descendants [a minimum change model (42)]. If increase in genome size were much more likely than decrease, then inferences about ancestral genome size will change accordingly.
Use of the Phylogeny in Plant Genome Initiatives
Currently most mapping effort has been expended on the major crops, which conveniently fall into the panicoid, pooid, and oryzoid groups. If additional genomic work—e.g., constructing expressed sequence tag (EST) databases and physical maps—is targeted to maize (panicoid), rice (oryzoid), and barley (pooid), a framework will emerge for understanding the entire family. Furthermore, maps constructed for additional members of these groups will determine how far we can generalize within and among subfamilies. However, to make strong inferences about ancestral genome structure and size, and about mechanisms of genomic change, requires study of species that are of less immediate agronomic importance. For example, if finger millet has any of the characteristic panicoid linkage groups, then we can immediately infer that that linkage group was formed at least as early as the common ancestor of Chloridoideae and Panicoideae (the base of the PACC clade, Fig. 1). Study of North American wild rice (Zizania) would immediately show which aspects of the rice genome were general (and ancestral) in the Oryzoideae and which are peculiar to Oryza itself. Any bambusoid would be intriguing (e.g., Dendrocalamus), although the polyploidy of the woody bamboos will make map construction somewhat more difficult. Teff (Eragrostis tef) is another chloridoid that would be a useful point of comparison with finger millet, in addition to its possible value as a grain crop.
Determining the ancestral genome arrangement would require comparison with monocots outside the grasses. The obvious point of comparison would be the sister genus, Joinvillea, but more readily accessible material might be in the more distantly related sedge family (Cyperaceae).
More important, understanding the genome of the grasses potentially makes the entire family—all 10,000 species—accessible for agronomic uses. Any polymorphism anywhere in the family can potentially be mapped and that map data transferred directly to rice, maize, or wheat. This offers the potential for finding new genes, for determining the function of genes known only from sequence data, and for uncovering new alleles of genes whose function is already known. The classifications and phylogenies already available can serve as guideposts and organizing principles in our developing understanding of the family.
Acknowledgments
This work was supported in part by National Science Foundation Grant DEB-9419748.
ABBREVIATIONS
- GBSSI
granule-bound starch synthase I
- ITS
internal transcribed spacer of rRNA
References
- 1.Clayton W D, Renvoize S A. Genera Graminum. London: Her Majesty’s Stationery Office; 1986. [Google Scholar]
- 2.Watson L. In: The Grass Family, Poaceae. Chapman G P, editor. Cambridge, U.K.: Cambridge Univ. Press; 1990. pp. 1–31. [Google Scholar]
- 3.Watson L, Dallwitz M J. The Grass Genera of the World. Wallingford, Oxon, U.K.: CAB International; 1992. [Google Scholar]
- 4.Wilson D E, Reeder D M. Mammal Species of the World. Washington, DC: Smithsonian Institution Press; 1993. [Google Scholar]
- 5.Monroe B L, Sibley C G. A World Checklist of Birds. New Haven, CT: Yale Univ. Press; 1993. [Google Scholar]
- 6.Bennetzen J L, Freeling M. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 7.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 8.Hatch M D. In: Photosynthesis: The Path of Carbon. Bonner J, Varner J E, editors. New York: Academic; 1976. pp. 797–844. [Google Scholar]
- 9.Edwards G E, Walker D A. C3, C4: Mechanisms and Cellular and Environmental Regulation of Photosynthesis. Oxford: Blackwell Scientific; 1983. [Google Scholar]
