1. Introduction
Lipids play an important role in physiology and pathophysiology of living systems. Until a few decades ago, the number of lipid molecules that were chemically characterized was a few hundred at most and were catalogued in monographs and compendia.1 Since the advent of the era of the genome and the proteome, there has been increasing recognition that other macromolecules like lipids and polysaccharides in living systems display considerable structural diversity and systematic efforts are underway to identify, characterize and catalog these molecules. With mass spectrometric techniques coming of age, several thousand distinct molecular species have been identified from living species and the roles of several of these are beginning to be characterized.2 Unlike genes and proteins, whose defined alphabets provide the framework for ontologies and classification at the sequence level, lipids and polysaccharides have been characterized for the large part by popular names, with no foundations for systematic classification.
The past two decades have witnessed two major advances in lipid biology. In the first, mass spectrometry has enabled the identification of thousands of lipid molecular species from cells and tissues and this has pointed to the important need for developing a systematic ontology that can rationally name and catalog the molecules. Second, the ability to investigate the functional roles of lipid molecules through systematic phenotypic studies has led to the identification of lipids as extremely important players in physiology and pathophysiology of living species.3 In combination with proteins and nucleic acids, lipids are integrally involved in biochemical networks that lead to phenotypes such as homeostasis, differentiation, and death of cells and tissues. Any approach to systems characterization of living systems, of necessity, has to include lipids along with other macromolecules and all complex cellular pathways involving lipid molecular species. Systems biology now extends in its scope to identify biosynthetic and metabolic lipid networks, cellular signaling networks that explicitly include lipid molecules and transcriptional and epigenetic networks where lipids play an integral role.4
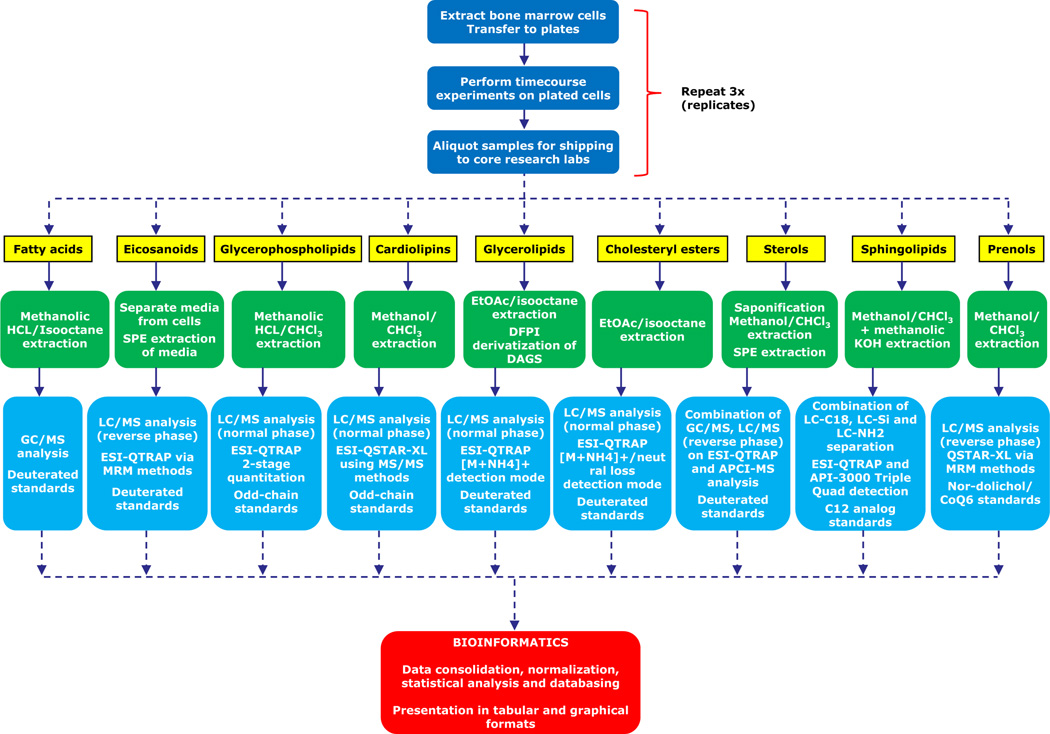
Several large scale projects to characterize lipids and their functional roles have been initiated as exemplified by the LIPID MAPS5 effort. The LIPID MAPS is an exemplar systems biology project that measures cell-wide lipid changes in an attempt to reconstruct biochemical pathways associated with lipid processing and signaling. The cell-wide measurements of components of these pathways include mass spectrometric measurements of lipid changes in response to stimulus in mammalian cells, changes in transcription profiles in response to stimulus and in select cases proteomic changes in response to stimulus. Figure 1 shows a schematic of the LIPID MAPS experiments related to different lipid categories/pathways and the subsequent processing of the experimental data generated. Network reconstruction efforts rely on organization, analysis and integration of these data and this requires a strong bioinformatics and systems biology effort. The former has to include development of a systematic and universal classification and nomenclature system, design and development of lipid and lipid-gene, lipid-protein databases with appropriate functional annotations, and efficient query and analysis systems that can be broadly useful to the biology research community. The latter has to include methods for analysis of large scale lipid measurements in cells, reconstruction of lipid metabolic and biosynthetic pathways, and quantitative models of lipid fluxes in cells under varied perturbations. In this review, we will provide a comprehensive summary of extant developments in lipid bioinformatics and systems biology and discuss the outlook for the future integration of lipidomics into cellular and organismic biology. The sections that follow are delineated into the informatics approaches specific to lipid biology followed by an overview and exemplar approach to analysis of large scale lipidomic data towards a systems description of mammalian cells.
Figure 1.
Overview of the process of performing a quantitative lipid analysis of macrophage cell sample (in this example, a time-course experiment using bone marrow derived macrophages). Extraction methods, LC/GC purification methods, MS acquisition strategies and quantitative standard approaches are optimized for each lipid class.2,70,72
2. Classification, Ontology, Nomenclature and Structure Representation of Lipid Molecules
The first step towards classification of lipids is the establishment of an ontology that is extensible, flexible and scalable. One must be able to classify, name and represent these molecules in a logical manner which is amenable to data basing and computational manipulation. Lipids have been loosely defined as biological substances that are generally hydrophobic in nature and in many cases soluble in organic solvents.6 These chemical features are present in a broad range of molecules such as fatty acids, phospholipids, sterols, sphingolipids, terpenes and others. In view of the fact that lipids comprise an extremely heterogeneous collection of molecules from a structural and functional standpoint, it is not surprising that there are significant differences with regard to the scope and organization of current classification schemes.
2.1. Classification, Ontology and Nomenclature
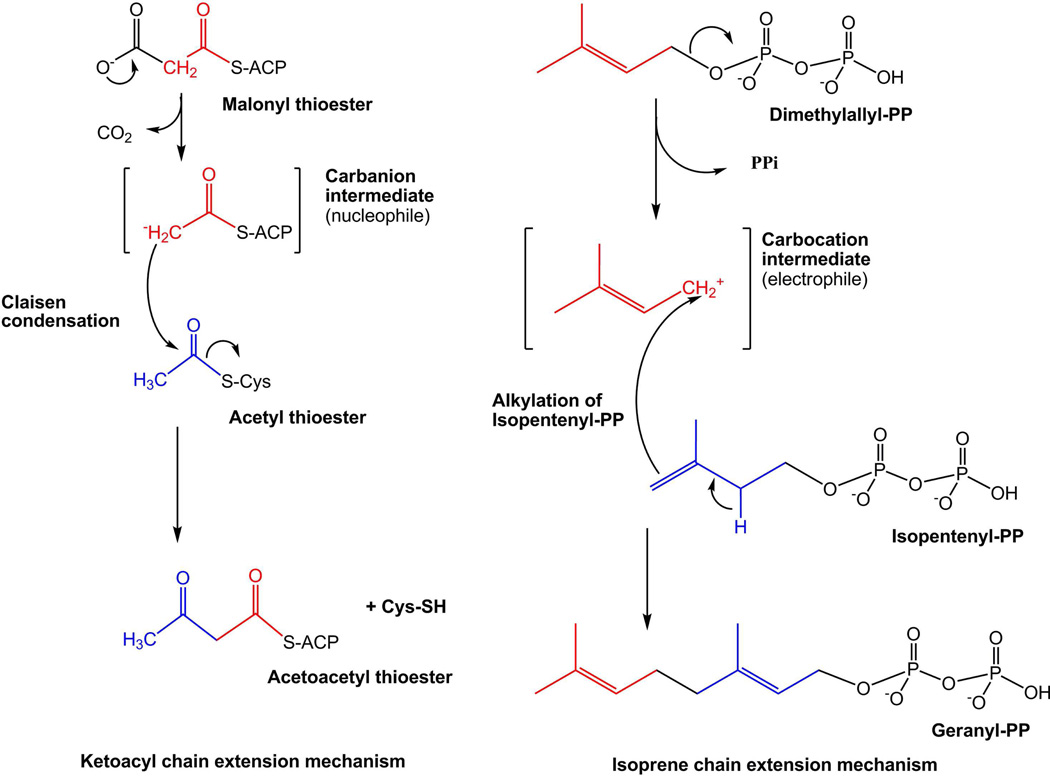
In order to address the lack of a consistent classification and nomenclature methodology for lipids, LIPID MAPS consortium members have developed a comprehensive classification system for lipids.7 The consortium has taken a more chemistry-based approach and defines lipids as hydrophobic or amphipathic small molecules that may originate entirely or in part by carbanion based condensations of thioesters (such as fatty acids and polyketides) and/or by carbocation based condensations of isoprene units (such as prenols and sterols). Figure 2 shows the mechanisms of lipid biosynthesis.8 Based on this classification system, lipids have been divided into eight categories: Fatty acyls, Glycerolipids, Glycerophospholipids, Sphingolipids, Sterol lipids, Prenol lipids, Saccharolipids, and Polyketides. Each category is further divided into classes and subclasses. Additionally, following the existing rules and recommendations proposed by the International Union of Biochemistry and Applied Chemists and the International Union of Biochemistry and Molecular Biology (IUPAC-IUBMB) commission on Biochemical Nomenclature, a consistent nomenclature scheme has also been developed to provide systematic names for various classes and subclasses of lipids.7
Figure 2.
Mechanisms of lipid biosynthesis. Biosynthesis of ketoacyl- and isoprene-containing lipids proceeds by carbanion and carbocation-mediated chain extension, respectively.8
All lipids in the LIPID MAPS Structure Database (LMSD) are classified and annotated using this comprehensive classification and nomenclature system developed by the LIPID MAPS consortium.
2.2. Structure Representation
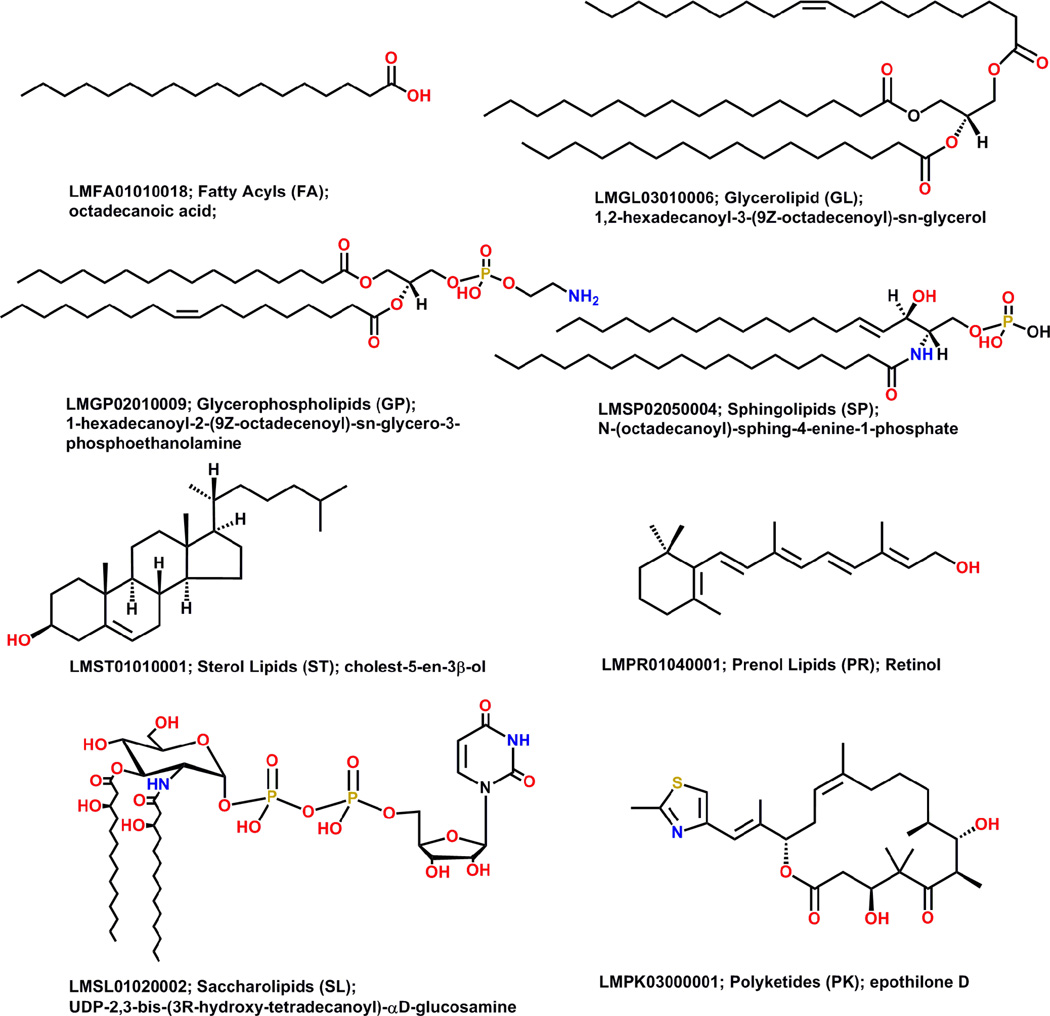
Currently, different members of the lipids community draw lipid structures in distinct ways. The same lipid structure in one lipid database can appear quite different in another database.9 Moreover, large and complex lipids are rather difficult to draw manually which leads to proliferation of shorthand and other abbreviations to represent lipid structures. In order to address these issues, the LIPID MAPS consortium proposed a consistent framework for representing lipid structures.7,10 In general, the acid/acyl group or its equivalent is drawn on the right side and hydrophobic chain is on the left. A number of structurally complex lipids – acylaminosugar glycans, polycyclic isoprenoids, and polyketides – cannot be drawn using these simple rules; these structures are drawn using commonly accepted representations. Structures of all lipids in LMSD adhere to the structure drawing rules proposed by the LIPID MAPS consortium. Figure 3 shows representative structures for each lipid category.
Figure 3.
Representative structures from each lipid category shown with LM ID, category name, category abbreviation, and systematic name (Reprinted with permission from Reference 10. Copyright 2007 Oxford University Press).
2.2.1. Structural Representation of Positional Isomers
LIPID MAPS core laboratories are engaged in identification, characterization and quantification of known and new lipids using liquid chromatography (LC) and mass spectrometry (MS) experimental techniques; Information about various lipid standards developed for these experiments, along with the protocols used, is available on the Lipidomics Gateway website.5 However, for some lipid categories such as glycerolipids and glycerophospholipids, it is not always straightforward to identify the positions of radyl (acyl, alkyl or alkenyl) hydrocarbon chains at the sn carbons on the glycerol group. For example, MS/MS experiments might be able to identify presence of three radyl hydrocarbons chains in a triacylglycerol but their positions on the glycerol backbone would be unknown. Combinatorial enumeration of the three radyl chains at sn carbons leads to six possible isomeric structures. These positional isomers are stored in LMSD as one structure and it is marked as a computationally generated structure. Structures for all other positional isomers are created on demand. To indicate the positional isomeric nature of the structure, a suffix “iso” followed by the number of isomers is also added to the abbreviation used as common name. For example, entry LMGL03010043 in LMSD, with common name TG(16:0/16:1(9Z)/18:1(9Z))[iso6] and systematic name 1-hexadecanoyl-2-(9Z-hexadecenoyl)-3-(9Z-octadecenoyl)-sn-glycerol, represents a lipid structure with six possible positional isomers.
2.2.2. Structural Representation of Glycans in Glycosphingolipids
For structural representation of lipids in neutral and acidic glycosphingolipids main classes under sphingolipids category, LMSD uses the symbol and text nomenclature as proposed by the Consortium for Functional Glycomics nomenclature committee on symbol and text representation of glycan structures.11 In addition to using symbol and text representation for glycans, the last four digits of LIPID MAPS identifier (LM ID) are further subdivided into two groups: The first two positions are used to differentiate glycan series within a subclass; the last two positions represent a unique ID. For the first two positions, only letters are used; the last two positions use combinations of numbers and letters.
2.3. Structure Drawing
The structures of large and complex lipids are difficult to represent in drawings, which leads to the use of many custom formats that often generate more confusion than clarity among members of the lipid research community. For example, usage of the Simplified Molecular Line Entry Specification (SMILES)12 format to represent lipid structures, while being very compact and accurate in terms of bond connectivity, valence and stereochemistry, does not contain information about atomic coordinates and causes problems when the structure is rendered. Different structure drawing tools end up generating different 2-dimensional structural layout corresponding to the same SMILES string for a lipid molecule. The structure drawing step is typically most time-consuming process in creating molecular databases of lipids. However, many classes of lipids lend themselves to automated structure drawing paradigms, due to their consistent 2-dimensional layout. The LIPID MAPS consortium has developed and deployed a suite of structure drawing tools13 that greatly increase the efficiency of data entry into lipid structure databases and permit “on-demand” structure generation. A consistent format is chosen for representing lipid structures7,10 where, in the simplest case of the fatty acid derivatives, the acid group (or equivalent) is drawn on the right and the hydrophobic hydrocarbon chain is on the left. Similarly for glycerolipids, glycerophospholipids and sphingolipids, the radyl hydrocarbon chains are drawn to the left and the headgroups are depicted on the right. This approach enables a more consistent, error-free approach to drawing lipid structures and has been used extensively in populating the LMSD, which currently contains over 30,000 molecules.10
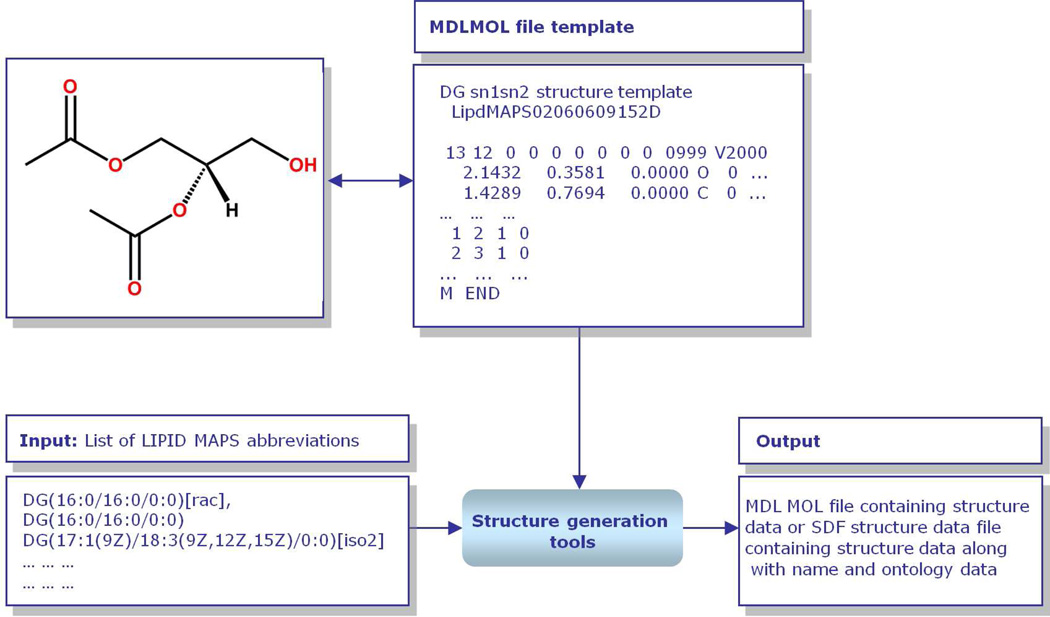
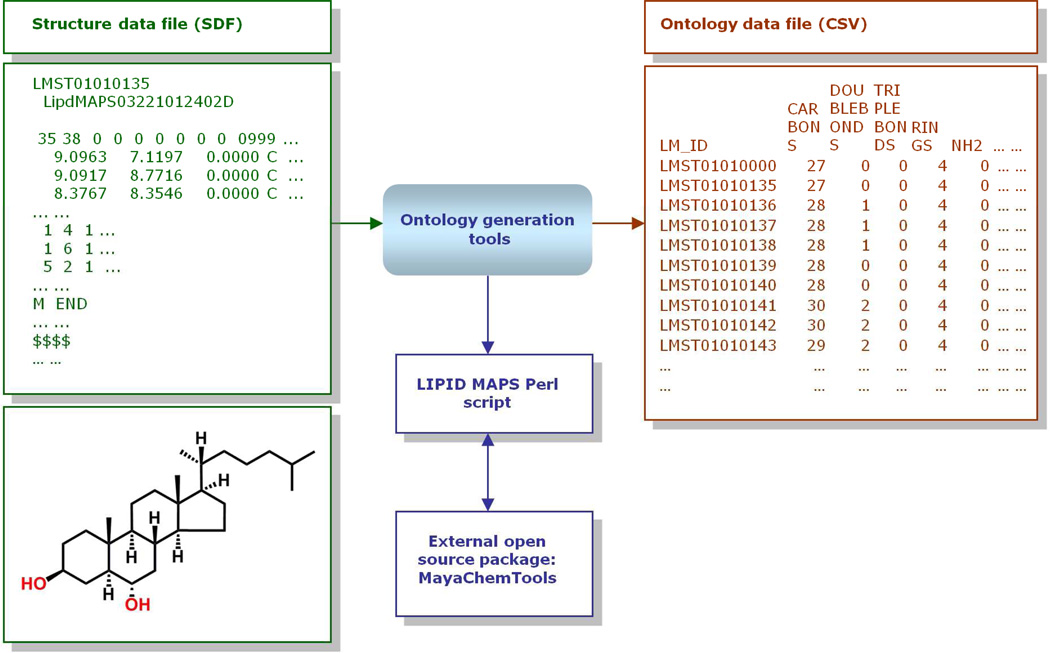
“Core” structures such as diacetyl glycerol (glycerolipids) and formic acid (fatty acyls) are represented as text-based MDL MOL files,14 and these MOL file templates are then manipulated to generate a variety of structures in MDL MOL files and Structure Data Format (SDF) files containing that core and other appropriate modifications (Figure 4). This manipulation is carried out by command-line or online programs written in the Perl15 programming language.
Figure 4.
Overview of LIPID MAPS structure data generation methodology. Starting from specified abbreviations for lipids corresponding to LIPID MAPS format, the structure generation tools select an appropriate lipid structure template internally stored in MDL MOL file format, attach appropriate radyl chains, enumerate appropriate lipid structures and generate a MDL MOL structure file or SDF file containing structural data along with name and other ontology data (Reprinted with permission from Reference 13. Copyright 2007 Oxford University Press).
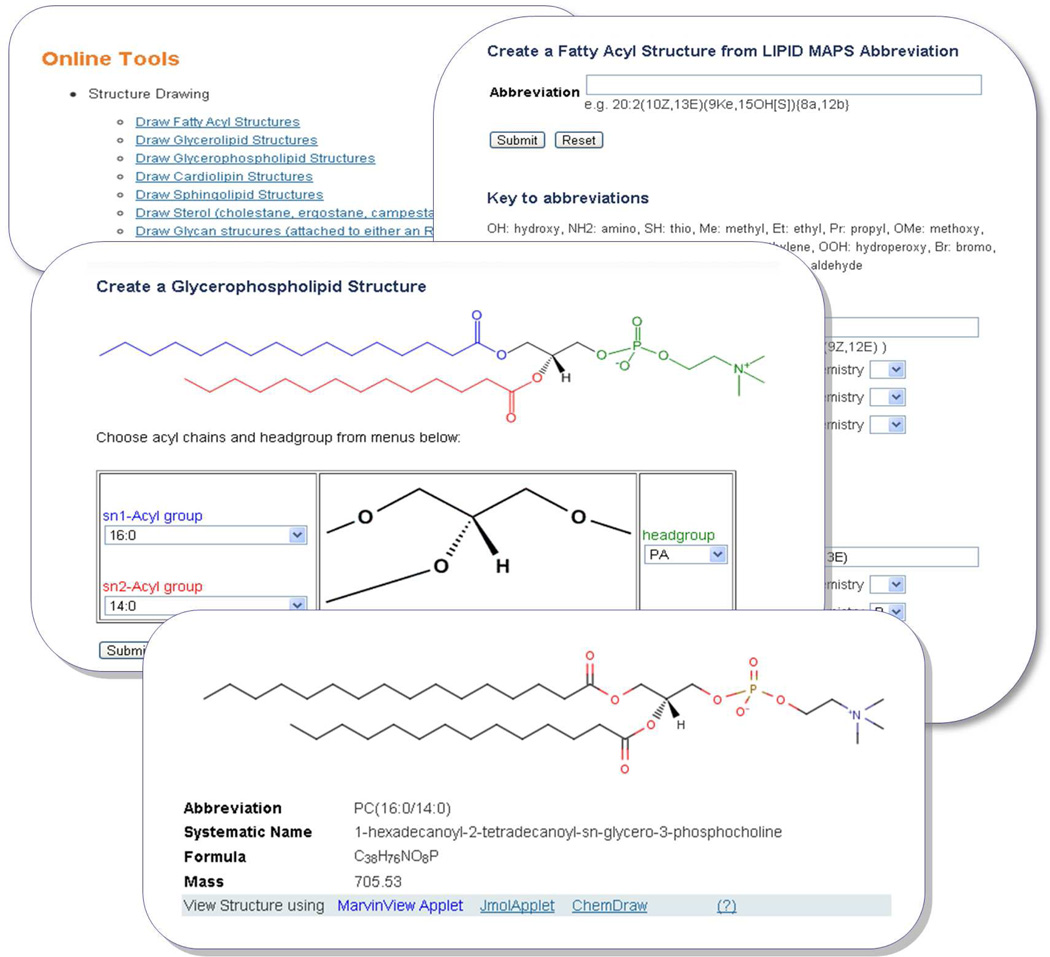
The Lipidomics Gateway website5 currently contains a suite of structure drawing tools for the following lipid categories: fatty acyls, glycerolipids, glycerophospholipids, cardiolipins, sphingolipids, sterols, and sphingolipid glycans. The online layout (Figure 5) consists of a “core” structure and pull-down menus arranged in locations appropriate for that structure. For example, in the case of the glycerophospholipid drawing tool, a central glycerol core is surrounded by pull-down menus allowing the end-user to choose from a list of head groups and sn1 and sn2 acyl side-chains. The list of acyl chains represents the more common species found in mammalian cells, and could easily be modified to include additional chains. The selected lipid structure is then generated via a server-side Perl script. The structure is rendered in the web browser as a Java®-based MarvinView applet16 or Jmol17 applet. Additionally, the structure may be viewed online with the Chemdraw ActiveX/Plugin18 by users who have this component installed on their system. Current versions of the fatty acyl drawing tools are now capable of drawing chiral centers and ring structures. Molecules with correct stereochemistry are drawn by implementing the following method: (1) Usage of custom developed module to define atoms, bonds and neighbors; (2) A recursive algorithm which applies Cahn-Ingold-Prelog (CIP)19 rules to a chiral center; (3) A scoring system to estimate substituent priority to assign chirality.
Figure 5.
A montage of screen shots showing LIPID MAPS structure drawing tools. The top left box shows structure drawing tools available on the LIPID MAPS website. The top right box shows options available for generating a fatty acyl structure starting from either a complete abbreviation or individual specification of chain and substituent from a with its position and stereochemistry. The middle box shows an example of structure generation using acyl chains and head groups for glycerophospholipids. A comprehensive list of commonly occurring acyl chains and head groups is provided as a pull down list. The button box shows an example of a structure generated for a glycerophospholipid along with other ontological information.
Concurrently, a generalized lipid abbreviation format7 has been developed which enables structures, systematic names and ontologies to be generated automatically from a single source format. Using this approach, a text file containing a list of lipid abbreviations may be submitted in batch mode to a drawing application which then generates structures (as MDL MOL files or SDF files), systematic names and ontological information such as formula, molecular weight, number of rings, number of double/triple bonds, hydroxyl, amino, keto groups, etc. In this way, thousands of lipid structures have been generated in a consistent fashion and deposited in the LMSD with considerable savings in time. Furthermore, the associated ontological information has been databased and used in various online search interfaces where, end-users may search for structures by presence (or number) of a functional group or other features.
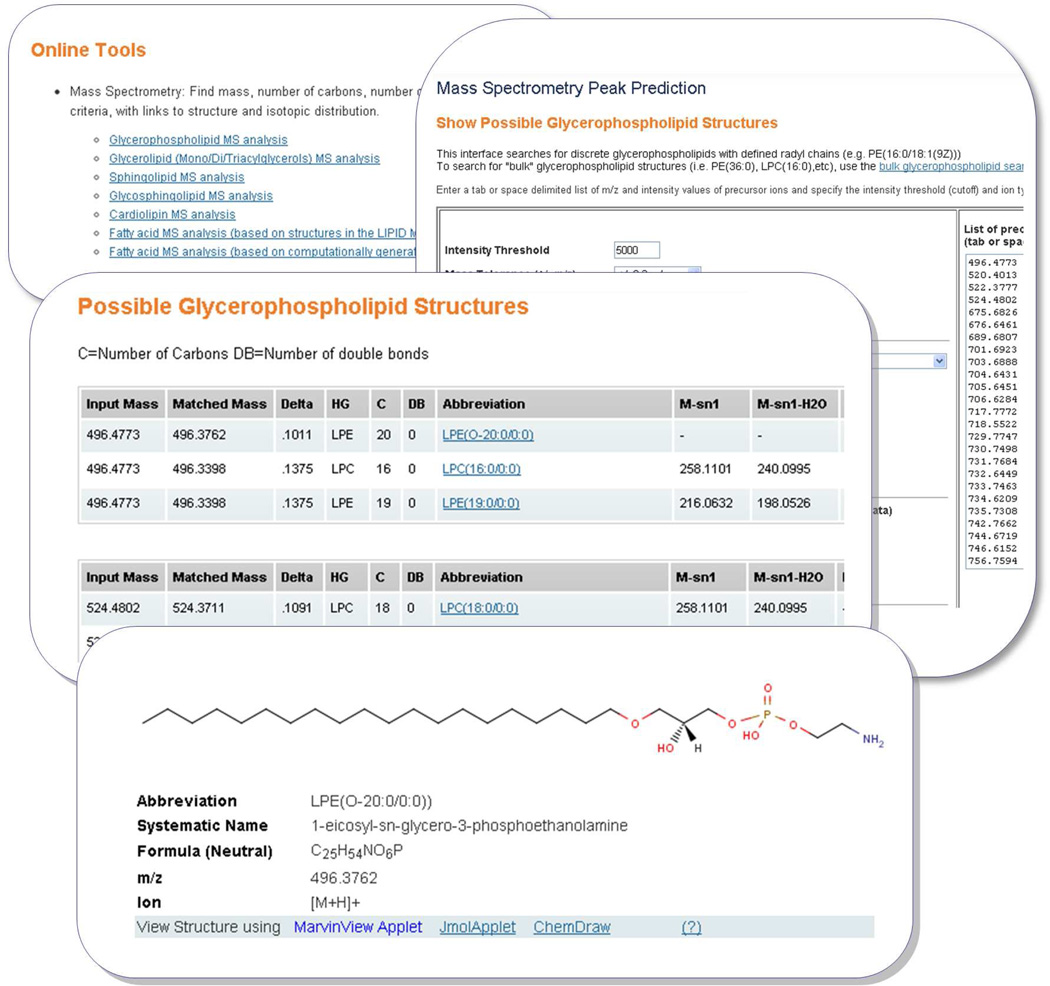
2.3.1. On-line Tools
A set of simple online interfaces have been developed to enable an end-user to rapidly generate a variety of lipid chemical structures, along with corresponding systematic names and ontological information. These are available in the “Tools” section of the Lipidomics Gateway website. The user interface is implemented using combination of Perl and Hypertext Preprocessor (PHP)20 scripts.
The lipid categories covered are fatty acyls, glycerolipids, glycerophospholipids (including cardiolipins as a special case), sphingolipid and sterols. Using the glycerophospholipids structure drawing tool as an example, the user selects from a pull-down list of radyl chain abbreviations for sn1 and sn2 position and also from a list of head groups. The corresponding lipid structure is then generated in MDL MOL format and rendered in the web browser using MarvinView applet16 which may alternatively be viewed using JMol17 applet or Chemdraw ActiveX/Plugin18. The fatty acyl structure drawing tool has a different user-input format where the user enters a valid fatty acyl LIPID MAPS abbreviation representing acyl chain length, presence of double or triple bonds and substituents on the acyl chain. Examples are “18:1(9Z)” (oleic acid) and “20:4(5Z,8Z,11E,14Z)(11OH[S])” (11S-hydroxy-5Z,8Z,11E,14Z-eicosatetraenoic acid).
The sterol drawing tools currently support the generation of structures derived from cholestane, ergostane, campestane, and stigmastane sterol cores. In addition to double bond position specification, the user can choose to substitute atoms in the cholestane core by C, N, O, and H along with the stereochemistry specification of alpha or beta for the substituted atom. Pull-down lists for position, stereochemistry and atom specification are provided for up to four simultaneous substitutions.
All major lipid categories contain glycosylated forms whose glycan substituents can be challenging to draw in full chair conformation. The LIPID MAPS glycan structure drawing tools support the generation of a wide variety of glycan structures by specifying the constituent sugars using the Consortium for Functional Glycomics nomenclature.11 The following sugar residues are supported: Glucose (Glc), Galactose (Gal), Mannose (Man), N-Acetylglucosamine (GlcNAc), N-Acetylgalactosamine (GalNAc), Xylose (Xyl), Fucose (Fuc), Acetylneuraminic acid (NeuAc), Glycolylneuraminic acid (NeuGc), Deaminated neuraminic acid (KDN) as either α or β anomers. Matched parentheses inside glycan chain specification indicate branched glycan chains; for example: GalNAca1-3GalNAcb1-3(Galb1-3GalNAcb1-4)Gala1-4Galb1-4Glcb.
2.3.2. Standalone Command Line Tools
A suite of structure drawing tools in the form of Perl scripts have been developed which can generate a large number of structures relatively quickly using a command-line interface. These command-line tools are particularly useful in the area of bioinformatics because structures and related information such as formulae, masses and abbreviations may be generated rapidly for large permutations of side-chain substituents. The tools are available from the Lipidomics Gateway website along with detailed documentation on the methods and functions used by these programs.
In addition to consistent structure representations from lipid abbreviations, the command line tools developed by the LIPID MAPS consortium also generate ontological information such as number of double bonds, chain lengths at different positions on the glycerol backbone, number of various functional groups, and other structural characteristics. The ontological information is also loaded into LMSD. The IUPAC International Chemical Identifier21 (InChI) string and InChIKeys for lipid structures are also generated using command line executable available from InChI website and loaded into LMSD database tables. Table 1 provides a list of tools available from LIPID MAPS.
Table 1.
Publicly available LIPID MAPS tools and resources discussed in this review.
| Name | URL |
|---|---|
| Pathway Editor | www.lipidmaps.org/pathways/pathwayeditor.html |
| Structure Database (LMSD) | www.lipidmaps.org/data/structure/ |
| Proteome Database (LMPD) | www.lipidmaps.org/data/proteome/index.cgi |
| On-line structure drawing tools | www.lipidmaps.org/tools/ |
| On-line mass spectrometry (MS) tools | www.lipidmaps.org/tools/ |
| Command line structure drawing tools package | www.lipidmaps.org/downloads/ |
| Command line ontology generation | www.lipidmaps.org/downloads/ |
| package Standalone windows MS prediction tool | www.lipidmaps.org/downloads/ |
| LMSD and LMPD data download | www.lipidmaps.org/downloads/ |
| Lipidomic and microarray data download | www.lipidmaps.org/data/index.html |
| Lipidomic Pathways download | www.lipidmaps.org/pathways/ |
| Experimental protocols | www.lipidmaps.org/protocols/ |
2.4. Ontology Generation
An issue of major importance in dealing with lipid structures is the huge diversity of chemical functional groups. This presents problems in explicitly classifying certain lipids containing multiple functional groups since assignment of a structure to a particular subclass may be somewhat subjective. For example, a fatty acid containing both epoxy and hydroxyl groups could be assigned to either the epoxy or hydroxy fatty acids subclass. To address this problem, the LIPID MAPS bioinformatics group has developed command line tools which calculate the number of functional groups, number of rings and other structural information from a MDL MOL file representation of a molecular structure (Figure 6). These tools are available for download from Lipidomics Gateway website. This approach may be performed in batch mode on the entire lipid structure database, thereby creating an “ontology” table which may then be incorporated into the database infrastructure. This in turn enables the use of an ontology-based search where a user may choose to search for lipids containing certain functional groups, number of carbons, rings, etc., irrespective of their classification designation. A web-based implementation of this type of ontology-based search has been implemented on the Lipidomics Gateway website.
Figure 6.
Overview of LIPID MAPS ontology data generation methodology from structure data.
3. Lipidome, Lipid Genome and Lipid Proteome Databases
3.1. Lipid Databases and Other Small Molecule Databases Containing Lipids
Lipids are generally hydrophobic in nature and soluble in organic solvents. However, lipid molecules show a remarkable structural and combinatorial diversity unlike other biological molecules such as nucleic acids and proteins. Chemical structures of lipids across different lipid categories are quite different and cover a wide range of chemical space. For example, sterol lipids are characterized by a four fused ring template consisting of three six membered rings and one five membered ring; Glycerolipids, on the other hand, typically do not contain any rings and contain radyl chains attached to sn carbons on glycerol group. The radyl chains may be further unsaturated with varied double bond positions and geometry adding to the structural heterogeneity of lipids. Additionally, a large number of possible radyl chains at various sn carbons on glycerol group along with different head groups lead to combinatorial isomeric positional diversity of lipid structures for various lipid categories such as glycerolipids, glycerophospholipids and sphingolipids. Given the structural diversity of lipids and the importance of their role in the regulation and control of cellular function and disease, it is essential to have a database of lipids which not only facilitates the storage, retrieval and dissemination of existing lipid structures and associated physiochemical properties data for the lipidomics community but is also extensible, flexible and scalable to handle the vast amount of data being generated by new lipidomic studies. A well-designed lipids database must include a defined ontology which incorporates classification, nomenclature, structure representations, definitions, related biological/biophysical properties, cross-references and physicochemical properties (formula, molecular weight, number of carbon atoms, number of various functional groups, etc.) of all objects stored in the database. This ontology can then be transformed into a well-defined schema that forms the foundation for a relational database of lipids. A large number of repositories (e.g. GenBank,22 SwissProt,23 ENSEMBL24 and GlycomeDB25) exist to support nucleic acids, proteins and carbohydrate databases; however, there are only a few specialized databases and resources (e.g. LMSD, LipidBank,9c,d LIPIDAT,9a,b Lipid Library9e and Cyberlipids9f) that are dedicated to cataloging lipids. A variety of other small molecule public and commercial databases (e.g. Human Metabolome Database (HMDB),26 DrugBank,27 Therapeutic target database (TTD),28 Chemical Entities of Biological Interest (ChEBI),29 ChemBank,30 PubChem,31 ZINC,32 ChemSpider,33 Chemical Abstract Service (CAS),34 eMolecules,35 Beilstein36 and Kyoto Encyclopedia of Genes and Genomes (KEGG) LIGAND37) also exist which provide information about lipid structures and their associated physicochemical properties.
While there has been no prior effort at systematic and comprehensive classification and nomenclature of lipid molecules, there are several small databases as mentioned in the previous paragraph which contain some or several lipid molecules. The LMSD database being developed by LIPID MAPS consortium is one of the latest databases dedicated to lipids and provides comprehensive information about lipids. We provide an overview of the LMSD database, other lipid specific databases and small molecule databases (Table 2) containing lipids in the rest of this section followed by detailed description of the LMSD database.
Table 2.
Resources and databases containing information about lipids.
| Name | URL | Comments/Description |
|---|---|---|
| LMSD | www.lipidmaps.org | LIPID MAPS lipids structure database |
| LIPID BANK | www.lipidbank.jp/ | Database of Japanese Conference on the Biochemistry of lipids |
| LIPIDAT | www.lipidat.tcd.ie | Database of thermodynamic and associated information on lipids |
| Lipid Library | www.lipidlibrary.org | Information about lipid chemistry, biology, technology and analysis |
| Cyberlipids | www.cyberlipid.org | Resource for lipid studies |
| HMDB | www.hmdb.ca | Human metabololome database |
| DrugBank | www.drugbank.ca | Drug data with target and action information |
| TTD | xin.cz3.nus.edu.sg/group/ttd/ttd.asp | Therapeutic target database along with drug information |
| ChEBI | www.ebi.ac.uk/chebi | Database and ontology for chemical entities of biological interest |
| ChemBank | chembank.broad.harvard.edu | Small molecule screening and cheminformatics resource database |
| PubChem | pubchem.nci.nih.gov | Public repository for biological properties of small molecules including assay and screening data |
| ZINC | zinc.docking.org | Commercially available compounds for virtual screening |
| ChemSpider | www.chemspider.com | Chemical information resource |
| CAS | www.cas.org | Small molecule databases and associated information |
| eMolecules | www.emolecules.com | Commercially available small molecules |
| Beilstein | www.reaxys.com/ | Small molecules structures and other information |
| KEGG LIGAND | www.genome.jp/kegg/ligand.html | Database of chemical compounds and reactions in biological pathways |
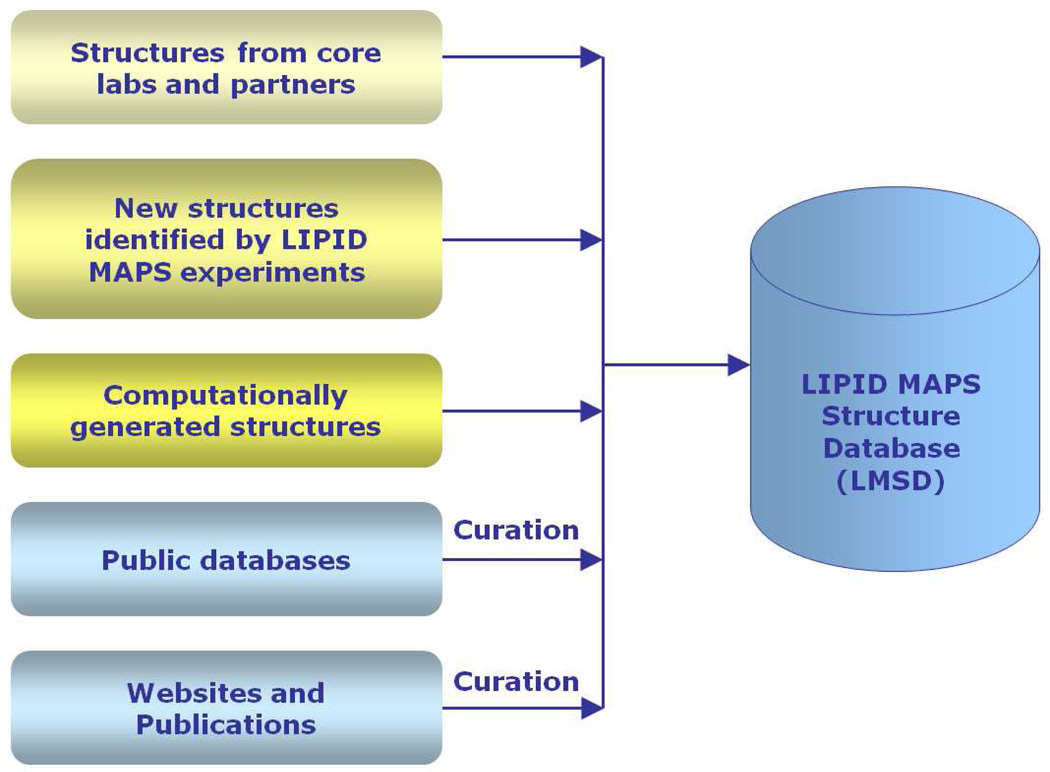
The LMSD10 is a relational database containing structures and annotations of biological relevant lipids. It is being developed and maintained by LIPID MAPS consortium, and currently contains over 30,000 structures which are obtained from the following sources (Figure 7): LIPID MAPS Consortium's core laboratories and partners; lipids identified by LIPID MAPS experiments; computationally generated structures for appropriate lipid classes; biologically relevant lipids manually curated from LIPID BANK, LIPIDAT and other public databases; peer-reviewed journals and book chapters describing lipid structures.
Figure 7.
Overview of LIPID MAPS Structure Database (LMSD) generation methodology.
The LIPID BANK is a lipid database of Japanese Conference on the Biochemistry of Lipids (JCBL). It contains over 7,000 lipids corresponding to the following main lipid classes: acylglycerol, bile acid, derived lipid, eicosanoid, ether type lipid, fat soluble vitamin, glycolipid, isoprenoid, lipid peroxide, lipoamino acid, lipopolysaccharide, lipoprotein, mycolic acid, phospholipid, steroid, and wax. In addition to classification-based browsing of lipids, the LIPID BANK supports text-based search and retrieval of lipids data using name and other physicochemical properties; the structure-based search is not available. The search results along with structure and other basis information such as molecular weight, molecular formula, name, and common name provide the following additional information about a lipid: biological activity, physical and chemical properties, spectral data (Ultra violet (UV), Infrared (IR), Nuclear magnetic resonance (NMR), Mass spectrometry (MS)), chromatogram data, chemical synthesis, metabolism, genetic information, and references.
The LIPIDAT is a relational database of thermodynamic and associated physicochemical properties information on lipids. It contains over 20,000 lipids. The users can search the database using various physicochemical properties through more than 2 dozens available text-based query pages. The detailed search results page about a lipid includes the following information: structure, name, and formula along with other basic information; bibliographic information; experimental results and methods.
The LIPID LIBRARY is not a database of lipids but an online resource about chemistry, biology, technology, and analysis of lipids. The online pages provide information about lipids organized into the following sections: basic information, biochemistry and nutrition, lipid analysis, oils and fats, and latest news. The basic information section covers structures, definitions, composition, biochemistry, and functions of these lipid categories: fatty acids and eicosanoids, simple and complex glycerolipids and phospholipids, sphingolipids, and sterols. The biochemistry and nutrition section covers only plant lipid biochemistry. The lipid analysis section provides descriptions of both chromatographic and spectroscopic techniques used for analysis of lipids along with literature surveys of analytical methodologies. The oils and fats section cover the chemistry and technology of oils and fats along with the history of science and technology. The detailed information available for lipids covered in basic information section provides the following details for each lipid: structure, name, source and occurrence, biochemistry and function along with appropriate literature references.
The Cyberlipids is an online resource for studies of lipids. It provides information about definitions, source, compositions, and physicochemical properties of lipids along with detailed review of various lipid analysis techniques. The users can retrieve detailed information about a lipid using its name for more than 900 lipids or get a list of all lipids with links to detailed information.
The Human Metabolome Database (HMDB) is a database containing information about small molecule metabolites, including lipids, found in the human body. It contains over 7,900 metabolites entries with links to over 7,200 protein and deoxyribo nucleic acid (DNA) sequences. The database provides links to three kinds of data: chemical data, clinical data, and molecular biology/biochemistry data. The users can search HMDB using text, chemical structure, and arbitrary relationships of available data fields. The database searching using spectral and chromatography data (MS, MS-MS, GC-MS, and NMR) is also available. Additionally, a variety of different data browsing options are provided: class-based browsing, pathway, disease, and so on. The detailed information about each molecule is presented as a MetaboCard containing over 110 different data fields with 2/3rd of the data fields containing information about chemical/clinical data and the rest about enzymatic and biological data. The links to other external data sources are also provided.
The DrugBank database provides detailed information about drugs, including lipids, along with the drug targets. The detailed drug information consists of chemical, pharmacological, and pharmaceutical information; the targets information corresponds to sequence, structure, and pathway. The database contains over 6,800 drug entries covering the following types of drugs: over 1,400 food and drug administration (FDA)-approved small molecule drugs, over 130 FDA-approved biologics drugs, over 83 nutraceuticals, and over 5,000 experimental drugs. Additionally, information for over 4000 non-redundant protein target sequences is linked to drug entries. The users can search the DrugBank database using text, chemical structure, and arbitrary combination of available data fields. A variety of different data browsing options are also available: drug name, pathway, class name, and so on. The detailed information about each drug is presented as a DrugCard containing over 150 data fields with half the information covering drug/chemical data and the rest corresponding to drug target.
The Therapeutic Targets Database (TTD) provides information about known targets along with information for associated disease, pathways, and drugs for these targets. The TTD database contains information for over 1,900 targets and over 5,000 drugs with over 3,000 small molecule drugs. The drugs information covers over 1,500 approved drugs, over 1,100 drugs in clinical trials, and over 2,300 experimental drugs. The text-based database search provides searching using target/disease name, drug name, function and classification. The detailed search results page contains information about target and disease, drug name and its function, and links to other external database containing information about targets and drugs.