- 10.Edwards G E, Huber S C. In: The C4 Pathway, Photosynthesis. Hatch M D, Boardman N K, editors. Vol. 8. New York: Academic; 1981. pp. 238–281. [Google Scholar]
- 11.Brown R. General Remarks, Geographical and Systematical, on the Botany of Terra Australis. London: G. and W. Nicol; 1814. [Google Scholar]
- 12.Brown R. Prodromus Florae Novae Hollandiae. London: J. Johnson & Co.; 1810. [Google Scholar]
- 13.Hitchcock A S. Manual of the Grasses of the United States. Washington, DC: U.S. Government Printing Office; 1935. [Google Scholar]
- 14.Hitchcock A S, Chase A. Manual of the Grasses of the United States. 2nd Ed. Washington, DC: U.S. Government Printing Office; 1950. [Google Scholar]
- 15.Stebbins G L, Crampton B. Advances in Botany (Lectures and Symposia, IX International Botanical Congress) 1961;1:133–145. [Google Scholar]
- 16.Kellogg E A, Linder H P. In: Monocotyledons: Systematics and Evolution. Rudall P J, Cribb P J, Cutler D F, Humphries C J, editors. Kew: Royal Botanic Gardens; 1995. pp. 511–542. [Google Scholar]
- 17.Campbell C S, Kellogg E A. In: Grass Systematics and Evolution. Soderstrom T R, Hilu K W, Campbell C S, Barkworth M E, editors. Washington, DC: Smithsonian Institution Press; 1987. pp. 217–224. [Google Scholar]
- 18.Doyle J J, Davis J I, Soreng R J, Garvin D, Anderson M J. Proc Nat Acad Sci USA. 1992;89:7722–7726. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duvall M R, Morton B R. Mol Phylogenet Evol. 1996;5:352–358. doi: 10.1006/mpev.1996.0030. [DOI] [PubMed] [Google Scholar]
- 20.Mathews S, Sharrock R A. Mol Biol Evol. 1996;13:1141–1150. doi: 10.1093/oxfordjournals.molbev.a025677. [DOI] [PubMed] [Google Scholar]
- 21.Kellogg E A, Campbell C S. In: Grass Systematics and Evolution. Soderstrom T R, Hilu K W, Campbell C S, Barkworth M E, editors. Washington, DC: Smithsonian Institution Press; 1987. pp. 310–322. [Google Scholar]
- 22.Davis J I, Soreng R J. Amer J Bot. 1993;80:1444–1454. [Google Scholar]
- 23.Doebley J, Durbin M, Golenberg E M, Clegg M T, Ma D P. Evolution. 1990;44:1097–1108. doi: 10.1111/j.1558-5646.1990.tb03828.x. [DOI] [PubMed] [Google Scholar]
- 24.Barker N P, Harley E, Linder H P. Syst Bot. 1995;20:423–435. [Google Scholar]
- 25.Clark L G, Zhang W, Wendel J F. Syst Bot. 1995;20:436–460. [Google Scholar]
- 26.Cummings M P, King L M, Kellogg E A. Mol Biol Evol. 1994;11:1–8. doi: 10.1093/oxfordjournals.molbev.a040084. [DOI] [PubMed] [Google Scholar]
- 27.Barker N P. Ph.D. thesis. Cape Town, South Africa: Univ. of Cape Town; 1995. [Google Scholar]
- 28.Nadot S, Bajon R, Lejeune B. Plant Syst Evol. 1994;191:27–38. [Google Scholar]
- 29.Hamby R K, Zimmer E A. Plant Syst Evol. 1988;160:29–37. [Google Scholar]
- 30.Linder H P. Kew Bull. 1987;42:297–318. [Google Scholar]
- 31.Crepet W L, Feldman G D. Amer J Bot. 1991;78:1010–1014. [Google Scholar]
- 32.Gale M D, Devos K M. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellogg E A, Watson L. Bot Rev. 1993;59:273–343. [Google Scholar]
- 34.Hsiao C, Chatterton N J, Asay K H, Jensen K B. Theor Appl Genet. 1995;90:389–398. doi: 10.1007/BF00221981. [DOI] [PubMed] [Google Scholar]
- 35.Catalan P, Kellogg E A, Olmstead R G. Mol Phylogenet Evol. 1997;8:150–166. doi: 10.1006/mpev.1997.0416. [DOI] [PubMed] [Google Scholar]
- 36.Bennett M D, Smith J B. Philos Trans R Soc London B. 1991;334:309–345. [Google Scholar]
- 37.Bennett M D, Smith J B, Heslop-Harrison J S. Proc R Soc London Ser B. 1982;216:179–199. [Google Scholar]
- 38.Bennett M D, Smith J B. Philos Trans R Soc London B. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 39.Bennett M D, Leitch I J. Ann Bot. 1995;76:113–176. [Google Scholar]
- 40.Kellogg E A, Appels R, Mason-Gamer R J. Syst Bot. 1996;21:321–347. [Google Scholar]
- 41.Doebley, J. (1990) Econ. Bot. 44, Suppl. 3, 6–27.
- 42.Maddison W P. Syst Zool. 1991;40:304–314. [Google Scholar]
- 43.Maddison W P, Maddison D R. MacClade, Analysis of Phylogeny and Character Evolution, Version 3. Sunderland, MA: Sinauer; 1992. [Google Scholar]