The Chemical Entities of Biological Interest (ChEBI) database provides structural and ontological information about molecular entities focused on small molecule compounds including lipids. The molecular entities are either natural products or synthetic products used for biological intervention; nucleic acids are not included. The ChEBI database contains over 19,000 small molecules. The information about small molecules in ChEBI comes from these four key sources: IntEnz38 – the integrated relational enzyme database of the European Bioinformatics Institute (EBI); KEGG COMPOUND;39 PDBeChem;40 and ChEMBL41 The users can search ChEBI database using text, chemical structure, and arbitrary combination of available data fields. The structure-based search also supports similarity and substructure searching. The detailed search results along with structure and other basic information such as molecular weight, molecular formula, name, and common name provide the following additional information about a small molecule: ChEBI ontology, brand name, references to other databases, registry numbers corresponding to external sources (CAS, Beilstein, and Gmelin), and literature references.
The ChemBank is a relational database containing data derived from small molecules, including lipids, and small molecule screens along with tools for analyzing these data. The database contents include chemical structures and names, calculated molecular descriptors, human curated information about small molecules activities, raw experimental results from high-throughput biological assays, and metadata describing the screening experiments. The ChemBank database contains data for over 1.7 million compound samples with over 1.2 million unique small molecule structures screened against more than 2,500 assays covering more than 180 projects. Additionally, it contains information for over 1,000 proteins, 500 cell lines and 70 species associated with various assays. The users can search ChemBank using text, chemical structure, and arbitrary relationships of available data fields. The structure-based searching, in addition to substructure and exact match, also supports similarity searching. The database searching using information about high-throughput screens and small molecule assays is also available. Additionally, a number of tools for analysis and visualization of small molecule screening results are provided. The detailed search results along with structure and other basic information such as molecular weight, molecular formula, name, and common name provide the following additional information for a small molecule: a large number of calculated physicochemical properties; compound sample information; screening information including project name, assay name, assay type, plate, well and z-score.
The PubChem database is a database of chemical molecules and biological activities of molecules screened against various assays. It also contains information about lipids as LIPID MAPS consortium upload its LMSD database of lipids into PubChem on a regular basis. The PubChem database is divided into three main categories: Compound database with over 32 million entries contains unique chemical substances derived from substance depositions; Substance database with over 74 million entries consists of chemical compounds submitted by depositors corresponding to mixtures, extracts, and complexes; BioAssay database containing biological activity results from over 1,600 high-throughput screening projects with several million measured values. The PubChem data deposition is open to the scientific community. The growing list of over 140 substance and 47 assay depositors represent all major sources including commercial vendors, public non-profit organizations, pharmaceutical companies, and individual contributors. The users can search PubChem compounds, substances, and bioassay databases using text, chemical structure, and arbitrary relationships of available data fields. The text-based searching supports the usage of a wide variety of parameters including name, formula, physiochemical properties, stereochemistry specifications, elements, and so on. The structure-based searching provides support for substructure/superstructure search, and identity/similarity search. The detailed search results page for compound along with structure and other basic information such as molecular weight, molecular formula, name, and common name provide the following additional information for a compound: synonyms, calculated physicochemical properties, substance information, biomedical annotation, pharmacological action and classification, chemical classification, safety and toxicology, links to exiting literature, and so on. The substance detailed results page, in addition to basic information such as chemical structure, name, and formula contains the following additional information: link to data depositor, links to any bioactivity information and other structurally related substances, and links to other databases maintained by the National Center of Biotechnology Information (NCBI). A variety of analysis tools such as bioactivity structure activity analysis and chemical structure clustering are also provided for the analysis of bioassay screening data.
The ZINC database contains commercially available small molecules for virtual screening. It contains over 13 million purchasable compounds including lipids. The users can search the ZINC database using compound name, chemical structure/substructure, physicochemical properties, vendor catalog number/source, and so on. The compound detailed search page includes chemical structure, name, formula, various calculated physicochemical properties, vendor and purchase information, and availability.
The ChemSpider is a chemical database and an online resource linking together compound information across the web. The compound information includes physical and chemical properties, chemical structure, systematic nomenclature spectral data, synthetic methods, known reactions, and safety information. The ChemSpider contains over 25 million unique chemical compounds sourced and linked to over 400 separate data sources including LIPID MAPS for lipids. The compound data is collected from over 50 difference sources. Additionally, the ChemSpider supports the uploading and curation of chemical structure and spectra data by the scientific community. The users can search ChemSpider database using text, chemical structure, and arbitrary relationships of available data fields. The text-based searching supports the usage of a wide variety of parameters including name, formula, physiochemical properties, literature search, and so on. The structure-based search supports chemical structure/substructure search along with arbitrary combinations of calculated physicochemical properties. The detailed search results page for a compound along with structure and other basic information such as molecular weight, molecular formula, name, and common name provide the following additional information: links to Wikipedia articles; associated data sources and commercial suppliers; patents; literature articles; calculates physicochemical properties; medical subject headings classification; pharmacological data; spectra; and inks to other literature data. The ChemSpider online resource also hosts a variety of web services such as chemical names to structure conversion, generation InChI strings, and calculation of various physicochemical properties.
The Chemical Abstract Service (CAS) is a comprehensive resource of chemical information combining databases with search and analysis tools available as chemical abstracts and chemical databases. The CAS provides two main chemical databases: CAplus and CAS Registry. The CAplus database consists of summaries and indices of scientific literature covering chemistry and chemistry related topics such as proteomics, genomics, and so on. The CAplus database contains over 33 million references and its coverage of scientific literature starts from early 1,800s and spans across 10,000 journals, technical reports, conference proceedings, and books in more than 60 languages; it also covers patent literature from over 60 countries. The CAS REGISTRY database contains over 52 million organic and inorganic chemical substances, and over 62 million sequences. Its coverage of chemical substances also starts from early 1,800 and covers substances from patents, chemical catalogs, and various web sources; the sequence data is retrieved from GenBank. In addition to basic compound information such as structure, name, formula, and molecular weight, the chemical substance record contains the following additional information: a unique CAS number, experimental and calculated physicochemical properties, ring analysis, and literature references. The CAS databases are searched using SciFinder which support both text-based and structure-based searching along with usage of other parameters during the search. In addition to CAplus and CAS REGISTRY, the CAS provides the following three databases: CASREACT, CHEMLIST, and CHEMCATS. The CASREACT and CHEMLIST databases contain information about chemical synthesis and regulated chemicals respectively. The CHEMACTS database contains over 44 million commercially available substances covering over 1,200 catalogs from 1,100 suppliers; it has over 12 million chemical substances with unique CAS numbers.
The eMolecules is an online resource for commercially available chemical molecules including lipids. It contains over 8 million unique molecules from variety of commercial catalogs and other on-line data sources such as National Institute of Standards and Technology (NIST), PubChem, DrugBank, and LIPID MAPS. The users can search eMolecules database using molecule name, molecule structure/substructure, suppliers, and various physicochemical properties. In addition to basic molecule information such as structure, name, formula, and molecular weight, the molecule record contains the information about suppliers and links to ordering chemicals.
The Beilstein database provides experimentally validated information about millions of chemical compounds uniquely identified by Beilstein Registry Numbers and chemical reactions compiled from scientific literature starting from 1,771. The original database was created using Beilstein's Handbook of Organic Chemistry and contains information about reactions, chemical substances, chemical structures, and physiochemical properties. The record for each substance has over 350 data fields corresponding to chemical and physical data along with appropriate literature references. The users can search the database using Reaxys system using one of the following three search options: reactions searching, substances and properties searching, and text searching. During reaction searching, a variety of other parameters such as starting materials, product, reaction conditions, and so on can also be specified. The substance and properties searching provides structure/substructure search along with specification of various physical and chemical properties. The text-based search allows the users to retrieve appropriate data using substance name, authors, and variety of other parameters. The detailed search results page for substance along with structure and other basis information such as molecular weight, molecular formula, name, and common name provide the following additional information: calculated physicochemical properties, physical and spectral data, synthesis information, and links to literature.
The KEGG LIGAND is a database of chemical compounds and reactions involved in biological pathways. It is a composite database consisting of three other databases: KEGG COMPOUND, KEGG ENZYME, and KEGG REACTION. The KEGG COMPOUND database contains information for over 7,000 metabolites and biologically relevant chemical compounds including lipids which are classified according to LIPID MAPS classification system and made available through KEGG BRITE database. The KEGG REACTION database contains information for over 5,000 reactions corresponding to metabolic and other reactions. The KEGG ENZYME database has information for over 3,800 enzymes involved in various transformations. The users can search KEGG LIGAND databases using text and chemical structures. The structure-based search supports structure/substructure search along with similarity searching. The detailed search results page for a compound along with structure and other basic information such as molecular weight, molecular formula, name, and common name provide the following additional information: links to ENZYME and REACTION databases, links to external data sources such as PubChem and CAS numbers.
3.1.1. Populating the Structure Database
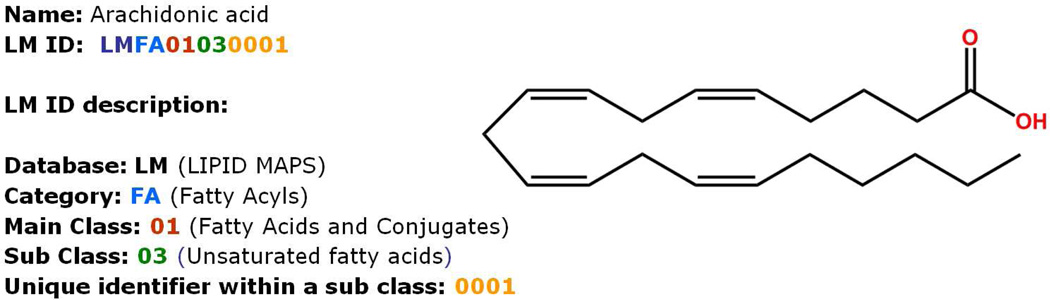
An object-relational database of lipids containing structural, biophysical and biochemical characteristics is available on the Lipidomics Gateway website with browsing and searching capabilities. The LMSD currently contains over 30,000 structures which are obtained from a variety of sources: LIPID MAPS Consortium's core laboratories and partners; Lipids identified by LIPID MAPS experiments; computationally generated structures for appropriate lipid classes; biologically relevant lipids manually curated from LIPID BANK, LIPIDAT and other public databases; peer-reviewed journals and book chapters describing lipid structures (Figure 7). All structures have been classified and redrawn according to LIPID MAPS guidelines. After lipids have been selected for inclusion into LMSD, they are classified following the LIPID MAPS classification scheme as explained earlier under the classification, ontology and nomenclature of lipid molecules section. Structures of the lipids are drawn either manually or generated automatically by computational structure drawing tools developed by the LIPID MAPS consortium; the structure representation is consistent and adheres to the rules proposed by LIPID MAPS consortium. Based on its classification, each lipid structure in LMSD is assigned a unique LM ID. The format of the LM ID (Figure 8) not only maintains uniqueness of ID but also provides the capability to add new categories, classes, and subclasses as the need arises.
Figure 8.
Description of the LIPID MAPS LM ID.
In addition to import and manual curation of biologically relevant lipids from other database sources, LMSD also stores their original IDs to enable cross-referencing. LMSD lipid structures are deposited into PubChem database periodically and a link to PubChem Substance ID (SID) is also maintained within LMSD. Access to complete set of LMSD lipid structures in the PubChem database is also available.42
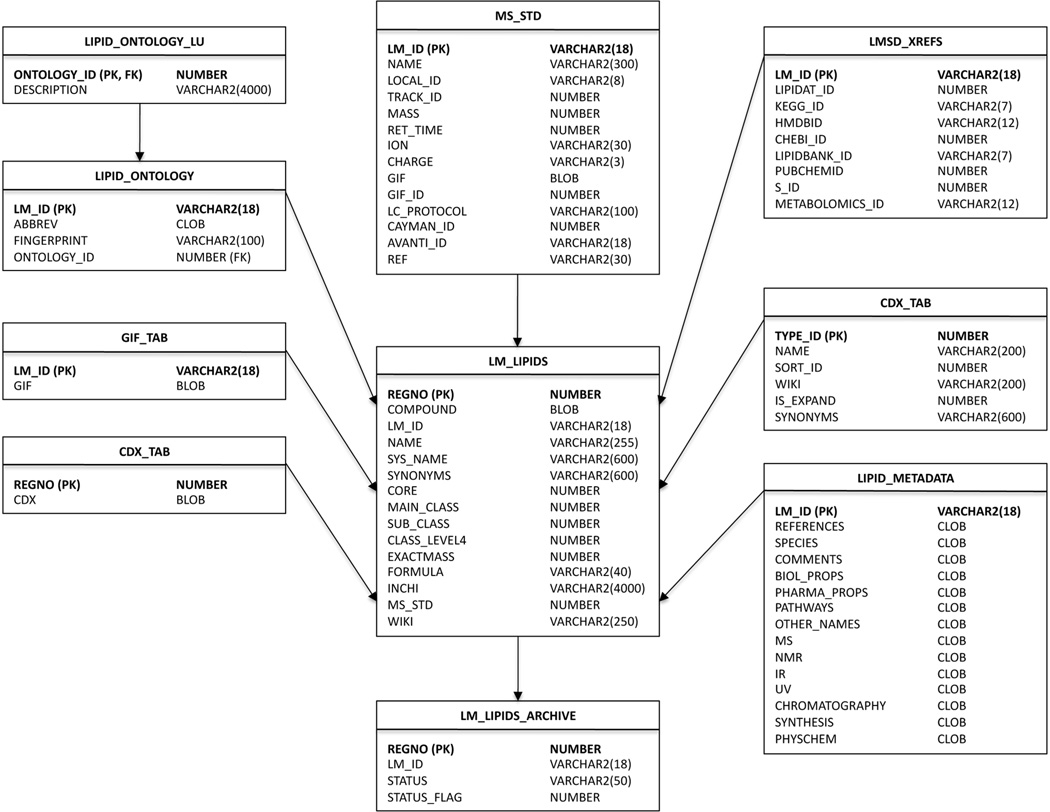
LMSD structures are either drawn manually using ChemDraw or generated automatically by structure drawing tools developed by LIPID MAPS consortium for various subclasses in fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, and sterols. The structure drawing tools are Perl scripts which can generate a large number of structures relatively quickly via command-line or web-based interface. In addition to consistent structure representations from lipid abbreviations, these scripts also generate ontological information such as number of double bonds, chain lengths at different positions on the glycerol backbone, number of various functional groups, and other structural characteristics. The ontological information is also loaded into LMSD. The InChI string and InChIKeys for lipid structures are also generated using command line executable available from InChI website and loaded into Oracle database43 tables. The database schema used for LMSD is outlined in an entity relationship diagram in Figure 9.
Figure 9.
Entity relationship diagram for LMSD showing the Oracle database tables containing structural and classification information as well as annotations and ontological data. The unique LM_ID identifier plays a central role as a primary key in this relational schema.
3.1.2. Searching the Structure Database
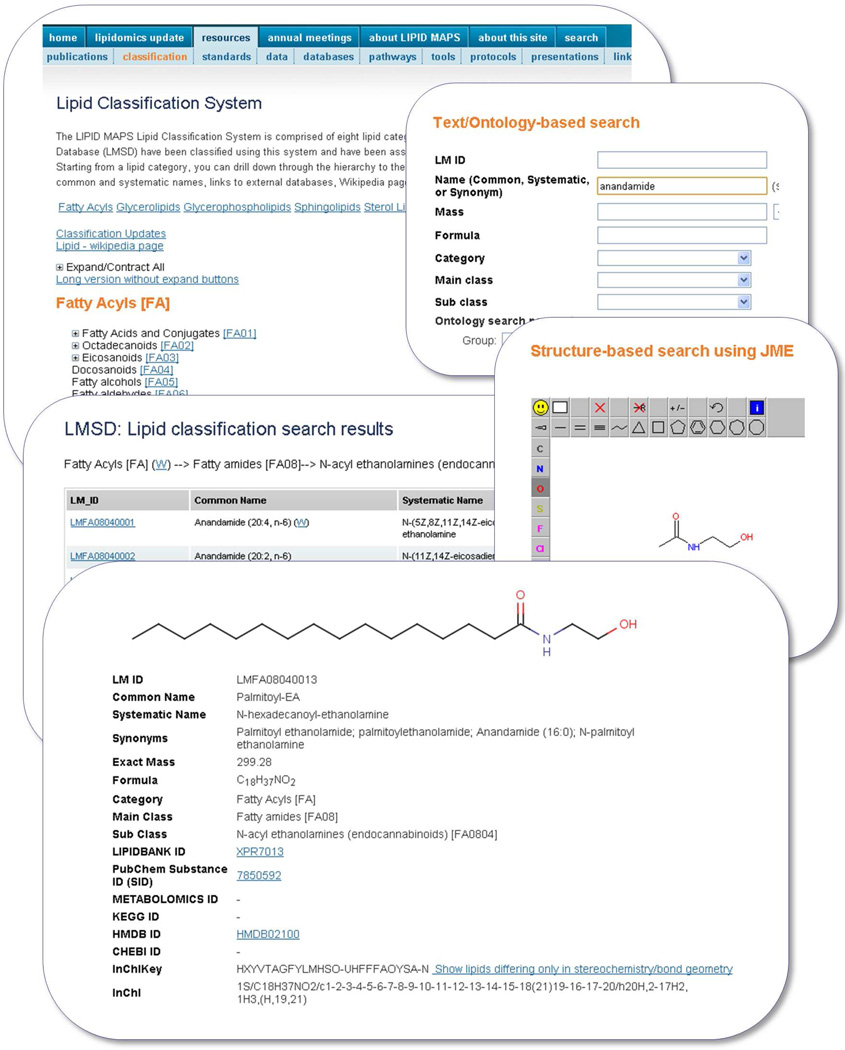
The Lipidomics Gateway website supports the searching of LMSD database in three different ways: classification-based, text/ontology-based, and structure-based search. The classification-based browsing provides the capability to retrieve lipids based on the LIPID MAPS classification scheme. After the user selects one of the main categories of lipids, a listing of all lipids present in the selected category, along with a link to the set of lipids in each main class and subclass, is provided. The user may then select all lipids which belong to either a main class or a subclass and display the results as a result summary page.
In case of lipids containing multiple functional groups, assignment of a structure to a particular subclass may be somewhat subjective. For example, a fatty acid containing both epoxy and hydroxy groups could be assigned to either epoxy or hydroxy fatty acids subclass. To address this situation, an ontology-based search is also provided. The user may choose to search for lipids containing similar functionality and all the lipids with the specific functionality, irrespective of their subclass designation, would be retrieved. The text/ontology-based query page allows the user to search LMSD by any combination of these data fields: LM ID, common or systematic name, mass along with a tolerance value, formula, category, main class, subclass, and various combinations of ontology parameters. The structure-based search page provides the capability to search LMSD by performing a substructure or exact match using the structure drawn by the user. Three supported structure drawing tools are MarvinSketch,16 JME,44 and ChemDrawPro.18 The first two of these structure drawing tools are Java applets and require only applet support in the browser. In addition to structure, the user can also specify LM ID and common or systematic name for the search.
The record details page, in addition to displaying the structure for the selected lipid, also contains all relevant information for that molecule such as, common and systematic names, synonyms, molecular formula, exact mass, classification hierarchy, InChIKey, and cross-references (if any) to other databases.
The default lipid detail page uses a Graphics Interchange Format (GIF) image for representing structure of the lipid. The decision to use GIF format for representing lipid structures in the web browser was made due to its native support across all the browsers. The structure may also be viewed and manipulated using MarvinView,16 JMol,17 and the ChemDraw, ActiveX/Plugin18 formats where structures may be manipulated, scaled and saved in a number of high-resolution formats. Figure 10 shows screen shots of the LMSD user interface for lipid classification-based browsing, text-based and structure-based searching.
Figure 10.
A selection of screen shots showing various options for searching the LIPID MAPS Structure Database (LMSD) and result summary for a specific LM ID.
3.2. Lipid Proteome Databases
3.2.1. Populating the Proteome Database
To fully understand the roles of lipids, we must also understand the enzymes that catalyze lipid-related metabolic pathways, transcription factors and signaling agents involved in lipid regulation, and other proteins that affect lipid biochemistry by binding to or interacting with lipids. While Entrez Gene45 and UniProt46 provide annotations of proteins and their corresponding genes vis-à-vis their functional role, there was previously no database that comprehensively cataloged all lipid-associated proteins. The LIPID MAPS Proteome Database (LMPD)47 developed by LIPID MAPS serves such a purpose.5
UniProt and Entrez Gene contain a significant part of the annotations of proteins and genes respectively, and most of the known lipid-related proteins have been annotated in these databases. However, prior to the development of LMPD there was no unique database of lipid-associated proteins that contained comprehensive and context dependent annotations. LMPD was developed in order to fill this void by providing a catalog of genes and proteins involved in lipid metabolism and signaling. LMPD can be searched by database ID, keyword, KEGG pathway, or Gene Ontology (GO) term, and is publicly available from the Lipidomics Gateway website.
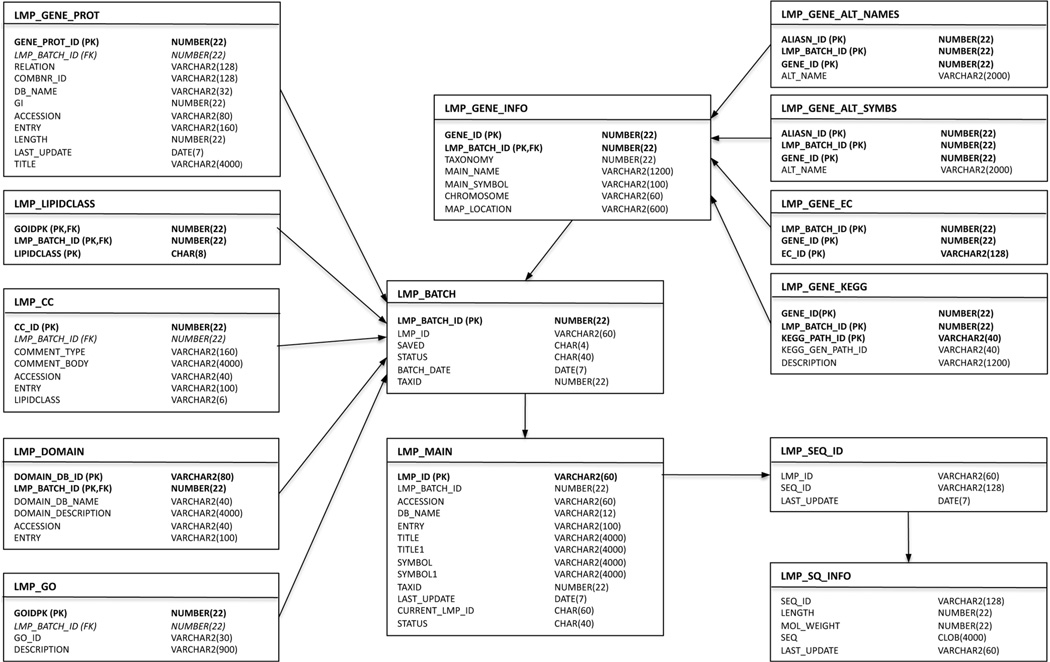
LMPD is constructed as an object-relational database of lipid-associated protein sequences and annotations. The database schema used for LMPD is outlined in an entity relationship diagram in Figure 11. The initial release of LMPD established a framework for creating a lipid-associated protein list, collecting relevant annotations, databasing this information and providing an online user interface. A similar approach was used previously for development of the MitoProteome database.48 The current release of LMPD contains approximately 1200 lipid-related proteins for each of human and mouse species.
Figure 11.
Entity relationship diagram for LMPD showing the Oracle database tables containing information pertaining to lipid-related genes and proteins for human and mouse species.
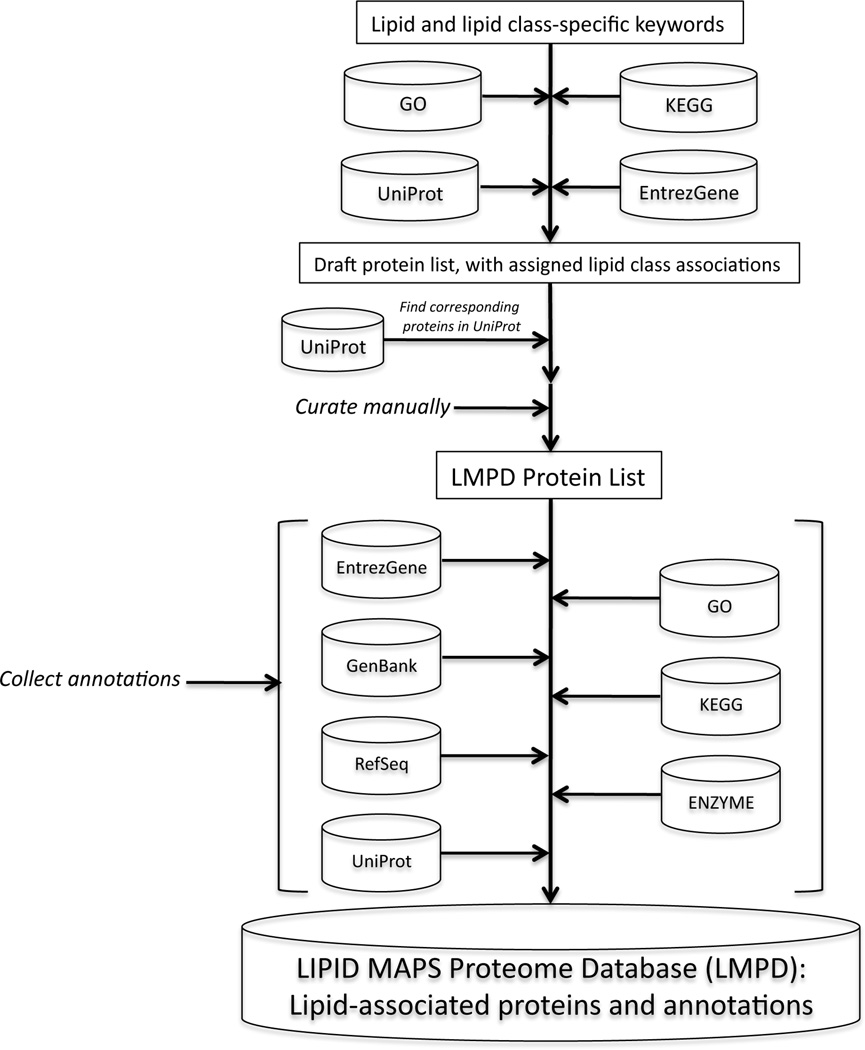
In order to construct LMPD, a curated set of lipid-related keywords was created for each of the 8 lipid categories. These keywords, containing terms such as “lipase”, “cyclooxygenase”, “ceramide” and “choline”, were then used to search name, description and annotation information in publicly available UniProt46, Entrez Gene,, GO49 and KEGG50 data repositories for mouse and human species in order to identify proteins, genes and related pathway and ontology information containing these terms. The GO terms identify proteins that are involved in particular anabolic, catabolic, and other metabolic processes, while proteins gathered from KEGG were identified as being involved in a lipid metabolic pathway. Experimental methods used in identifying these proteins included various enzyme assays, high performance liquid chromatography (HPLC), polyacrylamide gel electrophoresis, and mass spectrometry. All protein lists generated by these automated methods were then manually curated, erroneous entries were deleted, known lipid-related proteins not identified by the methods above were added and corresponding Entrez Gene ID’s and annotations were generated for all Uniprot records. This process is illustrated in Figure 12.
Figure 12.
Overview of the bioinformatics process for creating the LIPID MAPS Proteome Database (LMPD).
The Signaling Gateway Molecule Pages (SGMP) database, another database containing states of proteins involving lipids, is a repository derived from a comprehensive signaling protein ontology that covers functional states of a protein, the transitions between those states and the defined functions of a protein in a given cellular context.51 The SGMP data are exported to the Biological Pathway Exchange (BioPAX)52 and Systems Biology Markup Language (SBML).53 The SGMP database contains information on several lipid binding and modifying proteins (Table 3).
Table 3.
A representative list of lipid-related signaling proteins as Molecule Pages.
| SGMP ID | GenBank Accession | Molecule Page Name | Molecule Page Category |
|---|---|---|---|
| A001757 | AAH05636.1 | Phosphodiesterase 6D, cGMP-specific rod delta | Lipid binding protein |
| A003319 | NP_898977.2 | DFCP1 | Lipid binding protein |
| A000010 | NP_032892.1 | Acyl protein thioesterase 1 | Lipid modification, protein |
| A000095 | NP_032733.1 | Protein N-myristoyltransferase 1 | Lipid modification, protein |
| A001778 | NP_780565.1 | Phosphatidylinositol-4-kinase type III beta | Kinase, lipid |
| A002220 | NP_064395.2 | Sphingosine kinase 2 | Kinase, lipid |
| A001749 | AAC37702.1 | Phosphodiesterase 1C, calmodulin dependent | Phosphodiesterase |
| A001750 | NP_001008548.1 | Phosphodiesterase 2A, cGMP stimulated | Phosphodiesterase |
| A000046 | BAC00906.1 | Phospholipase C epsilon | Phospholipase |
| A001789 | AAH45156.1 | Phospholipase A2, group IIA | Phospholipase |
3.2.2. Searching the Proteome Database
Multiple LMPD query interfaces are available, enabling users to search LMPD by database ID or keyword; by KEGG pathway; or by GO term. From the search results, one can access annotations relevant to each protein of interest, cross-linked to external databases. Annotations are organized by record overview, Gene/GO/KEGG information, protein domain information, SwissProt/UniProt annotations, and related proteins and LIPID MAPS experimental data (if any). The record overview contains LMPD ID, species, description, gene symbols, lipid categories, enzyme code (EC) number, molecular weight, sequence length and protein sequence. Gene information includes Entrez Gene ID, chromosome, map location, primary name, primary symbol and alternate names and symbols; GO IDs and descriptions; and KEGG pathway IDs and descriptions. UniProt annotations include primary accession number, entry name and comments such as catalytic activity, enzyme regulation, function and similarity.
4. Lipid Experimental Protocols and Meta Data Management
The post-genome sequencing era has heralded the beginning of a new phase of scientific discovery that is based on massive volumes of data generated by high throughput technologies.54 This exploratory, data-driven approach represents a paradigm shift from the traditional scientific discovery where an individual laboratory’s effort is focused on a particular gene-product and the pathway in which the gene-product participates, i.e., a hypothesis-driven approach. Efforts to understand the detailed functioning of all the elements of the cellular machinery at the molecular level pose a major challenge that would require a large collective effort from a multidisciplinary organized team of scientists. If people working in academia were to engage in such an effort, the organization of the effort may perhaps require a consortium approach with laboratories having expertise in different areas such as cell biology, molecular biology, proteomics, functional genomics, and bioinformatics, contributing to a joint and well-integrated effort.
Each high-throughput technique generates a large body of data to be recorded. It brings two data management issues to the fore: first, how the sheer amount of data from heterogeneous but related experiments from various laboratories will be handled, secondly, how data will be shared and analyzed collectively among them and made available to the public at large. The laboratory notebook concept is insufficient to deal with the issues of data handling, structuring, and sharing.55 For such a research endeavor, utilization of high-throughput techniques to explore complex biological systems is the norm rather than an exception. In a high-throughput setup the output from one experiment is the input of another. Situations like these create another set of issues to be dealt with, since samples will be passed from one laboratory to another in bulk quantities for subsequent handling and analysis. The samples are all necessarily coded such that the recipient laboratory could recover the information about the history of each received sample. Laboratory notebooks could be replaced by a relational database, which would facilitate data deposition from various laboratories to a common repository and at the same time data could also be viewed by authorized personnel. The data structuring could be achieved by an appropriate database schema design, which could also enforce linking of the data from heterogeneous biological experiments, thus offering easy access to the data analysis en masse. The role of the pen will be replaced by graphical user interfaces (GUIs) and a keyboard; the GUI would enable the experiment to document the samples and their handling and directly deposit data to the database. There will be a separate GUI for each type of experiment, so the use can be guided as to what needs to be done. The GUI should be designed to check data validity prior to deposition into the database; this will minimize the manual data entry errors inherent in a notebook system. Data should be regularly backed up to guard against any kind of system failure. This scheme essentially represents a paper-free and scalable structured electronic notebook for data cataloging, and automated incorporation of timestamps to record the data entry. After successful deposition of the experimental parameters to the database through a GUI, the user must be provided with a label to identify the sample container, which in biological experiments is often a tube or flask. The label should uniquely identify each experiment and contain meaningful information to facilitate deciphering its contents.
The data structuring, handling, and data management requirements could be met by the use a laboratory information management system (LIMS). Use of LIMS is widespread in diverse industrial settings; they are used in pharmaceutical companies, forensic laboratories, environmental agencies, and food and beverage industries that have to follow strict quality assurance (QA)/quality control (QC) standards. Dozens of LIMS are available in the market from commercial vendors; they are generally expensive and may not meet the specific needs of a particular project.
Apart from organizing data, a more important reason for laboratory information management systems in lipidomics is to minimize inherent variability in experimental data, as procedures, time, and personnel can all cause significant variation in results. A LIMS should be organized in such a way as to minimize this variability and properly annotate the specific reagents and procedures utilized in a given experiment for future reference.
A LIMS must be usable by lab technicians and other personnel with limited bioinformatics experience. As much as possible, user interfaces must be engineered to provide important informational and contextual pointers for how they are intended to be used. Constraints on entries and readily understandable feedback messages should be provided in meaningful ways. In some cases, there may be no substitute for person-to-person interaction in providing assistance, and a person may be dedicated to providing help to other personnel. These features can foster the goal of achieving widespread user acceptance.
The LIPID MAPS project modified an earlier, highly developed LIMS system that had been constructed for the Alliance for Cell Signaling (AfCS).56 The principles of lipidomics involve many of the same concepts as those associated with the broader category of metabolomics. That is, metabolomics studies often involve inducing perturbations to the ongoing state of living systems and subsequently monitoring changes at specific time points.1b The various lipid species are measured at different time points and quantities are systematically determined. This may be performed within a single laboratory, or a number of laboratories may collaborate in the endeavor. In support of these aims, agreement must be reached among the persons performing the work on the experimental protocols at each step, and protocols and documents must be stored and made available to all. To accomplish transfer, centralized storage, and sharing of data among LIPID MAPS member laboratories, we have developed a LIMS to submit data to a central database and to obtain data from the same source.57 To handle the large amounts of data, a relational database is an essential requirement. The information entered into the system is best entered by individual users or laboratories. A 2- or 3-tier platform may be deployed and data entry forms may be presented in the form of a dedicated program or website.
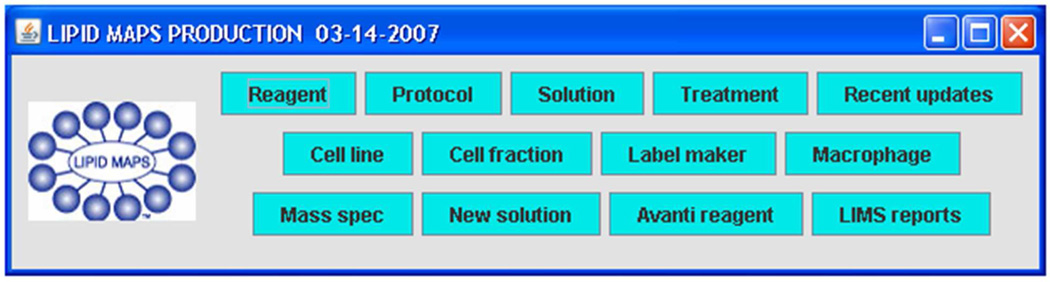
The user interface of the LIPID MAPS LIMS consists of a number of discrete GUIs representing modules of functionality that are accessed from a single main window interface (Figure 13). The entire application is downloaded from a web site as a Java Web Start application at the time of each use. These individual modules allow users to enter information and browse the LIMS database. After entering information, the user clicks a button to send information to a central Oracle database. The LIMS also allows tracking of laboratory materials and protocols via printed labels that may be scanned into modules using barcode readers, thus minimizing typing errors.
Figure 13.
Main user interface of LIPID MAPS LIMS (Reprinted with permission from Reference 57. Copyright 2007 Elsevier Limited).
The LIPID MAPS LIMS is organized around cellular treatments and mass spectrometry (MS) experiments. The LIMS enforces adherence to process controls in the form of exact control of experiments using strict solution and procedural protocols. A protocol ID is required by the majority of modules. The protocol ID refers to a document in the LIMS database that describes a laboratory procedure or solution composition. The user may use one of the protocol documents that are already within the LIMS for this purpose. In addition, any of the participating LIPID MAPS laboratories may upload a new protocol and generate a new protocol ID.
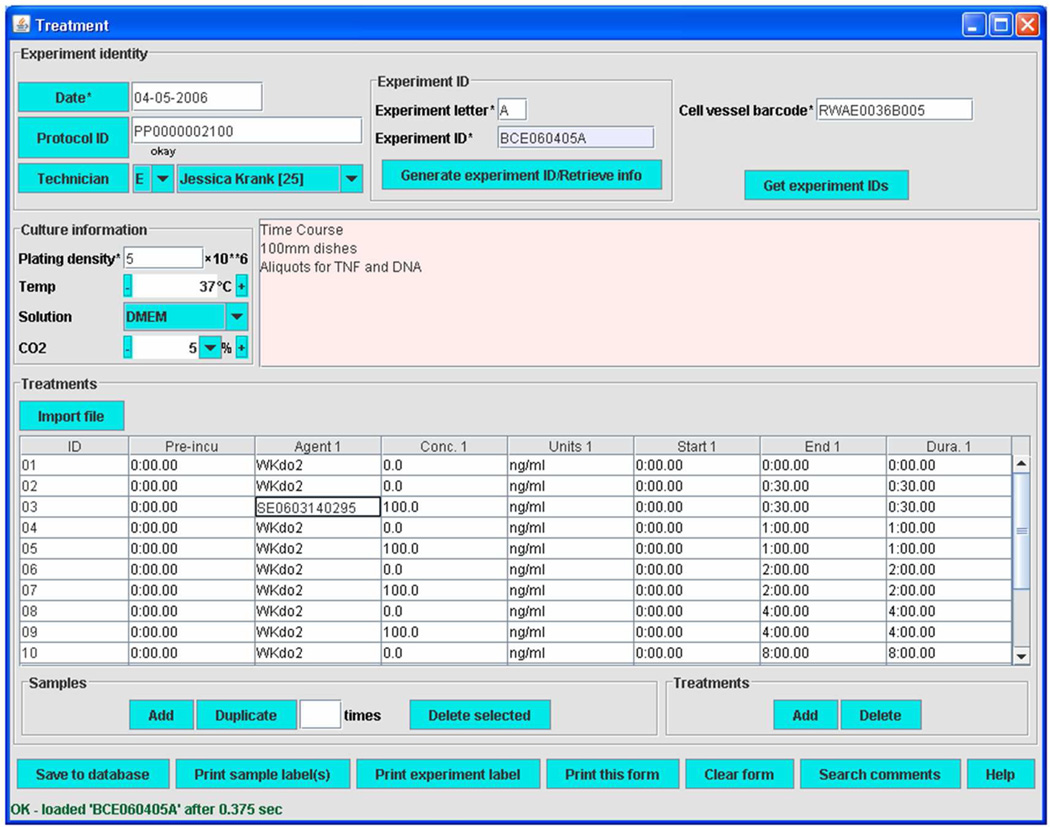
The Treatment module provides the essential lipidomics functionality of the LIMS (Figure 14). Into this form, details of treatment conditions are entered. These include reagent or solution IDs, concentrations, and the start time, end time, and durations of both current treatment and pre-treatment during an experiment with a particular cell preparation. These data are vital for studies of stimulus- and time-dependent alterations to lipid composition. Individual sample IDs are associated with cells receiving different treatments within an experiment.
Figure 14.
Treatment module of LIMS (Reprinted with permission from Reference 57. Copyright 2007 Elsevier Limited).
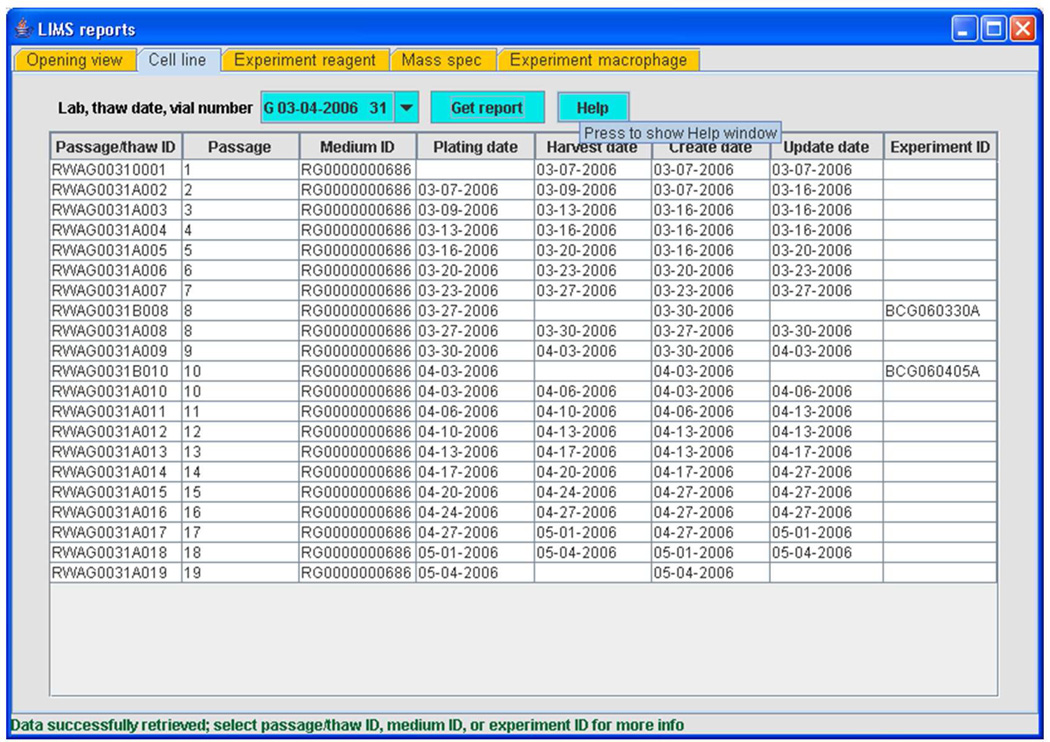
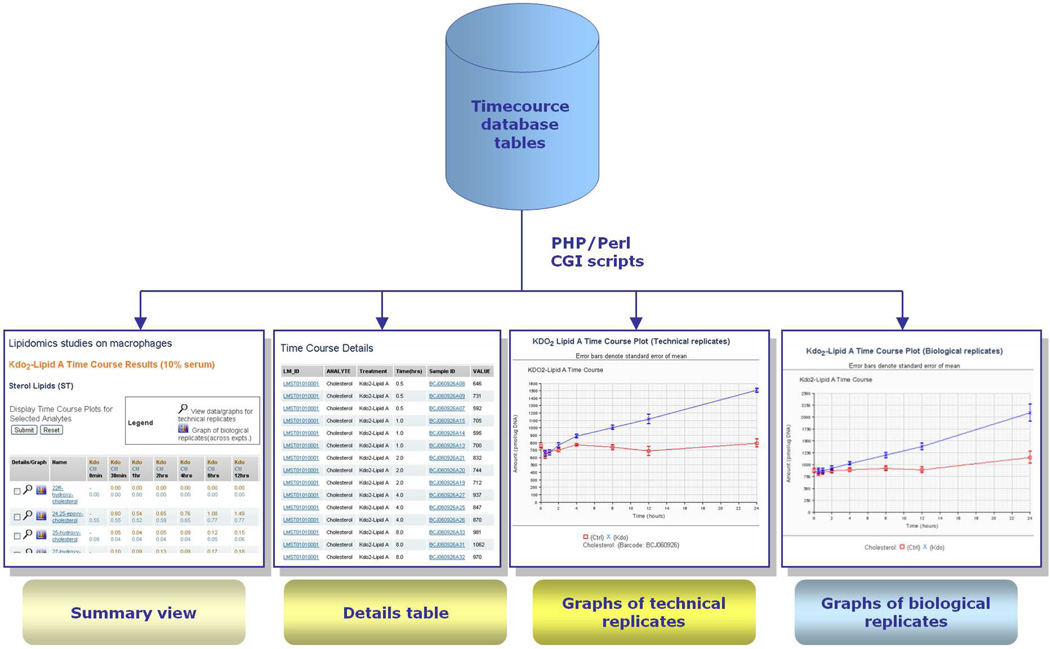
A significant contribution to the functionality in the LIMS arises from close integration of modules. Each module has search functions that search database tables for information entered by that module. Another implementation of searching and user interaction occurs in the case of the Reporter, or the LIMS Reports, module. The Reporter module allows the user to construct high-level reports summarizing overall database content using certain key parameters as search terms. For example, the user may obtain a summary table of cell vessel IDs that originate in a thaw of a particular vial of frozen cells used by a laboratory, along with the protocol ID that was used for thawing and passaging and the ID of any experiment in which a cell passage deriving from that vial was used (Figure 15). The history of a cell line from freezer to experiment is thus obtained.
Figure 15.
LIMS reporter (reporting tool) module (Reprinted with permission from Reference 57. Copyright 2007 Elsevier Limited).
The modules of the LIPID MAPS LIMS were intended to be used sequentially, with database identifiers from previous modules in list format made available to users for insertion into later modules. A flow chart published previously illustrates one potential usage sequence that begins with the Reagent module and ends with the Mass spec module.57
While most of these modules are generic in nature, others have been engineered that are specific for the needs of LIPID MAPS. For example, the Avanti reagent module allows the user to tracks reagents provided by our supplier of molecular standards with the aim of ensuring that materials used for quantitation purposes remained within quality specifications. Among other actions, users can download a current, updated certificate of quality for any lot of material previously shipped to a consortium laboratory. This can be an important consideration when using standards that may possess abbreviated shelf lives. In LIPID MAPS, only Avanti Polar Lipids can input such information, while all laboratories have access to downloading from this module.
On occasion, users may not have time to properly access all modules in succession. For example, the Solution module requires prior use of the Reagent module, along with the Protocol module to insert a protocol on solution composition. This step is of particular importance in mixing internal standards used in mass spectrometry. The New solution module allows bypassing both these modules, with only a brief sketch of solution content required. During later data analysis, performed after the conclusion of an experiment, acceptance or rejection of a questionable datum may hinge on whether the information trail that includes the information entered by either of these modules provides sufficient detail that its reliability can be affirmed. Consequently, the New solution module typically plays a role only in investigations that limited in scope to a specific laboratory.
Analysis and mining of the metadata and associated data obtained with the assistance of this LIMS is conducted off-line at the Bioinformatics core. LIMS metadata and the experimental data described by the metadata are available on the internet for browsing, and are directly linked to a public database of lipid structures that is curated by experts,10 and to a database of proteins known to be involved in lipid metabolism in mice and in humans.47 Both are available from the Lipidomics Gateway website.5 The availability of solution and procedure protocols as well as tools allowing searching and drawing of lipid structures are also featured at this site.
A widely publicized effort to standardize the content of metabolomics experiment informational resources to allow computerized searching has been proposed.58 However, such standardization efforts seem not to have been widely pursued in metabolomics projects, at least partly because of difficulties in adequately comparing experiments performed using disparate technologies, such as NMR spectroscopy and mass spectrometry.59
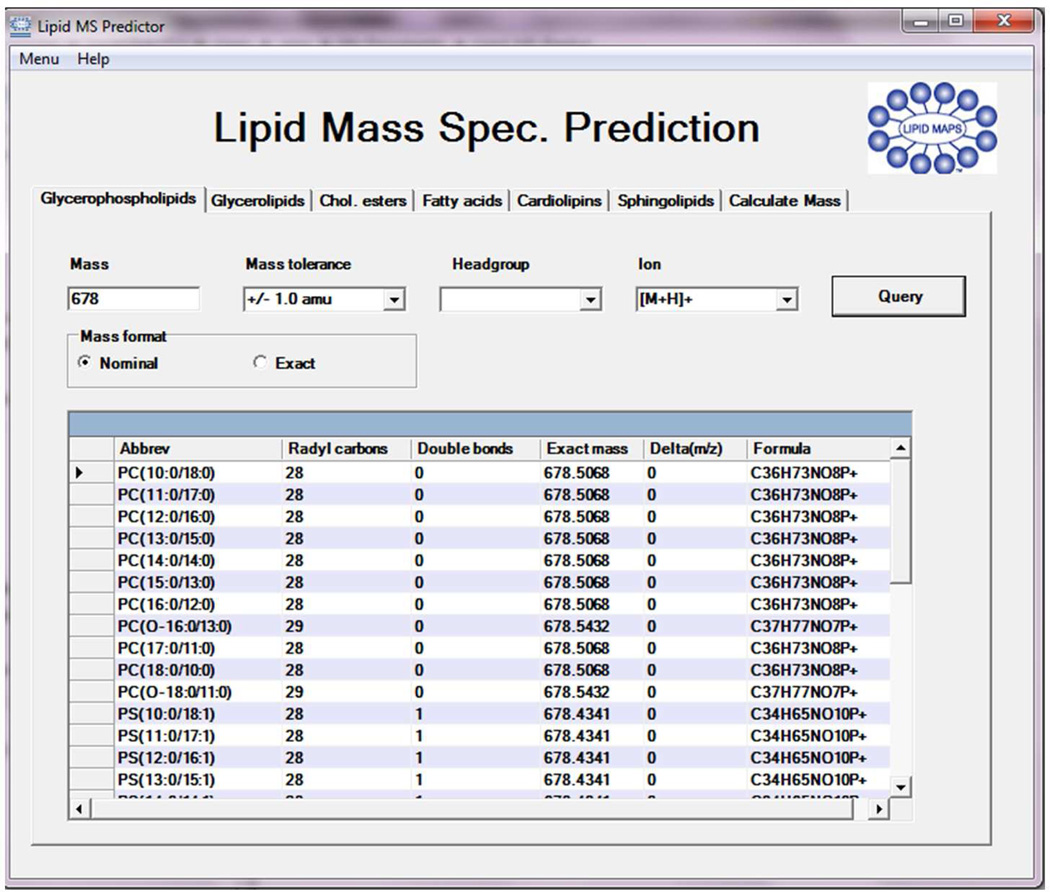
5. Analysis and Presentation of Lipid Mass Spectrometric Data
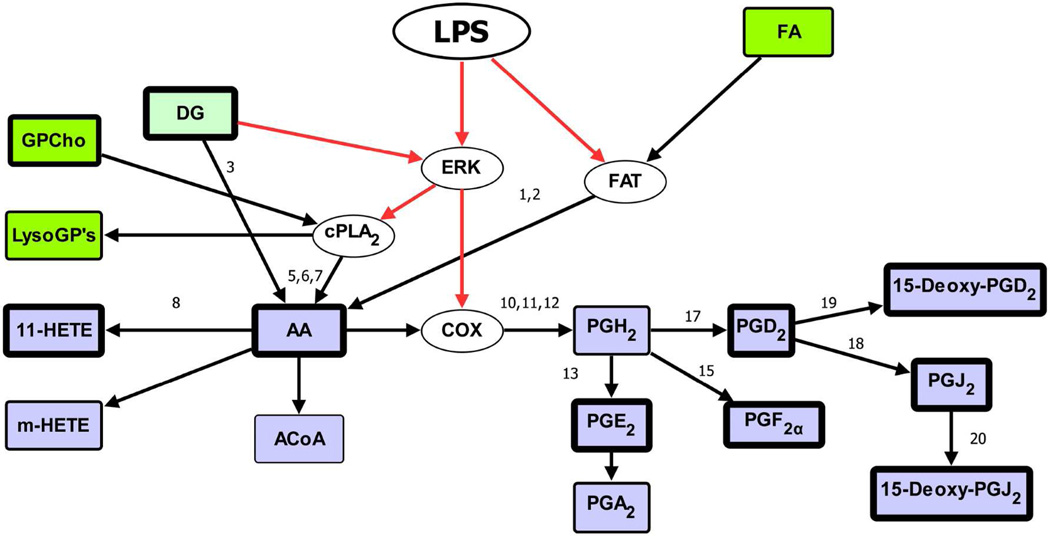
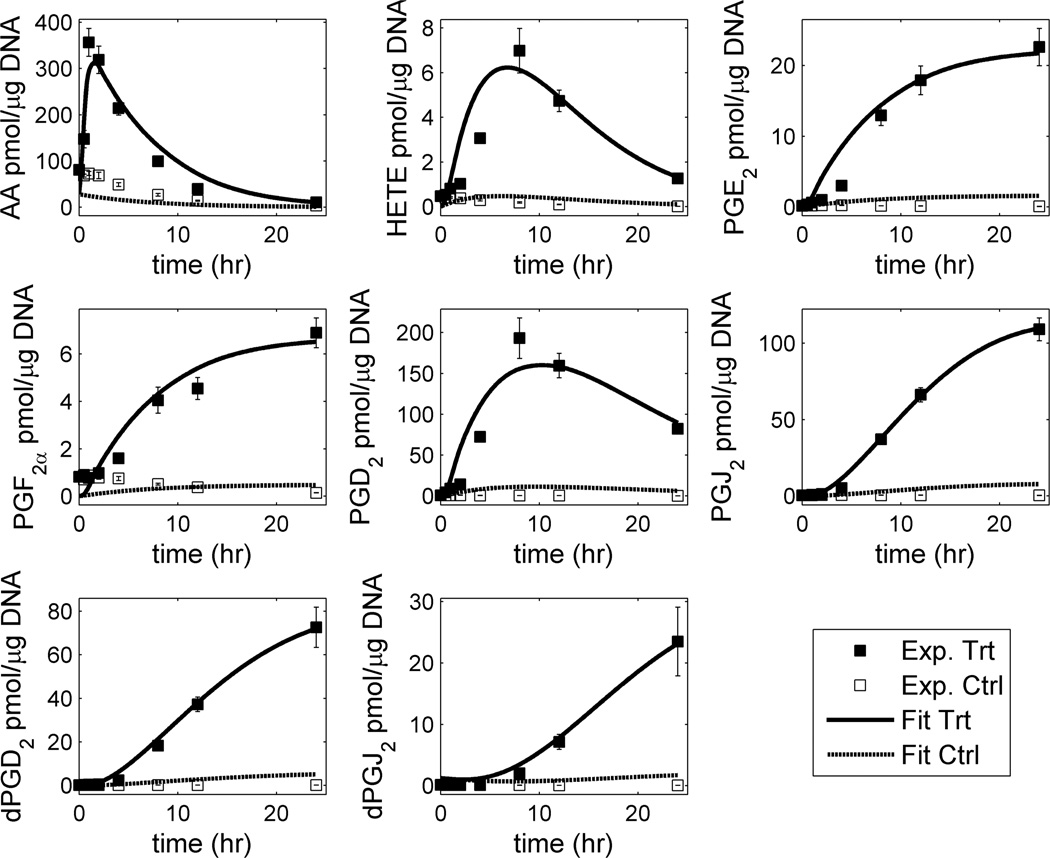
With the availability of sensitive analytical instrumentation such as mass spectrometry, it is now possible to obtain quantitative data on large numbers of lipid species under a variety of experimental conditions. MS methods for the characterization of lipid mixtures have also been published in recent years, most of them centered on the use of electrospray ionization (ESI) MS, atmospheric pressure chemical ionization (APCI) MS and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS.60 Currently, mass spectrometric analysis of lipids is mainly comprised of two complimentary approaches which employ either direct infusion (shotgun lipidomics)61 or use liquid chromatographic separations prior to mass spectrometric analysis (LC-MS). An advantage of shotgun lipidomics is that a mass spectrum displaying molecular ions of individual molecular species of a class of interest can be acquired at a constant concentration of the lipid solution during direct infusion. This unique feature of shotgun lipidomics allows researchers to perform precursor-ion scans of the particular fragment ions and/or neutral loss scans of the interested neutrally lost fragments for identification and quantitation of the individual molecular species of a lipid class or a category of lipid. On the other hand, customized LC-MS techniques tailored to a particular lipid class of interest have the ability to resolve complex lipid mixtures during the LC step, allowing for more reliable identification during the MS step. From a bioinformatics standpoint, MS data analysis can be divided into a number of distinct phases: (a) processing of raw data files which may involve peak averaging, normalization, integration, isotope correction and display of processed spectra; (b) peak identification using algorithms to match lipid ions against databases of known or computationally derived structures; (c) statistical analysis of MS data to quantify significant changes between different samples (lipidomic profiling), between different lipid species in the same sample (correlation analysis) or within the same species over time (temporal analysis); (d) modeling of lipid data onto biological pathways as part of a systems-biology approach.
5.1. MS Analysis Software
In recent years there has been an urgent need for informatics solutions to efficiently process the large amounts of MS data generated by lipidomics experiments and deal with the unique complexities of lipid structures. The number of software packages has expanded considerably over the last 5 years and include a number of freely available applications that are capable of handling multiple tasks in the analysis pipeline (see Table 4). The Java-based MZmine62 provides users with a modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data and is particularly useful for analyzing LC-MS experiments. Another recently released Java application is the Lipid Data Analyzer (LDA)63 in which the authors have developed new algorithms for detection and quantification of minor lipid analytes from LC-MS data. Examples of lipidomics software implemented as Microsoft Excel add-ons are the Fatty Acid Analysis Tool (FAAT)64 and LIpid Mass Spectrum Analysis (LIMSA).65 FAAT has been optimized for analysis of high-resolution MS data generated by Fourier Transform-Ion Cyclotron Resonance (FT-ICR) mass spectra. The LIMSA tool is capable of performing isotopic correction and peak integration as well as mass matching to a user-supplied list of expected lipids. Commercial MS instrument vendors such as AB-SCIEX (www.absciex.com) are developing their own platform-specific lipid analysis approaches such as Lipid Profiler and LipidView66 but suffer from the drawback that they must be used in conjunction with their proprietary Analyst software. A new open source Python programming language67 application called LipidXplorer68 is tailored toward the analysis of data from shotgun lipidomics experiments. LipidXplorer does not have a database of lipid masses for peak identification but instead enables the user to compose queries and constraints for lipid classes of interest using the novel concept of a Molecular Fragmentation Query Language (MFQL). The LIPID MAPS MS analysis tools (http://www.lipidmaps.org/tools/index.html) are a freely-available set of online resources and focus on the simpler task of matching peak lists of precursor ions to predicted structures under a variety of experimental conditions. Certain classes of lipids such as acylglycerols and glycerophospholipids composed of an invariant core (glycerol and head groups) and one or more acyl/alkyl substituents are good candidates for MS computational analysis. These molecules tend to fragment in a predictable fashion in collision-induced experiments leading to loss of acyl side-chains, neutral loss of fatty acids, and loss of water and other diagnostic ions69 depending on the nature of the head group. It is possible to create a virtual database of permutations of the more common side-chains for glycerolipids and glycerophospholipids and calculate “high-probability” product ion candidates in order to compare the experimental data with predicted spectra. The LIPID MAPS group has developed a suite of search tools13 that allows a user to enter an m/z value of interest and view a list of matching structure candidates, along with a list of calculated of neutral-loss ions and other “high-probability” product ions. The MS prediction tools are currently available for a number of different categories of lipids: glycerolipids, glycerophospholipids, cardiolipins, and sphingolipids. In each case, all possible structures corresponding to a list of likely head groups and acyl, alkyl-ether and vinyl-ether chains have been expanded and enumerated by computational methods to generate a table containing the nominal and exact mass for each discrete structure as well as additional ontological information such as formula, abbreviation and numbers of chain carbons and double bonds. This tabular data is then uploaded into category-specific database tables, making it amenable for online querying The MS prediction tools for glycerolipids and glycerophospholipids have been extended by computing production ion masses for commonly observed fragments corresponding to acyl chain ions, neutral loss of acyl chains, loss of water, head group-specific fragmentations and combinations of the above.
Table 4.
Examples of software for performing lipid MS analysis.
| Application | Platform | Comments/URL |
|---|---|---|
| Mzmine | Java | Comprehensive package for lipidomics profiling. http://mzmine.sourceforge.net/ |
| LDA | Java | Novel algorithms for detection of minor lipid species. http://genome.tugraz.at/lda |
| LipidXplorer | Python | Designed for shotgun lipidomics data. Uses novel query language http://sourceforge.net/projects/lipidxplorer/ |
| LIMSA | Excel add-on | Performs mass matching to a user-supplied list of expected lipids http://www.helsinki.fi/science/lipids/software.html |
| FAAT | Excel add-on | Designed for analysis of FT-ICR data. http://www.genomecenter.ucdavis.edu/leary |
| LipidView | Windows | Proprietary AB-SCIEX package used in conjunction with Analyst software. http://www.absciex.com/products/software |
| LIPID MAPS | Web interface | Set of online MS prediction tools tailored to different lipid classes. http://www.lipidmaps.org/tools/index.html |
The MS prediction tools for glycerolipids, cardiolipins and glycerophospholipids accept an m/z value from the user for the precursor ion and have a menu to allow selection of the ion mode ([M+H]+, [M+NH4]+, [M-H]−, etc.). In addition, a mass tolerance window and a head group (in the case of glycerophospholipids) may be specified to limit the number of matches. The list of matches may also be filtered by specifying a particular set of radyl chains (for example, only chains with even numbers of carbon atoms). On completion of a search, the output format (Figure 16) contains a list of structures that (a) satisfy the input criteria and (b) whose side-chains belong to the list of radyl chains used to populate the database. The predicted masses of the fragment ions are computed at run-time by the online application. All entries in the result set are hyperlinked to the structure-drawing application, enabling “on-demand” visualization of the molecular structures. Isotopic distribution profiles for each structure may also be viewed online. The online tools allow batch-mode searches of lists of precursor ions and intensity values which may be copied and pasted into the user interface. Users may perform searches where the matched ions are displayed in “bulk” format (e.g. PE(34:1), TG(54:2)) or as discrete molecular species (e.g. PE(16:0/18:1(9Z)), TG(18:0/18:1(9Z)/ 18:1(9Z))). Additionally, in the case of experimental samples where the relative amounts of the acyl groups of glycerolipids and glycerophospholipids are already known (e.g. from fatty acid methyl ester (FAME) analysis by GC), these data may be entered and a scoring algorithm then ranks the matched species based on the relative abundance of those acyl chains in each lipid. As mentioned above, the current versions of the LIPID MAPS MS prediction tools employ databases of mass permutations for the lipid classes of interest, but it is certainly possible to replace the database with user-specified lists of chains/head groups and perform all mass matching calculations in “real time”. This type of option would be useful in cases where the sample of interest contains lipids with rare or unusual side-chains such as those encountered in bacteria or invertebrates.
Figure 16.
A montage of screen shots showing LIPID MAPS mass spectroscopy tools.
A standalone Windows® application has also been developed (Figure 17) for predicting possible molecular species for a given MS ion. In contrast to the online tools which query a database table of masses corresponding to structural permutations for each lipid category, the standalone application (http://www.lipidmaps.org/tools/index.html) first computes these masses from first principles using a list of commonly occurring side-chains and head groups typically found in mammalian versions of glycerolipids, glycerophospholipids (including cardiolipins) and sphingolipids. This application enables a user to enter the m/z value of an unknown lipid ion and predict the most likely molecular species. There are separate user interfaces for: glycerolipids, glycerophospholipids, cardiolipins, sphingolipids, fatty acids and cholesteryl esters. There is also a user interface to calculate the exact mass of glycerophospholipid and glycerolipid ions with defined side-chains and head groups, along with a display of the isotopic distribution profile.
Figure 17.
A standalone Windows application, LIPID MAPS MS Prediction Tools, for predicting possible molecular species for a given MS ion. The application enables a user to enter the m/z value of an unknown lipid ion and predict the most likely molecular species. It is available for download at: www.lipidmaps.org/tools/index.html.
5.2. Presentation of MS data
The LIPID MAPS consortium has placed an emphasis on online presentation of MS data in order to maximize the level of interactivity with other web-based resources such as lipid/gene databases and experimental protocols. Recent studies by the LIPID MAPS consortium. have quantified over 550 different lipids from mouse macrophage cells2 and almost 600 lipids from human plasma70 using MS and statistical bioinformatics techniques. This ability to simultaneously assess the metabolic dynamics of hundreds of lipid species reveals a wealth of information regarding the cellular lipidome. On a more general scale, the LIPID MAPS consortium has embarked on a time–dependent study of a wide range of lipid classes in mouse macrophage cells, in response to stimulation by a number of agonists such as Kdo2-lipid A (KLA), Adenosine TriPhosphate (ATP) and 25-hydroxy-cholesterol. Large-scale integrated studies have been carried out on both cultured cells such as the RAW264.7 cell-line and on primary cells such as Thioglycolate-Elicited Peritoneal Macrophages (TGEM) and Bone-Marrow Derived Macrophages (BMDM). Quantitative data from these experiments are being used to validate existing lipid networks and elucidate novel interactions. MS quantitative measurements from time course experiments on the various categories of lipids are obtained from the individual LIPID MAPS cores in Microsoft Excel or text format. These heterogeneous formats are then imported into a common data format prior to processing and conversion into Oracle database tables. Data on different cell samples (biological replicates), and/or different MS runs (technical replicates) for each lipid species is consolidated. A middleware layer composed of a web server and PHP/Perl scripting has been deployed used to create a web-based user interface with the MS data stored in an Oracle database. All calculations used to display averages of technical and biological replicates, as well as all Standard Error of Mean (SEM) and standard deviation calculations are performed via Structured Query Language (SQL) code. All online data displays were integrated with the LIMS system (via sample barcodes) and the LIPID MAPS structure database (via LM_ID identifiers where applicable), allowing seamless navigation across both data and metadata. A software drawing component called dynamic graphics (GD, http://www.boutell.com/gd/) was used to generate online graphs “on-the-fly”, in response to user input. The database schema design was optimized for access speed and high data integrity. A set of online query and display tools was developed to allow the end-user to view MS time course data in a number of different formats (Figure 18). These include tabular and graphical displays of data as averages of technical and biological replicates, as well as “drill-down” links to the corresponding LIMS metadata (cell samples) and structure/classification information (analytes). All lipidomic and gene-array data generated by the LIPID MAPS consortium is available in the ‘Resources/Data’ section of the website.5 With a view to enabling lipidomics researchers to identify discrete lipid species, an online library of lipid standards, including tandem mass spectral data generated by the LIPID MAPS core facilities, has been made available on the LIPID MAPS website. This database currently consists of over 550 analytes spanning the 8 major lipid categories with annotated diagnostic product ion identifications and with links to molecular structures and MS acquisition protocols used to generate the raw spectra (http://www.lipidmaps.org/data/standards/index.html).
Figure 18.
Schema for online data display of timecourse results.
6. Models of Lipid Metabolism and Pathways
Pathways may be broadly described as models that characterize movement of material through a network of molecular species and processing steps. They serve as the basis upon which much of the new field of systems biology must build. Many tools have become available over the last 10 years for enabling biological pathway construction.71 Their construction has been stimulated by the growth in information resulting from adoption of new laboratory tools accompanying high-throughput data acquisition, such as mass spectrometry.1b,72 The process of constructing pathways requires ready access to information in the form of experimental data of a quantitative nature. The use of reference model pathways as starting points for new work, as well as inclusion of well-characterized compounds in pathway schemes, are also of great importance.
Lipids play central roles in energy storage, cell membrane structure, cellular communication and regulation of biological processes such as inflammatory response, neuronal signal transmission and carbohydrate metabolism. Organizing these processes into useful, interactive pathways and networks represents a great bioinformatics challenge. The KEGG consortium maintains a collection of manually drawn pathway maps73 representing current knowledge on the molecular interaction and reaction networks, several of which pertain to lipids including fatty acid biosynthesis and degradation, sterol metabolism and phospholipids pathways. Additionally, the KEGG Brite74 collection of hierarchical classifications includes a section devoted to lipids where the user can select a lipid of interest and view reactions and pathways involving that molecule. A number of category-specific lipid pathways have been constructed, notably the SphinGOMAP,75 a pathway map of approximately 400 different sphingolipid and glycosphingolipid species.
In general, the field of metabolomics involves inducing perturbations to the ongoing state of living systems and subsequently monitoring changes to compounds at specific time points. The interactions among components of a pathway are then inferred by a variety of techniques, including metabolite fingerprinting and profiling, and by comparison between organisms that have been genetically perturbed or subjected to altered nutritional states.71c,d
A recent review of pathway editing tools76 points out that a major function of pathway visualization tools is to enable new insights into biology. The choice of a program depends upon the task to be accomplished. For example, a tool may be selected based upon the nature of the data to be examined, or whether mathematical modeling or statistical analysis is to be performed.
An important function of pathway editor programs, in general, is to permit exchange of pathways. Different file format standards exist for this purpose. They include KEGG Markup Language (KGML),77 SBML,53b BioPAX,52 and CellML.78
To construct pathways, the LIPID MAPS Bioinformatics core is using two pathway editing tools: VANTED79 and the LIPID MAPS Pathway Editor, which is based upon a toolkit referred to as the BioPathways Workbench.53a,80 These tools read data from files and/or directly from databases and enable viewing of experimental data in the drawing panel. Most importantly, they enable setting node appearance on an individual basis, thus providing important visual clues as to the roles of the molecular species in the pathway. The Pathway Editor presents measurement data according to experiment and enables detailed viewing of data that may be selected based upon the treatment, reproducibility of the measurements, and other, more qualitative aspects, in the judgment of the user. Both Pathway Editor (Figure 19) and VANTED (Figure 20) have Java-based GUIs providing a comprehensive range of viewing and import/export formats.
Figure 19.
The Pathway Editor showing the mouse arachidonate pathway and timecourse data mapped in heatmap format displayed under the lipid and enzyme (gene) nodes. Samples of RAW264.7 cells (a tissue cell line derived from mouse macrophages) were treated with KLA for times ranging from 0 to 24 hr. Shown are ratios in pmol/ug DNA for metabolites and ratios of normalized intensity for RNA spots with respect to untreated control cells.2,80
Figure 20.
VANTED application showing the mouse arachidonate pathway and timecourse data mapped in heatmap format displayed under the lipid and enzyme (gene) nodes.
Various methods are employed when constructing pathways. For example, a user may position a node in a pathway on the basis of whether the measured data that is presented meets with expectations according to domain knowledge, including early or late responsiveness to a stimulus, and the magnitude of the response. Automated selection and layout, including filtering nodes based on quantitative or qualitative features, are also commonly used. The LIPID MAPS project has manually adapted mouse and human pathways relating to lipid metabolism from various sources and made them available for downloading through the Pathway Editor for viewing and modification.
7. Statistical Analysis, Correlations and Integration of Genomic and Lipidomic Data in Macrophages
From a systems perspective, the genome, metabolome and proteome provide the complete parts-list which can be used to reconstruct networks. However, in a given context, the entire parts-list may not be of relevance. Hence, context-specific data, such as gene-microarray or other types of genomic data, metabolomic data and proteomic data obtained from specific experiments, can be used to obtain a refined (sub)-parts-list using various statistical analyses such as identification of significantly regulated genes and analysis of variance (ANOVA). Such a refined parts-list serves as the starting-point for network reconstruction by integration of experimental data and legacy knowledge.81 The tools for network reconstruction include pathway enrichment analysis for studying pathway-level sub-global changes, motif-discovery for co-regulated genes and correlation analysis for comparing different gene, proteins or metabolites. Nextgen sequencing methods are now beginning to provide very accurate transcript measurements and will no doubt be used in gene expression studies. Once the transcriptomic changes are deciphered from the mapping of sequence tags, strategies as the ones described for analog microarray experiments can be used.82 In this section, various bioinformatics tools used for analyzing different types of data and their integration are discussed. Where appropriate, the data and studies from mouse macrophage RAW264.7 cells in LIPID MAPS have been used for illustrative purposes.
7.1. Identification of Significantly Regulated Genes
Gene microarray experiments provide a cost-effective way of studying the whole-genome level response of the cell or tissue system. While there are about 30,000 genes in mouse and human, in any experimental/treatment condition, only a small fraction of these genes show significant changes as compared to the normal (un-treated) condition. The naïve approach to identify which genes are significantly regulated would be to use a cut-off on the ratio of the intensities for treatment versus control conditions. However, due to the differences in the hybridization efficiency of different probes for the genes, a wide-range of image intensity values are obtained across the whole genome. Coupled with the measurement noise and other effects, the large intensity range makes it difficult to use a single threshold for different genes on the ratio of the intensities between the treatment and control conditions. Hence, in the last fifteen years, several approaches have been developed for the analysis of transcriptomic data to account for the wide intensity range across the gene-chip. Variance modeling with prior exponentials (VAMPIRE),83 CyberT,84 and LInear Models for MicroArray data (LIMMA)85 techniques are commonly used to identify the significantly regulated genes. VAMPIRE involves modeling the global variance structure of array data in the context of a Bayesian framework. CyberT employs statistical analyses based on regularized t-tests that use a Bayesian estimate of the local variance among gene measurements. Both VAMPIRE and CyberT are available as web application. LIMMA uses linear models for the analysis of differentially expressed gene and is available as a part of Bioconductor project (http://www.bioconductor.org/) in R programming language (http://www.r-project.org/). These methods are able to detect gene expression changes with only two array replicates. In the analysis of LIPID MAPS microarray data in RAW264.7 cells upon KLA and Compactin (a HMG-CoA reductase inhibitor86) treatment, CyberT was applied.2 Figure 21 shows the number of significantly regulated (up - or down-regulated) genes at various time-points. In this analysis, a gene is identified as significantly regulated if its p-value is less than 0.01. Generally, multiple testing correction methods such as false discovery rate and Bonferroni correction are used for further refinement87. In this dataset, compactin showed mild transcriptomic response. Bonferroni and FDR corrections were too stringent for this dataset and resulted in no significantly regulated genes. Thus to find the top significantly regulated genes, no further correction was applied. For further analysis, one may also use a cut-off of 2.0 on the fold-change to generate a refined list of significantly regulated genes.
Figure 21.
Time course of the number of regulated genes in RAW264.7 cells stimulated with KLA, Compactin and KLA+Compactin.
Ultimately, the utility of any combination of microarray platform and analytical method is determined by how well statistical predictions are matched by experimental validation. For expression analysis, Quantitative - Polymerase Chain Reaction (QT-PCR) assays are performed. LIPID MAPS investigators have several hundred validated PCR primers for genes that are of particular interest to them. These primers are used to validate results of microarray experiments. While not comprehensive, sufficient probes are available to determine whether different analysis methods provide reliable results. Validation of microarray experiments in RAW264.7 cells for several genes using QT-PCR is discussed in a recent study.2
7.2. Analysis of Variance (ANOVA)
T-test is sufficient to compare between two conditions, namely, control or untreated samples and stimulated or treated samples. Hence, methods such as VAMPIRE, CyberT and LIMMA can identify the differentially regulated genes between two conditions corresponding to a single treatment. However, in the case of multiple treatment experiments or experiments at several time points, the above approaches cannot delineate the effect of different treatments on a particular gene or other measurements. It is necessary to separate the effect of different treatments or the time-component to draw rational conclusions from the data. This task is accomplished by the analysis of variance (ANOVA) approach which has been widely used to deconvolute the effect of different treatments. In ANOVA, the observed variance in the measured data is partitioned into the effect of individual factors or treatments.88 If necessary, terms corresponding to the interactions among different factors can also be included in the variance partitioning model. Similar to statistical tests such as t-test (used by VAMPIRE, CyberT, etc.), in ANOVA, a p-value is assigned to the effect of different factors included in the model. ANOVA can be used to factor out the significance of different treatments or time-effects on any experimental measurements such as genes,89 proteins90 and metabolites.2 ANOVA can also be used for the identification of significantly regulated genes as well89 because for the case of one-factor with only two possible values for the factor (e.g., control vs. single treatment), ANOVA (called 1-way ANOVA) and unpaired t-test are equivalent although this cannot account for the effect of intensity range on the measure of variance, a hall-mark of techniques such as CyberT and VAMPIRE. In LIPID MAPS studies, ANOVA was applied to transcriptomic and lipidomic data from RAW264.7 cells upon KLA and Compactin treatment to separate the effect of KLA and Compactin on the genes or lipids.2 In a previous study relating to network reconstruction, ANOVA was applied on the measurement of phosphorylation states of signaling proteins and cytokines to find putative lumped connections from the stimuli to the signaling pathways or cytokine regulation.90 Another study suggests that there is potential for further analysis of the ANOVA results by performing multivariate analyses such as principal component analysis (PCA) on the interaction terms for different factors91 to find out if such interactions may be significant under certain conditions. Bi-plots from PCA may also aid in visualization and interpretation of results. More recently, the combined approach of ANOVA-PCA has gained considerable attention from the statisticians, especially when three or more factors need to be analyzed.92 Their utility for analyzing data with only two treatments may be limited.
7.3. Gene Ontology and Pathway Enrichment Analysis
The differentially-regulated features obtained from any statistical test must be interpreted biologically. In this direction, gene ontology (GO) and pathway enrichment analysis is prevailing significantly. These analyses identify which processes and pathways are affected significantly as compared to what would be expected by chance in the experiment. There are many tools available as software or web applications. For example, AmiGO,93 Goby (part of VAMPIRE suit)83a and Database for Annotation, Visualization and Integrated Discovery (DAVID)94 are available as web application. SubpathwayMiner is available as a part of Bioconductor project in R programming language.95 This database-driven application stores annotation data from several sources, namely, GO, KEGG, TRANSFAC96 and Biocarta.97 In addition, it can be easily updated with user-defined annotation lists.83a Most of these applications use hypergeometric distribution or Fisher exact test to compute the enrichment likelihoods.
Goby was used extensively in the analysis of gene expression data in RAW264.7 macrophages.2 Some of the results for the microarray data from RAW264.7 cells in the KLA/Compactin study are listed in Table 5 which shows that majority of the genes from the KEGG Toll-like Receptor (TLR) pathway are upregulated. Other pathways relevant to inflammation, such as Jak-Stat, NF- κB and cytokine-cytokine receptor interaction KEGG pathways are also significantly enriched.
Table 5.
Global KEGG pathways (mouse) enrichment analysis of RAW264.7 cells treated with KLA, Compactin or KLA+Compactin. Number of genes regulated (UP/DOWN) are listed for each timepoint for the three experiments (p <= 0.05).
| S. No |
KEGG PathID |
Pathway Name |
Total Genes |
Ligand | Reg. Type |
30 min |
1 hr |
2 hr |
4 hr |
8 hr |
12 hr |
24 hr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 04060 | Cytokine-cytokine receptor interaction | 237 | KLA | UP | 10 | 24 | 40 | 56 | 61 | 53 | 39 |
| DOWN | 1 | 2 | 7 | 11 | 8 | 9 | 13 | |||||
| Compactin | UP | - | - | - | - | 0 | 2 | 13 | ||||
| DOWN | - | - | - | - | 7 | 4 | 3 | |||||
| KLA+Compactin | UP | 10 | 25 | 32 | 51 | 62 | 51 | 40 | ||||
| DOWN | 0 | 1 | 4 | 8 | 10 | 11 | 13 | |||||
| 2 | 04620 | Toll-like receptor signaling pathway | 96 | KLA | UP | 9 | 17 | 25 | 30 | 33 | 27 | 24 |
| DOWN | 1 | 2 | 7 | 4 | 5 | 2 | 8 | |||||
| Compactin | UP | - | - | - | - | 0 | - | 7 | ||||
| DOWN | - | - | - | - | 3 | - | 1 | |||||
| KLA+Compactin | UP | 9 | 19 | 19 | 31 | 32 | 26 | 25 | ||||
| DOWN | 0 | 3 | 5 | 3 | 4 | 3 | 6 | |||||
| 3 | 04010 | MAPK signaling pathway | 261 | KLA | UP | 17 | 26 | 30 | 34 | 51 | 45 | 45 |
| DOWN | 1 | 6 | 8 | 19 | 21 | 13 | 15 | |||||
| Compactin | UP | - | - | - | - | - | - | 15 | ||||
| DOWN | - | - | - | - | - | - | 1 | |||||
| KLA+Compactin | UP | 14 | 27 | 27 | 32 | 57 | 42 | 48 | ||||
| DOWN | 0 | 6 | 7 | 13 | 12 | 10 | 12 | |||||
| 4 | 04920 | Adipocytokine signaling pathway | 64 | KLA | UP | 5 | 11 | 13 | 11 | 13 | 13 | 17 |
| DOWN | 0 | 0 | 3 | 5 | 2 | 3 | 3 | |||||
| Compactin | UP | - | - | - | - | - | - | - | ||||
| DOWN | - | - | - | - | - | - | - | |||||
| KLA+Compactin | UP | 3 | 11 | 10 | 11 | 15 | 14 | 15 | ||||
| DOWN | 0 | 1 | 0 | 4 | 2 | 1 | 3 | |||||
| 5 | 04621 | NOD-like receptor signaling pathway | 59 | KLA | UP | 8 | 15 | 21 | 23 | 26 | 22 | 20 |
| DOWN | 0 | 1 | 1 | 1 | 2 | 1 | 2 | |||||
| Compactin | UP | - | - | - | - | - | - | - | ||||
| DOWN | - | - | - | - | - | - | - | |||||
| KLA+Compactin | UP | 7 | 15 | 16 | 25 | 24 | 21 | 20 | ||||
| DOWN | 0 | 1 | 1 | 0 | 0 | 1 | 2 | |||||
| 6 | 04630 | Jak-STAT signaling pathway | 145 | KLA | UP | 5 | 11 | 26 | 34 | 37 | 31 | 31 |
| DOWN | 1 | 2 | 6 | 7 | 1 | 2 | 5 | |||||
| Compactin | UP | - | - | - | - | - | 1 | - | ||||
| DOWN | - | - | - | - | - | 2 | - | |||||
| KLA+Compactin | UP | 4 | 10 | 19 | 28 | 42 | 35 | 30 | ||||
| DOWN | 0 | 4 | 2 | 6 | 2 | 1 | 4 | |||||
| 7 | 04514 | Cell adhesion | 146 | KLA | UP | - | - | 16 | 22 | 24 | 24 | 26 |
| DOWN | - | - | 1 | 6 | 8 | 6 | 8 | |||||
| Compactin | UP | - | - | 2 | - | - | 3 | 10 | ||||
| DOWN | - | - | 0 | - | - | 0 | 1 | |||||
| KLA+Compactin | UP | - | - | 9 | 24 | 27 | 25 | 30 | ||||
| DOWN | - | - | 1 | 6 | 7 | 8 | 8 | |||||
7.4. Sequence Motif Discovery
Identification of transcription factor binding sites (TFBSs) or motifs has been a challenge in the area of bioinformatics. The de novo discovery of the motifs requires the availability of TFBS databases and state of the art software tools. JASPAR98 and TRANSFAC96 have been good resources for obtaining the Position Weight Matrices (PWMs) for several hundreds of transcription factors (TFs). There have been two approaches in the use of alignment for motif discovery. The first approach compares the TFBSs alignment on the promoter sequence with the alignment on random sequence based on Adenine(A), Thymine (T), Cytosine (C) and Guanine (G) composition of the genome.99 The second approach compares the enrichment of TFBSs alignment in target set with background set.100
Based on the second approach, a novel computational method to identify regulatory motifs in co-regulated genes was developed. The method builds on previous efforts to find DNA motifs that discriminate between the foreground (i.e., co-regulated) and background promoter sequences, allowing to harness both positive and negative binding information. The algorithm attempts to find a motif that has maximal enrichment in foreground sequences relative to background sequences. Enrichment is found by considering the overlap of genes in the foreground with genes that contain the motif, using the hypergeometric distribution to calculate the probability of this overlap by chance. The algorithm works by exhaustively checking short motifs of a given length for enrichment between foreground and background promoter sequences, keeping the highest scoring motifs. The highest scoring motifs are then used as seeds to a greedy optimization algorithm that creates degenerate probability matrices that maximize the enrichment of the motif in the positive set of sequences. This formulation requires surprisingly few assumptions, offering a natural description of motif quality that is applicable to a variety of problems such as finding binding sites that are associated with changes in gene expression or Chromatin immunoprecipitation (ChIP)-chip results. An application of this method to identify enriched motifs in the promoters of genes induced by KLA in RAW264.7 cells from the time course experiment is shown in Figure 22.
Figure 22.
Identification of NF-κB, ISRE and AP-1/ATF/CREB binding sites in KLA-stimulated genes in RAW macrophages. Blue color indicates significant enrichment of the motif in promoters of positively regulated genes for each time point.
Three of the most highly enriched motifs identified by this method correspond to binding sites for transcription factors that were previously established to mediate responses to TLR4 activation; NF-κB, Interferon Response Factors (IRFs), and Activator Protein (AP)-1/ Activating TF (ATF)/cAMP response element-binding (CREB) family members. Furthermore, many of the genes identified as having NF-κB, interferon-responsive sequence element (IRSE), or AP-1/CREB sites were shown to be direct targets of these TFs by conventional assays, providing one line of validation for this method. In contrast, conventional motif discovery methods failed to identify NF-κB or AP-1/ATF-1/CREB binding sites in transcriptionally activated genes. One of the interesting features of the enrichment plot illustrated in Figure 22 is the temporal windows in which IRF3/ISRE motifs and AP-1/ATF/CREB motifs appeared. These data have implications for understanding how the complex transcriptional response to TLR4 activation is regulated in a time-dependent manner. Several other sequence motifs are identified by this motif method in the set of Lipopolysaccharide (LPS) responsive genes, and provide the basis for a series of new studies to identify roles of other classes of transcription factors in regulating the genome-wide response to TLR4 signaling.
7.5. Processing and Analysis of Proteomic Data
A novel, MS-based approach for the relative quantification of proteins, relying on the derivatization of primary amino groups in intact proteins using isobaric Tags for Relative and Absolute Quantitation (iTRAQ) was used to measure relative protein intensities in RAW264.7 cells in the presence or absence of KLA. The technique is based on chemically tagging the N-terminus of peptides generated from protein digests that were isolated from different samples, e.g., KLA-treated cells and control cells101. The two labeled samples are then combined, fractionated by nanoLC, and analyzed by tandem mass spectrometry. Database searching of the peptide fragmentation data allows identification of the labeled peptides and hence the corresponding proteins. Due to the isobaric mass design of the iTRAQ reagents, differentially labeled proteins do not differ in mass; accordingly, their corresponding proteolytic peptides appear as single peaks in MS scans. Fragmentation of the tag attached to the peptides generates a low molecular mass reporter ion that is unique to the tag used to label each of the digests. Measurement of the intensity of these reporter ions enables relative quantification of the peptides in each digest and hence the proteins from which they originate. The iTRAQ method was used to measure relative protein levels in three samples of RAW264.7 cells treated with KLA for 24 hours and three corresponding control cell samples. Protein KLA/Control ratios were then compared to messenger ribonucleic Acid (mRNA) ratios generated from gene array experiments on RAW264.7 cells. Statistical analyses using covariance plots of the 24-hour protein ratio data with mRNA ratios at multiple time points established a maximal correlation at 18 hours, as would be expected when one considers the time-lag between transcription and translation (Figure 23).
Figure 23.
Covariance plot between 24 hour protein data from iTRAQ measurements and gene array data at various time points (only proteins with K/C ratio >1.5 were chosen).
A high correlation between mRNA and protein KLA/Control ratios was observed for those proteins whose ratios were increased or decreased 2-fold or more. A disadvantage of the “shotgun” LC-MS approach used in these iTRAQ experiments is the lack of sensitivity for detection of low abundance proteins. A tagged tryptic digest of the entire cell extract is applied to the LC column and, due to sample complexity and a large range in protein concentrations, only about 25% of proteins (as compared to gene array experiments) are detected. A consequence is that many enzymes involved in lipid metabolism are not detected by the iTRAQ method. This disadvantage could be overcome by employing additional purification steps prior to LC-MS, such as subcellular fractionation and affinity chromatography. In addition, this methodology is capable of detecting proteins with post-translational modifications, providing another level of information with regard to function and activity.
7.6. Correlation Analysis
Pearson correlation is widely used to find which variables show similar changes across different experiments or time-points.102 Pearson correlation coefficients can also be used to perform hierarchical clustering103 and generate correlation networks.104 Such networks may capture some aspects of the causality among variables or factors. A more elaborate discussion on the issue of correlation versus causality is presented elsewhere.105 Pearson correlation has also been used, at least conceptually, in various ways in data-driven network reconstruction106 using approach such as least-squares or principal component regression90 and partial least-squares.107 Correlation analysis has been applied to various biological system to elucidate how different molecular components function in a network and to understand their phenotypic similarities and differences. Some examples are succinctly described below.
Fiehn and Weckwerth108 have presented an interesting review on how the data on gene, protein and metabolite measurements are correlated resulting in complex networks. A related min-review is presented by Steuer et al.109 They have also used metabolite-metabolite correlation analysis-based clustering and principal component analysis (PCA) to develop and visualize data-derived metabolic networks.110 The visualization approach also includes a clique finding algorithm for improved interpretation. Recently, they have used PCA and partial least squares analysis for feature extraction to differentiate between the responses of different metabolites in rice to a bacterial pathogen.111 Schmitt et al.112 have used correlation between time-lagged data on genes to develop gene interaction networks. They have used gene expression time-course data under different light conditions and were able to find several gene groups containing light-stimulated gene clusters, such as Synechocystis sp. photosystems I and II and carbon dioxide fixation pathways. Numata et al.113 have used mutual information as a non-linear correlation metric. They have shown that the mutual information-based analysis was able to uncover some non-linear relationships undetectable by the Pearson coefficient-based analysis in a data set from Arabidopsis thaliana. Fukushima et al.104a have also used correlation networks and a graph-clustering approach to find modules using data from 3 Arabidopsis genotypes, namely, Col-0 wild-type, methionine over-accumulation 1, and transparent testa4 in samples of roots and aerial parts.
To analyze the LIPID MAPS data, Pearson correlation was used to find the similarity between two time-courses.102 In RAW264.7 cell experiments, the time-course for gene data or lipid data consisted of 8 time points (including the value at t = 0 hr). Correlation value can be thought of as the cosine of the angle between the normalized time-course curves (z-scores). Some details previously used in such analyses are presented below.
Gene and lipid data
For the genes, the ratio of the value under the treatment condition to the value for the control condition was used at each time point. In order to compute the correlation between the time course for the lipids and the time courses for the genes in the same pathway (curated list of genes for each lipid pathway as listed on the LIPID MAPS website (http://www.lipidmaps.org/pathways/vanted.html) or the list of genes from KEGG pathways) the ratios to control values were used for the lipids as well. The time points for the lipids and the genes were [0 0.5 1 2 4 8 12 24] hr.
Consideration of time-delay
Since it is the enzyme or the protein level that may affect the time course of the lipid, in the absence of specific knowledge for individual genes, a time-delay of 4 hrs corresponding to the time taken for mRNA translation, post-translational modification and protein translocation was used for gene data.
Weighted correlation
Since the measurements are taken at non-uniform time-intervals (more frequently at the beginning and less frequently at the later time-points), a weighted correlation in which the time-points were weighted proportional to the time-interval is more appropriate than the raw correlation described above. Assuming a weight vector, W = [w1, w2, w3, w4, w5, w6, w7, w8,], the weighted correlation was computed as follows:2
First the weighted mean, weighted standard deviation and weighted z-score (n = 8, the number of time-points) were computed, and then the weighted dot-product was computed:
| (1) |
In the above, for convenience, the weight-vector W was normalized to unit sum as at the beginning so that the division by is not explicitly required in the above expressions.
The above equations are easily extended for two data matrices X and Y with several rows in each where rows correspond to different gene or lipids and the columns correspond to different time points as in the above equations.
The data values to be used with the weighted correlation
Linear interpolation of data in each time interval was used as an approximation to the scenario where data was measured at equal time intervals. Hence the mean value of the data in the time interval (i.e., (xk + xk+1))/2 for the kth time-interval) was used.
Lipid and Gene categories
Lipid-gene correlation was performed for five different lipid pathways, namely eicosanoids in the media, sphingolipids, sterols, glycerolipids, glyrecophospholipids and unsaturated fatty acids inside the cells. For the LIPID MAPS specific curated gene list, the pathways, used included: eicosanoid biosynthesis, sphingolipid biosynthesis, cholesterol biosynthesis, glycerolipid/glycerophospholipid biosynthesis and fatty acid biosynthesis. In each selected pathway, only those genes which show significant regulation (differential expression) at one or more time points, computed using CyberT 84 were used. More details can be found elsewhere.2
Display of the Data and Correlations
For the display of the data and the correlation, correlation-based hierarchical clustering 103 was used to layout the variables (lipids and/or genes) so that the rows corresponding to the variables with high correlation were displayed near each other in the heat map for the data. The Statistics/Bioinformatics toolbox of Matlab®114 was used to perform the computations. Using the hierarchical clustering tools, clusters were identified (distance-method = user-specified weighted correlation (Eq. 1), linkage-method = average, cut-off criterion = distance; cut-off = 0.75). It can be noted that a correlation range of [−11] corresponds to the equivalent distance range of [2 0] (d = 1-r). So, the cut-off of 0.75 on the distance corresponds to a cut-off of 0.25 on the correlation. When applied on the lipid-gene data sets, each cluster may have one or more of genes and lipids. Some clusters may include no genes or no lipids (but not empty).
The interesting clusters are those which have at least one gene and one lipid since they indicate that such genes and lipids are changing together and serve as a target for investigating causal relationships. In the case of lipid-gene correlations, the information flow was from the genes (proteins/enzymes) to the lipids (after accounting for the time-delay). Using this strategy it would be possible to generate correlation-based directed graphs. The links between two lipids or two genes would then be bidirectional.
For illustrative purposes, the heat map for the data for the eicosanoids (measured in the media) is shown in Figure 24. The prostaglandin lipids (e.g., prostaglandin (PG) E2 (PGE2), PGJ2 and PGF2α) and the prostaglandin synthase genes (Ptgs2, Ptges) changed in a similar manner resulting in strong correlation between them. The mechanistic relationship between these genes/enzymes and their corresponding products is shown in the pathway diagram of Figure 20; e.g., production of PGE2 was catalyzed by the enzyme corresponding to the gene prostaglandin E synthase (Ptges). Similarly, the correlation analysis between various sterols and the genes for cholesterol biosynthesis suggested that its precursors and its several derivatives co-vary with the mRNA of HMG CoA reductase (Hmgcr) and cholesterol 25-hydroxylase (Ch25h). Correlation analysis between the sphingolipids and related genes has shown that several sphingolipids are co-clustered with the important genes in the pathway including serine palmitoyltransferase (Sptlc1, Sptlc2) and ceramide synthases (CerS) Lass4 and Lass62. At a semi-systemic level, these results had suggested that the joint-correlation analysis can potentially uncover such underlying physical mechanisms.
Figure 24.
Heat map for the data for eicosanoids (measured in the media) and eicosanoid biosynthesis related genes (Reprinted with permission from Reference 2. Copyright 2010 American Society for Biochemistry and Molecular Biology). The four panels correspond to: (1) Data based on ratio of values for treatment with KLA to the values for control experiments, (2) ratio of compactin treatment to control, (3) ratio of treatment with both KLA and compactin to control, and (4) ratio data from (1)–(3) combined. The data in each row is scaled and offset by the t = 0 value. The names of the lipids/genes displayed on the y- and/or x-axis are listed in different colors to indicate the clusters.
8. Quantitative Kinetic Models of Lipid Metabolism
All biological processes are inherently dynamical systems. Thus the use of systems biology approaches is becoming common in the study of metabolic and other networks to elucidate their functions and roles in human health and diseases. Towards this end, several software have been developed which allow various types of modeling and analysis, such as steady state analysis, kinetic modeling, parameter estimation, sensitivity analysis, metabolic control analysis, stochastic simulation and consideration of spatial variation (partial differential-equation-based modeling). An extensive list of such software is available at the SBML website.53 Some of them are: CellML (http://www.cellml.org/,115 JSim (http://nsr.bioeng.washington.edu/jsim/docs/overview.html), VCell (http://www.nrcam.uchc.edu/;116), Systems Biology Workbench (http://sys-bio.org/;117), COPASI (http://www.copasi.org/;118) and MCEll (http://www.mcell.cnl.salk.edu/;119). Their salient features are summarized in Table 6. All these software have some capability to plot and visualize the results of simulation. This comparison, although simple and concise, can help the modeler choose the appropriate software application. Majority of the software allow the modeling of signaling and metabolic pathways as a biochemical reaction system. Most of them have SBML import/export capability although the information related to pathway/network visualization may be lost during SBML export, a common problem relating to the interoperability of most such software applications.
Table 6.
A representative list of different software for quantitative simulation and analysis of biological systems.
| Software | Tasks allowed (All these software have capability of plotting/visualization of results) | ||||||
|---|---|---|---|---|---|---|---|
| Deterministic simulation | Stochastic simulation | Sensitivity analysis |
Parameter scan/ estimation |
Sub-model import |
|||
| Well-mixed system (ODEs) |
Spatial- modeling (PDEs) |
Well mixed system |
Spatial (reaction- diffusion) |
||||
| CellML/ OpenCell | √ | √* | √ | ||||
| JSim | √ | √ | √ | √ | |||
| Virtual cell | √ | √ | √ | √ | √ | ||
| COPASI | √ | √ | √ | √ | |||
| MCell | √(3D) | ||||||
| SBW | √ | √* | √ | √ | √ | ||
Abbreviations: SBW: Systems Biology Workbench.
This capability is available by interfacing with other software.
Among many metabolic pathways, there has been tremendous progress in modeling of glucose metabolic networks. Several researchers have developed genome-scale metabolic networks for different organisms such as Saccharomyces cerevisiae, Escherichia coli and human.120 There have been efforts in the modeling of signaling pathways as well. Some of the examples include modeling of Mitogen Activated Protein (MAP) kinase pathway,121 regulation of cell-cycle122 and calcium signaling.123 Some of the above approaches are also being used to study plant metabolism. Fiehn et al. have worked extensively on metabolite profiling and their analysis for Arabidopsis thaliana.124
Due to the complexity of lipid metabolism, and the paucity of data for its many metabolites, there are only a few models of lipid metabolism available in the literature. For example, Callender et al. have developed a model of diacylglycerol dynamics in the RAW264.7 macrophage.125 Yang et al. have developed a model of arachidonic acid (AA) metabolism in human polymorphonuclear leukocytes.126 Only two models of sphingolipid metabolism are found in the literature, one by Alvarez-Vasquez et al.127 for yeast and one by Henning et al.128 (cell system was not specified). All of these models suffer from the unavailability of sufficiently large datasets. Though there are several enzymes for which activity data is/are available (Table 7), their number is still significantly smaller than numbers of enzymes in the pathways.
Table 7.
A representative list of lipid metabolism related enzymes for which kinetic data is available in BRENDA database.136
| KEGG Pathway | Enzyme Name (EC Number) | Specific Activity (µmol/min/mg) |
|---|---|---|
| Arachidonic acid | prostaglandin-D synthase (5.3.99.2) | 14.9–434.0 |
| metabolism | leukotriene-A4 hydrolase (3.3.2.6) | 0.185-0.49 |
| Sphingolipid | 3-dehydrosphinganine reductase (1.1.1.102) | 0.000121 |
| metabolism | serine C-palmitoyltransferase (2.3.1.50) | 0.000044 |
| ceramide glucosyltransferase (2.4.1.80) | 0.0000082 | |
| sphinganine kinase (2.7.1.91) | 2.0E-6-2.4E-4 | |
| ceramide kinase (2.7.1.138) | 4.0E-9-1.41E-6 | |
| Steroid biosynthesis | sterol O-acyltransferase (2.3.1.26) | 2.22E-5-1.56E-4 |
| Glycerolipid metabolism | glycerol-3-phosphate O-acyltransferase (2.3.1.15) | 1.3E-4-0.192 |
| diacylglycerol O-acyltransferase (2.3.1.20) | 3.0E-4-0.169 | |
| Glycerophospholipid | choline kinase (2.7.1.32) | 0.000139 |
| metabolism | acetylcholinesterase (3.1.1.7) | 7700 |
Towards a comprehensive study of lipid metabolism, the LIPID MAPS consortium69 has quantified the global changes in lipid metabolites ("lipidomics"). Using LIPID MAPS data, context specific pathway models were developed for several lipid categories by integrating the legacy knowledge and experimental data on lipid changes in macrophages upon KLA stimulation.4a,129 A central question that can be addressed through quantitative measurements of lipids as a function of time is the flux of metabolites through the cellular network. This is possible as the rate of change of the metabolite concentrations, which can be computed directly from the time-course data, is related to its fluxes corresponding to the different reactions. This enables the development of kinetic models for several lipid pathways. Once the kinetic model is developed and the rate-parameters are estimated, the reaction fluxes (and their relative distribution in different branches of the network) can be computed. It is useful to note that in most kinetic modeling studies on biochemical pathways, generic values for the rate parameters are used because system- and context-specific values are lacking. As we have illustrated in a previous review,105 lack of such specific rate-parameter values is a major challenge in computational systems biology. However, in the LIPID MAPS study, due to the availability of a large amount of data (about 5-data points per unknown rate-constant), the rate-constants were estimated with good accuracy129. A matrix-based approach and optimization was used to estimate the rate constants using experimental data and known network topology from the literature while ensuring that the rate constants are positive. Modeling of the eicosanoid pathway is presented as an example. More details can be found elsewhere.129 The network model used, which includes only the measured metabolites, is presented in Figure 25.
Figure 25.
LPS/ KLA stimulated eicosanoid metabolism and signaling pathway. The numbers above the arrows are reaction numbers (Table 8) and default degradation reactions are not labeled. Black lines represent lipid metabolism and red lines indicate signaling pathways. Metabolites and enzymes are represented as rectangular and ovals boxes, respectively. The measured and unmeasured metabolites are differentiated by thick and thin borders, respectively. Purple color is used for eicosanoids and green for glycerolipids and glycerophospholipids (Reprinted with permission from Reference 129. Copyright 2009 Elsevier Limited).
8.1. Kinetic Model and Parameter Estimation
A kinetic model was developed for the simplified lipid network involving AA metabolism.129 The reaction rates were described by linear or law of mass action kinetics. Thus, the flux expressions obtained from this scheme were linear in rate parameters and nonlinear in metabolite concentrations. The matrix-based approach to estimate the rate constants is described below in terms of the reaction numbers labeled in Figure 25 and listed in Table 8. The metabolite concentrations were known and the rate parameters were unknown. Hence, the ordinary differential equations (ODEs) describing the rate of change of concentrations of metabolites can be rearranged in a matrix format as shown in Eq. 2 for [PGH2] and [PGD2].
Table 8.
The estimated parameter values for eicosanoids model (Reprinted with permission from Reference 129. Copyright 2009 Elsevier Limited).
| No. | Reactions | Parameter Names |
Values |
|---|---|---|---|
| 1 | a[LPS] FA → AA | k1 | 355.637 |
| 2 | FA→ AA | k2 | 10−15 |
| 3 | DG → AA | k3 | 10−15 |
| 4 | b,cAA → | k4 | 10−15 |
| 5 | a[DG] GPCho → AA | k5 | 10−15 |
| 6 | a,d[LPS] GPCho → AA | k6 | 0.330 |
| 7 | GPCho → AA | k7 | 10−15 |
| 8 | AA → HETE | k8 | 0.007 |
| 9 | HETE → | k9 | 0.187 |
| 10 | e[DG] AA → PGH2 | k10 | 0.024 |
| 11 | [LPS] AA → PGH2 | k11 | 0.111 |
| 12 | AA → PGH2 | k12 | 0.098 |
| 13 | PGH2 → PGE2 | k13 | 0.204 |
| 14 | PGE2 → | k14 | 10−15 |
| 15 | PGH2 → PGF2a | k15 | 0.061 |
| 16 | PGF2a → | k16 | 10−15 |
| 17 | PGH2 → PGD2 | k17 | 3.116 |
| 18 | PGD2 → PGJ2 | k18 | 0.054 |
| 19 | PGD2 → dPGD2 | k19 | 0.029 |
| 20 | dPGD2 → | k20 | 0.014 |
| 21 | PGJ2 → dPGJ2 | k21 | 0.034 |
| 22 | dPGJ2 → | k22 | 0.116 |
[DG] and [LPS] indicates the effect of signaling (molecules) in the reaction.
X → means default degradation of the metabolite X.
Unit of first order reaction is 1/hr.
Unit of second order reaction is 1/hr when it involves either FA or LPS as one of the metabolites as we have used scaled profile for these variables.
Unit of second order reaction is µg DNA/(Ratio Int * hr) when it involves DG as one of the metabolites.
where the rate constants ki (i = 10, 11, 12, 13, 15, 17, 18, 19) are as defined in Table 8.
| (2) |
X is completely known. The left hand side of the equations (matrix Y) was computed using discretization and the experimental data. To avoid singularity during matrix inversion and to require positive values of the rate parameters, a constrained least-squares approach was used (Matlab®114 function lsqlin). The parameter values thus obtained were used as good initial values for further refinement by using generalized constrained nonlinear optimization (Matlab® function fmincon). The objective function for use with fmincon was:
| (3) |
where, nt is the number of time-points and nsp is the number of species. The first-term represents the fit-error between the experimental and predicted concentrations and the second term represents the fit-error between their experimental and predicted derivatives. Different weights (wi) can be assigned to these two terms to improve the fit. The initial concentrations of the metabolites were also optimized in a narrow range around the experimental values. When data on more than one condition was available, then all the data was used to compute the fit-error by simulating the model several times individually and minimizing the objective function collectively.
Table 8 lists the reactions and the corresponding estimated reaction-rate parameters included in the model. Figure 26 shows the simulation results.129 For most time points, the difference between the predicted and experimental data was within the standard-error of the mean (SEM) (Figure 26). Thus, good fit to the data from both treatment and control conditions suggested that the topology of the simplified network was correct and captured the important metabolic and signaling effects. The model was validated by excluding the data on one of the intermediate metabolites from objective function minimization. The rate-parameters were estimated and the predictions were compared with the actual experimental data. There are two intermediate metabolites present in the network: PGD2 and PGJ2. The validation was performed on both of the metabolites and satisfactory results were obtained. Parametric sensitivity analysis was also performed.129 In short, for each parameter and each metabolite, monotonic increase, decrease or no change was observed depending upon the respective location of parameter and the metabolite chosen in the network. The change in the parameters belonging to the upper part of the network produced a larger change in almost all metabolites as compared to those for the parameters belonging to lower part of the network.
Figure 26.
Simulation of kinetic modeling of simplified lipid network: fit of the predicted response (control and treatment with KLA) to the corresponding experimental data. In the legend, ‘Ctrl’ refers to control and ‘Trt’ refers to KLA treatment of RAW264.7 cells. The error-bars shown on the experimental data are standard-error of mean (Reprinted with permission from Reference 129. Copyright 2009 Elsevier Limited).
8.2. Time-Scale Analysis
Time-scale characterization is important to understand the metabolite dynamics and its response time.129 The analysis for the AA metabolism model was performed by computing eigen-values and eigen-vectors of the Jacobian matrix of ordinary differential equations at the steady-state conditions. Time-scale analysis has been used previously to find the slow and fast modes in nonlinear dynamical systems.130 Characteristic time-constants (time-scales) are the inverse of the eigen-values since the dynamic response of the system for small perturbation from the steady state consists of exponential terms such as exp(−λt), λ being an eigen-value.131 As a consequence, if all the eigen-values have negative real-part then the dynamic system would be stable and also, if some of the eigen-values are complex then the system would exhibit sustained or un-sustained oscillatory response for small perturbations. In the time-scale analysis of the AA metabolism, the eigen-values were split into three broad ranges. For each eigen-value, the metabolites with substantial contribution to the corresponding eigen-vector were identified. Depending upon the eigen-values and metabolites significantly contributing to the corresponding eigen-vectors, these metabolites were divided into three categories as listed in Table 9. Medium time scale metabolites go up and return to the basal levels in 24 hr time; however the slow time scale metabolites show monotonic increases up to 24 hr (Figure 26).
Table 9.
Results of Eigen-value based time-scale analysis of the metabolites (Reprinted with permission from Reference 129. Copyright 2009 Elsevier Limited).
| Fast (~1 hr) | Medium (~10 hr) | Slow (~50 hr) |
|---|---|---|
| PGH2 | AA | PGE2 |
| PGD2 | PGF2α | |
| HETE | PGJ2 | |
| dPGD2 | ||
| dPGJ2 |
8.3. Comparison of Rate Parameters for the Enzymes
The values for the rate constant for the enzymes Cyclooxygenase (COX) reported in the literature were based on in-vitro measurements with partially purified proteins.132 Thus, it was assumed that the literature values represented its basal activity and compared these activities (flux through the enzyme) with predicted activities of these enzymes in the “control” simulation. The computed value (10−13 µM/min/cell) and reported value (10−14 µM/min/cell) for COX are within one order-of-magnitude.133
8.4. Stable Isotope Labeling for Improved Characterization of Fluxes
Stable isotope labeling of one key metabolite in a given metabolic pathway introduces point (species)-wise perturbation in the network. For system identification purposes, labeling is equivalent to exciting the system which helps decipher the network topology. Stable isotope labeling can be used to differentiate, in the production of metabolites in the downstream parts of the above network (Figure 25), the contribution of the metabolite that is labeled from the contribution by other metabolites. The propagation network of the labeled metabolite is less complex than the original propagation network. Thus, using labeled data, the reaction rate parameters can be estimated with better accuracy. Labeled data helps identify alternate/new pathways.134 Further, it provides a more direct approach of computing fluxes and estimating the split ratios at branch points. Mass balance can be used to detect the leakage through unmodeled pathways and potential connections between two different parts of the pathway can be detected. Deconvoluting the spectra in the context of lipid metabolites to identify peaks has been discussed previously.134a The main source of complexity in modeling labeled data is the presence of feedback loops.135 When reactions result in elongation or breakdown of one or more chains of labeled carbon atoms or result in other structural changes then labeling of multiple carbon atoms changes even if all the carbon atoms in the original labeled metabolite were 13C. These complexities need to be taken into account in using labeled data in kinetic modeling studies.
9. Perspective and Future of Lipidomics
Although the field of lipidomics is relatively young, quantitative estimation of lipids over a wide dynamic range is already possible and comparative analysis of lipid compositions and concentrations between normal and pathological tissues is beginning to yield rich insights into lipid-associated mechanisms of pathology. With next generation mass spectrometers, methods for quantitative identification of lipid molecular species and context-specific association of lipid species with proteins involved in biosynthesis and metabolism and the concomitant genes encoding these proteins, several lipid specific pathways will be reconstructed in the future. These pathways will help delineate physiological function of cells and tissues, in conjunction with associated cellular signaling and transcriptional changes, in normal and pathological conditions. The early efforts serve as a harbinger for the integration of lipids as important molecular players in physiology and pathophysiology leading to integrative systems biology approaches to describing function.
The challenges for lipidome bioinformatics and systems biology are manifold. With increasing ability to catalog lipids, the number and diversity of lipid species will increase dramatically. The classification of these lipids, their organization and most importantly characterizing their functional role will form a significant part of the lipidomics future. Most importantly, the quantification of lipids in a contextual manner, i.e. identifying small differences between lipids under two different conditions, normal and pathological or untreated and treated tissues, will form a significant challenge even with the availability of standards. Characterization of lipids in vivo is a daunting task and despite advances in imaging mass spectrometry, image and data analysis to quantify specific lipids will require novel methods.
To study differences between normal and pathological samples it is not adequate to merely measure and quantitate lipid species. It will be important to decipher and study the biochemical pathways associated biosynthesis and metabolism of lipids and to study the fluxes associated with lipid changes with disease or treatment. The fluxes will also reveal hitherto uncharacterized pathways. Isotopomer experiments are one route to deciphering the unknown pathways. Using labeled data, the reaction rate parameters can be estimated with better accuracy. Labeled data helps identify alternate/new pathways.134 Further, it provides a more direct approach of computing fluxes and estimating the split ratios at branch points.
Proteins, genes and lipids act in combination in pathways to create biological function. The key challenge for systems biology lies in the integration of proteomics, genomics, regulatory genomics and metabolomics data to provide a context-specific systems-level perspective on phenotypic responses of living systems to stimuli. Identifying all the parts lists, such as the cell or tissue-wide lipidome, is only a first step and needs to be significantly extended to identify interactions, mechanisms, and pathways. While traditional statistical methods can be applied to each type of data, e.g. gene expression, proteomics or lipidomics, the integration across these data to provide mechanistically meaningful models continues to be a difficult challenge. Correlation methods and analyses suggest mechanistic connections, but have no foundation for causal relationships. Use of prior knowledge can provide useful constraints in developing network models, but also has the potential to bias the analyses of data to yield false connections and pathways. Dynamic measurements, when analyzed in context, can provide causal links, but for these to be accurate the density of measurements across time needs to be very high. Synergistic measurements of all components and “ome-integrated” reconstruction of pathways is essential for providing a mechanistic model. Even then, this model needs to have the dynamic element, which can only be obtained by time-varying measurements at necessary and sufficient granularity. Once such a dynamic model is created, the scope exists for quantitative modeling using physical principles to obtain predictive input-response relationships.
In developing computational models of biological processes, there is a growing realization that given the enormous complexity of biochemical interactions and paucity of data (as compared to how much data is required to uniquely identify the networks and parameters), unique networks would be seldom obtained in data-driven network identification. When manageable, this degeneracy in network reconstruction is not necessarily bad because it provides new and alternate hypotheses that can be further tested by knockout and pathway inhibition (intervention) studies, thus leading to the refinement of the network models. To date, most approaches to incorporate prior knowledge into network modeling are based on Bayesian network or its variants. Can prior knowledge be systematically included in deterministic approaches (e.g. state-space formulation) as well? In all likelihood, the answer is yes. Such a framework must be able to operate on the network topology and the parameters simultaneously. It will require the ability to manipulate the topology, the complex expressions for the postulated cause-effect relationships and the corresponding model parameters simultaneously. It is imperative that such an approach will require nonlinear optimization methods. Given the complexity of nonlinear optimization, stochastic-search based approaches are expected to be more practical for such an application. 105
It is anticipated that in the coming decades several models of lipid metabolic and signaling networks will be developed and systems biology approaches will provide predictive approaches to input-response relationships in cellular function. The tools of informatics and systems biology will be valuable in this research landscape.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Collaborative Grant U54 GM69338-04 LIPID MAPS (SS), the National Institute of Diabetes and Digestive and Kidney Diseases NIDDK Grant P01-DK074868 (SS), the National Heart, Lung and Blood Institute (NHLBI) grant 5 R33 HL087375-02 (SS), National Science Foundation (NSF) grant DBI-0641037 (SS), the NSF collaborative grant DBI-0835541 (SS), the NIH/NIGMS grant GM078005-05 (SS) and the NSF collaborative grant STC-0939370 (SS). We wish to thank the LIPID MAPS core directors Drs. H Alex Brown (Vanderbilt University), Edward A Dennis (University of California, San Diego), Christopher K Glass (University of California, San Diego), Alfred H Merrill Jr. (Georgia Institute of Technology), Robert C Murphy (University of Colorado, Denver), Christian RH Raetz (Duke University), David W Russell (University of Texas Southwestern Medical Center, Dallas), Walter A Shaw (Avanti Polar Lipids, Inc., Alabaster, AL), Michael S VanNieuwenhze (Indiana University), Stephen H White (University of California, Irvine), Nicholas Winograd (Pennsylvania State University) and Joseph L Witztum (University of California, San Diego). We would like to thank and acknowledge Dr. Xiang Li for generating the figure for motif enrichment (Figure 22).
Biographies
Shankar Subramaniam

Shankar Subramaniam is the Joan and Irwin Jacobs Endowed Chair in Bioengineering and Systems Biology and a Professor of Bioengineering, Bioinformatics and Systems Biology, Cellular and Molecular Medicine, Chemistry and Biochemistry, and Nanoengineering at the University of California, San Diego (UCSD). He was the founding director of the Bioinformatics and Systems Biology Program at UCSD. He received his B.S. and M.S. degrees from Osmania University in India and a Ph.D. in Chemistry from Indian Institute of Technology Kanpur in 1982. He was a Professor of Biophysics, Biochemistry, Molecular and Integrative Physiology, Chemical Engineering and Electrical and Computer Engineering at the University of Illinois at Urbana-Champaign prior to moving to UCSD. He has written over 200 articles and is on the editorial board of several journals. His awards include a Genome Technology All Star Award, Smithsonian Institution Award for Innovation, and a Faculty Research Excellence Award. Research in his laboratory spans several areas of bioinformatics, systems biology and medicine. In bioinformatics he is involved in developing novel strategies for identifying protein interaction networks, and identification of functional networks in cells. In systems biology he is involved in deciphering mammalian cellular networks from high throughput and phenotypic data and in developing strategies for modeling cellular signaling networks. In systems medicine, he is interested in mapping the circuitry of cells to mechanisms and phenotypes in physiology and pathology and to develop quantitative models of cellular pathways.
Eoin Fahy

Eoin Fahy has a B.Sc. in Biochemistry from University College Galway and a Ph.D. in Chemistry from the University of British Columbia, specializing in the structure elucidation of marine natural products. He completed a post-doctoral appointment at the Scripps Institution of Oceanography in University of California, San Diego (UCSD). He has over 15 years experience in the biotechnology industry in the areas of organic chemistry, drug target discovery, molecular biology, proteomics, genomics and informatics. He joined the LIPID MAPS consortium in 2003 and serves as project coordinator for the Bioinformatics core at UCSD. His current interests include development of a lipid classification system as a member of the International Lipids Classification and Nomenclature Committee (ILCNC), design and development of database infrastructures for lipidomics, development of mass spectrometry software for lipid research, design of novel lipid structure drawing tools and development of integrated pathway tools and resources.
Shakti Gupta

Shakti Gupta received his bachelor degree in Chemical Engineering from the India institute of Technology, Kanpur, India, in 1999. He joined the Department of Chemical Engineering, University of Florida, in 2000 and received his Ph.D. in 2005. He worked at the Center for Disease Control, Atlanta for one year before joining the University of California, San Diego (UCSD) in 2006 as a postdoctoral fellow. Presently, he is working as a Research Scientist at the San Diego Supercomputer Center and the Department of Bioengineering at UCSD. His research interests include bioinformatics analysis of high-throughput data, data-driven network reconstruction and nonlinear modeling of metabolic and signaling pathways.
Manish Sud

Manish Sud is currently involved with LIPID MAPS project at San Diego Supercomputer Center (SDSC)/University of California, San Diego (UCSD). His interests include research, development and application of computational discovery tools. He has been working on development and usage of computational discovery tools at various small and large software development and drug discovery companies for over two decades.
Robert W. Byrnes

Robert W. Byrnes is a Research Programmer in the San Diego Supercomputing Center and the Department of Bioengineering, University of California at San Diego. He maintains databases for the LIPID MAPS Pathway Editor program and the LIPID MAPS LIMS, writes software code and provides user support for these programs. Previously, he worked on a grid portal interface for TeraGrid applications. He has a B.S. in Physics from the University of Rochester, New York, an M.S. in Natural Sciences and Mathematics from SUNY-Buffalo, and a Ph.D. in Cellular and Molecular Biophysics from the Roswell Park Division, SUNY-Buffalo. Dr. Byrnes has also held a position of Staff Scientist at the Department of Chemistry, UW-Milwaukee, where he worked on metal biochemistry and oxidative DNA damage.
Dawn Cotter

Dawn Cotter is a Senior Computational Scientist at the San Diego Supercomputer Center, University of California, San Diego (UCSD). She received a BS in Economics from the University of Illinois, Urbana-Champaign (UIUC) in 1989, and entered the PhD program in Molecular and Integrative Physiology at UIUC in 1993. Dawn earned her MS in Physiology in 1996; her thesis title was "Dynamic Simulation Modeling of Changes in Human Body Composition". While pursuing her PhD, she also worked part time for the Automated Learning and Education Groups at the National Center for Supercomputing Applications (NCSA), UIUC. In 1999, she accepted a full-time position with Dr. Shankar Subramaniam at NCSA and moved to UCSD with Dr. Subramaniam that same year to continue work on the Biology Workbench. Dawn is interested in simulation modeling, data visualization, and metabolism, particularly as it pertains to regulation of human body composition.
Ashok Reddy Dinasarapu

Ashok Reddy Dinasarapu received his B.Sc in Chemistry and Biology from Andhra Loyola College, Vijayawada, India in 1997. Dr. Dinasarapu received Master's degrees, M.Sc in Biochemistry from University of Hyderabad, Hyderabad, India, in 2000 and M.Tech in Biotechnology from Anna University, Chennai, India, in 2002. Then Dr. Dinasarapu joined Shantha Biotechnics Pvt. Ltd., Hyderabad, India and worked on gene cloning, expression and purification of single chain antibodies. Later in 2003, Dr. Dinasarapu moved back to the Department of Biochemistry at the University of Hyderabad for his doctoral research where he focused on the bioinformatics analysis of the promoter sequences of eukaryotes and graduated in 2007. Since 2008, Dr. Dinasarapu is a postdoctoral researcher in the Department of Bioengineering at the University of California, San Diego. His research interests include bioinformatics analysis of signal transduction and regulation and semantic integration and visualization of the life sciences data.
Mano Ram Maurya

Mano Ram Maurya completed his B.Tech. in Chemical Engineering from IIT Bombay in 1998, M.E. in Chemical Engineering from City College of New York in 1999 and Ph.D. in Chemical Engineering from Purdue University in 2003. Dr. Maurya was a postdoctoral researcher for three years in the Department of Bioengineering and the San Diego Supercomputer Center at University of California, San Diego (UCSD). Then he worked in the Department of Bioengineering as an Assistant Scientist from October 2006 to November 2010. Since then, Dr. Maurya is a Research Scientist at the San Diego Supercomputer Center and the Department of Bioengineering at UCSD. In 2005, Dr. Maurya received the Best Paper Award jointly with his Ph.D. advisor Dr. Venkat Venkatasubramanian and co-advisor Dr. Raghunathan Rengaswamy for their paper published in the journal of Engineering Applications of Artificial Intelligence in 2004. In August 2011, he joined the editorial board of ISRN Biophysics. Dr. Maurya’s current research interests include the study of complex biochemical processes and pathways using systems engineering/biology and bioinformatics approaches and their applications to biomedicine.
Contributor Information
Shankar Subramaniam, Email: shankar@ucsd.edu.
Eoin Fahy, Email: efahy@ucsd.edu.
Shakti Gupta, Email: shaktig@gmail.com.
Manish Sud, Email: msud@sdsc.edu.
Robert W. Byrnes, Email: rbyrnes@sdsc.edu.
Dawn Cotter, Email: dcotter@sdsc.edu.
Ashok Reddy Dinasarapu, Email: adinasarapu@ucsd.edu.
Mano Ram Maurya, Email: mano@sdsc.edu.
References
- 1.(a) Watson AD. J. Lipid Res. 2006;47:2101. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]; (b) Wenk MR. Nat. Rev. Drug Discov. 2005;4:594. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 2.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman GT, Reichart D, Merrill AH, Sullards MC, Wang E, Murphy RC, Raetz CR, Garrett T, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. J. Biol. Chem. 2010;285:39976. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wymann MP, Schneiter R. Nat. Rev. Mol. Cell Biol. 2008;9:162. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 4.(a) Gupta S, Maurya MR, Merrill AH, Jr, Glass CK, Subramaniam S. BMC Syst. Biol. 2011;5:26. doi: 10.1186/1752-0509-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Garcia J, Shea J, Alvarez-Vasquez F, Qureshi A, Luberto C, Voit EO, Del Poeta M. Mol. Syst. Biol. 2008;4:183. doi: 10.1038/msb.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LIPID MAPS - Nature Lipidomics Gateway. doi: 10.1021/ed200088u. www.lipidmaps.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AD. Oxford dictionary of biochemistry and molecular biology. Oxford: Oxford University Press; 2000. Rev. ed. [Google Scholar]
- 7.(a) Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. J. Lipid Res. 2005;46:839. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]; (b) Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. J. Lipid Res. 2009;50(Suppl):S9. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahy E, Cotter D, Sud M, Subramaniam S. Biochim. Biophys. Acta. 2011 doi: 10.1016/j.bbalip.2011.06.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Caffrey M, Hogan J. Chem. Phys. Lipids. 1992;61:1. doi: 10.1016/0009-3084(92)90002-7. [DOI] [PubMed] [Google Scholar]; (b) LIPIDAT website. www.lipidat.tcd.ie.; (c) Watanabe K, Yasugi E, Oshima M. Trends Glycosci. Glyc. 2000;12:175. [Google Scholar]; (d) LIPID BANK website. www.lipidbank.jp.; (e) The Lipid Library website. http://lipidlibrary.org/; (f) Cyberlipid Center website. www.cyberlipid.org.
- 10.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. Nucleic Acids Res. 2007;35:D527. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Functional Glycomics Gateway. www.functionalglycomics.org. [Google Scholar]
- 12.(a) Weininger D. J. Chem. Inf. Comp. Sci. 1988;28:31. [Google Scholar]; (b) SMILES website. www.daylight.com/smiles/index.html.
- 13.Fahy E, Sud M, Cotter D, Subramaniam S. Nucleic Acids Res. 2007;35:W606. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Structure Data File (SDF) format. www.symyx.com/solutions/white_papers/ctfile_formats.jsp. [Google Scholar]
- 15.CPAN - Comprehensive Perl archive network. www.cpan.org. [Google Scholar]
- 16.ChemAxon website. www.chemaxon.com.
- 17.Jmol website. http://jmol.sourceforge.net.
- 18.CamridgeSoft website. www.cambridgesoft.com.
- 19.(a) Cahn RS, Ingold C, Prelog V. Angew. Chem. Int. Edit. 1966;5:385. [Google Scholar]; (b) Prelog V, Helmchen G. Angew. Chem. Int. Edit. 1982;21:567. [Google Scholar]
- 20.PHP: Hypertext Preprocessor us2.php.net
- 21.The IUPAC International Chemical Identifier (InChi) website. doi: 10.1186/s13321-015-0068-4. www.iupac.org/inchi. [DOI] [PMC free article] [PubMed]
- 22.(a) GenBank website. www.ncbi.nlm.nih.gov/genbank.; (b) Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. Nucleic Acids Res. 2008;36:D25. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swiss-Prot protein knowledgebase website. www.expasy.ch/sprot.
- 24.Ensemble website. www.ensembl.org.
- 25.Ranzinger R, Herget S, von der Lieth CW, Frank M. Nucleic Acids Res. 2011;39:D373. doi: 10.1093/nar/gkq1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Human Metabolome Database. www.hmdb.ca. [Google Scholar]
- 27.(a) DrugBank. www.drugbank.ca. [Google Scholar]; (b) Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. Nucleic Acids Res. 2011;39:D1035. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Chen X, Ji ZL, Chen YZ. Nucleic Acids Res. 2002;30:412. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhu F, Han B, Kumar P, Liu X, Ma X, Wei X, Huang L, Guo Y, Han L, Zheng C, Chen Y. Nucleic Acids Res. 2010;38:D787. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(a) Chemical Entities of Biological Interest (ChEBI) http://www.ebi.ac.uk/chebi/ [Google Scholar]; (b) de Matos P, Alcantara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Nucleic Acids Res. 2010;38:D249. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) ChemBank. http://chembank.broadinstitute.org/ [Google Scholar]; (b) Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA. Nucleic Acids Res. 2008;36:D351. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) PubChem. http://pubchem.ncbi.nlm.nih.gov/; (b) Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. Nucleic Acids Res. 2009;37:W623. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: Integrated Platform of Small Molecules and Biological Activities. In: Ralph AW, David CS, editors. Annu. Rep. Comput. Chem. Vol. 4. Elsevier; 2008. p. 217. [Google Scholar]
- 32.Irwin JJ, Shoichet BK. J. Chem. Inf. Model. 2005;45:177. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ChemSpider. http://www.chemspider.com/ [Google Scholar]
- 34.Chemical Abstracts Service. http://www.cas.org/ [Google Scholar]
- 35.eMolecules. http://www.emolecules.com/ [Google Scholar]
- 36.Beilstein database. www.reaxys.com. [Google Scholar]
- 37.KEGG LIGAND Database. http://www.genome.jp/kegg/ligand.html. [Google Scholar]
- 38.Integrated Enzyme Database. http://www.ebi.ac.uk/intenz/ [Google Scholar]
- 39.Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. Nucleic Acids Res. 2002;30:402. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PDBeChem. http://www.ebi.ac.uk/msd-srv/msdchem/cgi-bin/cgi.pl. [Google Scholar]
- 41.ChEMBL. https://www.ebi.ac.uk/chembldb/ [Google Scholar]
- 42.URL to retrieve all LMSD structures from PubChem database. www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pcsubstance&term=LipidMAPS[sourcename]
- 43.Oracle website. www.oracle.com.
- 44.JME sketcher. www.molinspiration.com/jme/index.html. [Google Scholar]
- 45.Maglott D, Ostell J, Pruitt KD, Tatusova T. Nucleic Acids Res. 2007;35:D26. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. Nucleic Acids Res. 2004;32:D115. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cotter D, Maer A, Guda C, Saunders B, Subramaniam S. Nucleic Acids Res. 2006;34:D507. doi: 10.1093/nar/gkj122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cotter D, Guda P, Fahy E, Subramaniam S. Nucleic Acids Res. 2004;32:D463. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.(a) Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. Nucleic Acids Res. 2004;32:D258. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gene Ontology website. www.geneontology.org.
- 50.Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.(a) Li J, Ning Y, Hedley W, Saunders B, Chen Y, Tindill N, Hannay T, Subramaniam S. Nature. 2002;420:716. doi: 10.1038/nature01307. [DOI] [PubMed] [Google Scholar]; (b) Saunders B, Lyon S, Day M, Riley B, Chenette E, Subramaniam S, Vadivelu I. Nucleic Acids Res. 2008;36:D700. doi: 10.1093/nar/gkm907. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dinasarapu AR, Saunders B, Ozerlat I, Azam K, Subramaniam S. Bioinformatics. 2011;27:1736. doi: 10.1093/bioinformatics/btr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, Schacherer F, Martinez-Flores I, Hu Z, Jimenez-Jacinto V, Joshi-Tope G, Kandasamy K, Lopez-Fuentes AC, Mi H, Pichler E, Rodchenkov I, Splendiani A, Tkachev S, Zucker J, Gopinath G, Rajasimha H, Ramakrishnan R, Shah I, Syed M, Anwar N, Babur O, Blinov M, Brauner E, Corwin D, Donaldson S, Gibbons F, Goldberg R, Hornbeck P, Luna A, Murray-Rust P, Neumann E, Reubenacker O, Samwald M, van Iersel M, Wimalaratne S, Allen K, Braun B, Whirl-Carrillo M, Cheung KH, Dahlquist K, Finney A, Gillespie M, Glass E, Gong L, Haw R, Honig M, Hubaut O, Kane D, Krupa S, Kutmon M, Leonard J, Marks D, Merberg D, Petri V, Pico A, Ravenscroft D, Ren L, Shah N, Sunshine M, Tang R, Whaley R, Letovksy S, Buetow KH, Rzhetsky A, Schachter V, Sobral BS, Dogrusoz U, McWeeney S, Aladjem M, Birney E, Collado-Vides J, Goto S, Hucka M, Le Novere N, Maltsev N, Pandey A, Thomas P, Wingender E, Karp PD, Sander C, Bader GD. Nat. Biotechnol. 2010;28:935. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.(a) The Systems Biology Markup Language. http://sbml.org. [Google Scholar]; (b) Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. Bioinformatics. 2003;19:524. [Google Scholar]
- 54.Ideker T, Galitski T, Hood L. Annu. Rev. Genomics Hum. Genet. 2001;2:343. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 55.(a) Avery G, McGee C, Falk S. Anal. Chem. 2000;72:57A. doi: 10.1021/ac0027082. [DOI] [PubMed] [Google Scholar]; (b) Goodman N, Rozen S, Stein LD, Smith AG. Bioinformatics. 1998;14:562. doi: 10.1093/bioinformatics/14.7.562. [DOI] [PubMed] [Google Scholar]
- 56.Madhusudan VI, Krause L, Ning Y, Lyon S, Morrison P, Sorrells L, Taussig R, Subramaniam S. Proceedings of the International Conference on Mathematical and Engineering Techniques in Medicine and Biological Sciences. METMBS ’04; Las Vegas, NV, USA. 2004. [Google Scholar]
- 57.Byrnes RW, Fahy E, Subramaniam S. The Association for Laboratory Automation. 2007;12:230. [Google Scholar]
- 58.(a) Jenkins H, Hardy N, Beckmann M, Draper J, Smith AR, Taylor J, Fiehn O, Goodacre R, Bino RJ, Hall R, Kopka J, Lane GA, Lange BM, Liu JR, Mendes P, Nikolau BJ, Oliver SG, Paton NW, Rhee S, Roessner-Tunali U, Saito K, Smedsgaard J, Sumner LW, Wang T, Walsh S, Wurtele ES, Kell DB. Nat. Biotechnol. 2004;22:1601. doi: 10.1038/nbt1041. [DOI] [PubMed] [Google Scholar]; (b) Lindon JC, Nicholson JK, Holmes E, Keun HC, Craig A, Pearce JT, Bruce SJ, Hardy N, Sansone SA, Antti H, Jonsson P, Daykin C, Navarange M, Beger RD, Verheij ER, Amberg A, Baunsgaard D, Cantor GH, Lehman-McKeeman L, Earll M, Wold S, Johansson E, Haselden JN, Kramer K, Thomas C, Lindberg J, Schuppe-Koistinen I, Wilson ID, Reily MD, Robertson DG, Senn H, Krotzky A, Kochhar S, Powell J, van der Ouderaa F, Plumb R, Schaefer H, Spraul M. Nat. Biotechnol. 2005;23:833. doi: 10.1038/nbt0705-833. [DOI] [PubMed] [Google Scholar]
- 59.Griffin JL, Steinbeck C. Genome Med. 2010;2:38. doi: 10.1186/gm159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blanksby SJ, Mitchell TW. Annu. Rev. Anal. Chem. 2010;3:433. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 61.Gross RW, Han X. Chem. Biol. 2011;18:284. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(a) Katajamaa M, Miettinen J, Oresic M. Bioinformatics. 2006;22:634. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]; (b) Pluskal T, Castillo S, Villar-Briones A, Oresic M. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hartler J, Trotzmuller M, Chitraju C, Spener F, Kofeler HC, Thallinger GG. Bioinformatics. 2011;27:572. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- 64.Leavell MD, Leary JA. Anal. Chem. 2006;78:5497. doi: 10.1021/ac0604179. [DOI] [PubMed] [Google Scholar]
- 65.Haimi P, Uphoff A, Hermansson M, Somerharju P. Anal. Chem. 2006;78:8324. doi: 10.1021/ac061390w. [DOI] [PubMed] [Google Scholar]
- 66.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Anal. Chem. 2006;78:6202. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 67.Python programming language. http://www.python.org/ [Google Scholar]
- 68.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murphy RC, Fiedler J, Hevko J. Chem. Rev. 2001;101:479. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 70.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. J. Lipid Res. 2010;51:3299. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.(a) Karp PD, Paley S, Romero P. Bioinformatics. 2002;18(Suppl 1):S225. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]; (b) Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. Nucleic Acids Res. 2005;33:W633. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ludemann A, Weicht D, Selbig J, Kopka J. Bioinformatics. 2004;20:2841. doi: 10.1093/bioinformatics/bth278. [DOI] [PubMed] [Google Scholar]; (d) Sorokin A, Paliy K, Selkov A, Demin OV, Dronov S, Ghazal P, Goryanin I. IBM J. Res. Dev. 2006;50:561. [Google Scholar]; (e) Shulaev V. Brief Bioinform. 2006;7:128. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]; (f) Junker BH, Klukas C, Schreiber F. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Baitaluk M, Sedova M, Ray A, Gupta A. Nucleic Acids Res. 2006;34:W466. doi: 10.1093/nar/gkl308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. Methods Enzymol. 2007;432:171. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 73.KEGG Pathway database. www.genome.jp/kegg/pathway.html. [Google Scholar]
- 74.KEGG BRITE database. www.genome.jp/kegg/brite.html. [Google Scholar]
- 75.SphinGOMAP pathways. sphingolab.biology.gatech.edu. [Google Scholar]
- 76.Gehlenborg N, O'Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, Kohlbacher O, Neuweger H, Schneider R, Tenenbaum D, Gavin AC. Nat. Methods. 2010;7:S56. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 77.KGML website. www.genome.jp/kegg/xml/docs.
- 78.Lloyd CM, Halstead MD, Nielsen PF. Prog. Biophys. Mol. Biol. 2004;85:433. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 79.Junker BH, Klukas C, Schreiber F. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Byrnes RW, Cotter D, Maer A, Li J, Nadeau D, Subramaniam S. BMC Syst. Biol. 2009;3:99. doi: 10.1186/1752-0509-3-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.(a) Papin JA, Hunter T, Palsson BO, Subramaniam S. Nat. Rev. Mol. Cell Biol. 2005;6:99. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]; (b) Papin J, Subramaniam S. Curr. Opin. Biotechnol. 2004;15:78. doi: 10.1016/j.copbio.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 82.(a) Ozsolak F, Milos PM. Nat. Rev. Genet. 2011;12:87. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang Z, Gerstein M, Snyder M. Nat. Rev. Genet. 2009;10:57. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.(a) Hsiao A, Ideker T, Olefsky JM, Subramaniam S. Nucleic Acids Res. 2005;33:W627. doi: 10.1093/nar/gki443. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hsiao A, Worrall DS, Olefsky JM, Subramaniam S. Bioinformatics. 2004;20:3108. doi: 10.1093/bioinformatics/bth371. [DOI] [PubMed] [Google Scholar]
- 84.Baldi P, Long AD. Bioinformatics. 2001;17:509. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 85.Smyth GK. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 86.Endo A, Kuroda M, Tsujita Y. J. Antibiot. (Tokyo) 1976;29:1346. doi: 10.7164/antibiotics.29.1346. [DOI] [PubMed] [Google Scholar]
- 87.(a) Benjamini Y, Hochberg Y. J. Roy. Stat. Soc. B Met. 1995;57:289. [Google Scholar]; (b) Glantz SA. 6th ed. New York: McGraw-Hill Companies; 2005. [Google Scholar]; (c) Storey JD. J. Roy. Stat. Soc. B Stat. Met. 2002;64:479. [Google Scholar]
- 88.Churchill GA. Biotechniques. 2004;37:173. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- 89.Draghici S, Kulaeva O, Hoff B, Petrov A, Shams S, Tainsky MA. Bioinformatics. 2003;19:1348. doi: 10.1093/bioinformatics/btg165. [DOI] [PubMed] [Google Scholar]
- 90.Pradervand S, Maurya MR, Subramaniam S. Genome Biol. 2006;7:R11. doi: 10.1186/gb-2006-7-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Haan JR, Wehrens R, Bauerschmidt S, Piek E, van Schaik RC, Buydens LM. Bioinformatics. 2007;23:184. doi: 10.1093/bioinformatics/btl572. [DOI] [PubMed] [Google Scholar]
- 92.Climaco-Pinto R, Barros AS, Locquet N, Schmidtke L, Rutledge DN. Anal. Chim. Acta. 2009;653:131. doi: 10.1016/j.aca.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 93.the Gene Ontology database. http://amigo.geneontology.org. [Google Scholar]
- 94.(a) Huang da W, Sherman BT, Lempicki RA. Nucleic Acids Res. 2009;37:1. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huang da W, Sherman BT, Lempicki RA. Nat. Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 95.Li C, Li X, Miao Y, Wang Q, Jiang W, Xu C, Li J, Han J, Zhang F, Gong B, Xu L. Nucleic Acids Res. 2009;37:e131. doi: 10.1093/nar/gkp667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.TRANSFAC. http://biobase-international.com/index.php?id=transfac. [Google Scholar]
- 97.BIOCARTA. http://www.biocarta.com. [Google Scholar]
- 98.JASPAR. http://jaspar.cgb.ki.se. [Google Scholar]
- 99.Bailey TL, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park, CA, USA. 1994. p. 28. [PubMed] [Google Scholar]
- 100.Halperin Y, Linhart C, Ulitsky I, Shamir R. Nucleic Acids Res. 2009;37:1566. doi: 10.1093/nar/gkn1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andreyev AY, Shen Z, Guan Z, Ryan A, Fahy E, Subramaniam S, Raetz CR, Briggs S, Dennis EA. Mol. Cell Proteomics. 2010;9:388. doi: 10.1074/mcp.M900432-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.(a) Egghe L, Leydesdorff L. J. Am. Soc. Inf. Sci. Technol. 2009;60:1027. [Google Scholar]; (b) Anderson TW. An introduction to multivariate statistical analysis. 2nd ed. New York: Wiley; 1984. [Google Scholar]
- 103.Langfelder P, Zhang B, Horvath S. Bioinformatics. 2008;24:719. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 104.(a) Fukushima A, Kusano M, Redestig H, Arita M, Saito K. BMC Syst. Biol. 2011;5:1. doi: 10.1186/1752-0509-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Johansson A, Loset M, Mundal SB, Johnson MP, Freed KA, Fenstad MH, Moses EK, Austgulen R, Blangero J. Hum. Genet. 2011;129:25. doi: 10.1007/s00439-010-0893-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Adourian A, Jennings E, Balasubramanian R, Hines WM, Damian D, Plasterer TN, Clish CB, Stroobant P, McBurney R, Verheij ER, Bobeldijk I, van der Greef J, Lindberg J, Kenne K, Andersson U, Hellmold H, Nilsson K, Salter H, Schuppe-Koistinen I. Mol. Biosyst. 2008;4:249. doi: 10.1039/b708489g. [DOI] [PubMed] [Google Scholar]; (d) Han L, Zhu J. Biosystems. 2008;91:158. [Google Scholar]
- 105.Maurya MR, Subramaniam S. Computational Challenges in Systems Biology. In: Liu ET, Lauffenburger DA, editors. Systems Biomedicine: Concepts and Perspectives. San Diego: Academic Press; 2009. p. 177. [Google Scholar]
- 106.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. Science. 2005;310:1646. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 107.Gupta S, Maurya MR, Subramaniam S. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000654. e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fiehn O, Weckwerth W. Eur. J. Biochem. 2003;270:579. doi: 10.1046/j.1432-1033.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 109.Steuer R, Kurths J, Fiehn O, Weckwerth W. Biochem. Soc. Trans. 2003;31:1476. doi: 10.1042/bst0311476. [DOI] [PubMed] [Google Scholar]
- 110.(a) Kose F, Weckwerth W, Linke T, Fiehn O. Bioinformatics. 2001;17:1198. doi: 10.1093/bioinformatics/17.12.1198. [DOI] [PubMed] [Google Scholar]; (b) Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A. Plant Cell. 2001;13:11. doi: 10.1105/tpc.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sana TR, Fischer S, Wohlgemuth G, Katrekar A, Jung KH, Ronald PC, Fiehn O. Metabolomics. 2010;6:451. doi: 10.1007/s11306-010-0218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmitt WA, Jr, Raab RM, Stephanopoulos G. Genome Res. 2004;14:1654. doi: 10.1101/gr.2439804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Numata J, Ebenhoh O, Knapp EW. Genome Inform. 2008;20:112. [PubMed] [Google Scholar]
- 114.The Mathworks, Inc.© 1994 – 2009. http://www.mathworks.com. [Google Scholar]
- 115.Miller AK, Marsh J, Reeve A, Garny A, Britten R, Halstead M, Cooper J, Nickerson DP, Nielsen PF. BMC Bioinformatics. 2010;11:178. doi: 10.1186/1471-2105-11-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Falkenburger BH, Jensen JB, Hille B. J. Gen. Physiol. 2010;135:99. doi: 10.1085/jgp.200910345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, Kitano H. OMICS. 2003;7:355. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 118.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. Bioinformatics. 2006;22:3067. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 119.Casanova H, Berman F, Bartol T, Gokcay E, Sejnowski T, Birnbaum A, Dongarra J, Miller M, Ellisman M, Faerman M, Obertelli G, Wolski R, Pomerantz S, Stiles J. Int. J. High Perform. Comput. Appl. 2004;18:3. doi: 10.1177/1094342004041290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.(a) Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome Res. 2003;13:244. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO. Mol. Syst. Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO. Proc. Natl. Acad. Sci. U S A. 2007;104:1777. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Burgard AP, Nikolaev EV, Schilling CH, Maranas CD. Genome Res. 2004;14:301. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhalla US, Ram PT, Iyengar R. Science. 2002;297:1018. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 122.Chen KC, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson JJ. Mol. Biol. Cell. 2000;11:369. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.(a) Hinch R, Greenstein JL, Tanskanen AJ, Xu L, Winslow RL. Biophys. J. 2004;87:3723. doi: 10.1529/biophysj.104.049973. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mishra J, Bhalla US. Biophys. J. 2002;83:1298. doi: 10.1016/S0006-3495(02)73901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Maurya MR, Subramaniam S. Biophys. J. 2007;93:709. doi: 10.1529/biophysj.106.097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Nat. Biotechnol. 2000;18:1157. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 125.Callender HL, Horn MA, DeCamp DL, Sternweis PC, Alex Brown H. J. Theor. Biol. 2010;262:679. doi: 10.1016/j.jtbi.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang K, Ma W, Liang H, Ouyang Q, Tang C, Lai L. PLoS Comput. Biol. 2007;3:e55. doi: 10.1371/journal.pcbi.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA. Nature. 2005;433:425. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- 128.Henning PA, Merrill AH, Wang MD. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2004;4:2913. doi: 10.1109/IEMBS.2004.1403828. [DOI] [PubMed] [Google Scholar]
- 129.Gupta S, Maurya MR, Stephens DL, Dennis EA, Subramaniam S. Biophys. J. 2009;96:4542. doi: 10.1016/j.bpj.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.(a) Okino MS, Mavrovouniotis ML. Chem. Eng. Commun. 1999;176:115. [Google Scholar]; (b) Strogatz SH. Nonlinear dynamics and chaos : with applications to physics, biology, chemistry, and engineering. 1st ed. Cambridge. MA: Westview Press; 2000. [Google Scholar]
- 131.(a) Jamshidi N, Palsson BO. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000177. e1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Strang G. Introduction to linear algebra. 4th ed. Wellesly, MA: Wellesley-Cambridge; 2009. Eigenvalues and Eigenvectors; p. 283. [Google Scholar]
- 132.(a) Roth GJ, Siok CJ, Ozols J. J. Biol. Chem. 1980;255:1301. [PubMed] [Google Scholar]; (b) Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. Nucleic Acids Res. 2004;32:D431. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chan G, Boyle JO, Yang EK, Zhang F, Sacks PG, Shah JP, Edelstein D, Soslow RA, Koki AT, Woerner BM, Masferrer JL, Dannenberg AJ. Cancer Res. 1999;59:991. [PubMed] [Google Scholar]
- 134.(a) Haynes CA, Allegood JC, Wang EW, Kelly SL, Sullards MC, Merrill AH., Jr J Lipid Res. 2011;52:1583. doi: 10.1194/jlr.D015586. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tserng KY, Griffin R. Anal. Biochem. 2004;325:344. doi: 10.1016/j.ab.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 135.(a) Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Nat. Biotechnol. 2008;26:1179. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Klapa MI, Aon JC, Stephanopoulos G. Eur. J. Biochem. 2003;270:3525. doi: 10.1046/j.1432-1033.2003.03732.x. [DOI] [PubMed] [Google Scholar]; (c) Park SM, Klapa MI, Sinskey AJ, Stephanopoulos G. Biotechnol. Bioeng. 1999;62:392. doi: 10.1002/(sici)1097-0290(19990220)62:4<392::aid-bit2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 136.BRENDA. http://www.brenda-enzymes.org. [Google Scholar]