Abstract
RIG-I like receptors (RLR) that recognize non-self RNA play critical roles in activating host innate immune pathways in response to viral infections. Not surprisingly, RLRs and their associated signaling networks are also targeted by numerous antagonists that facilitate viral pathogenesis. Although the role of RLRs in orchestrating antiviral signaling has been recognized for some time, our knowledge of the complex regulatory mechanisms that control signaling through these key molecules is incomplete. A series of recent structural studies shed new light into the structural basis for dsRNA recognition and activation of RLRs. Collectively, these studies suggest that the repression of RLRs is facilitated by a cis element that makes multiple contacts with domains within the helicase and that RNA binding initiated by the C-terminal RNA binding domain is important for ATP hydrolysis and release of the CARD domain containing signaling module from the repressed conformation. These studies also highlight potential differences between RIG-I and MDA5, two RLR members. Together with previous studies, these new results bring us a step closer to uncovering the complex regulatory process of a key protein that protects host cells from invading pathogens.
Introduction
Pattern recognition receptors (PRRs) known as the retinoic acid inducible gene-I (RIG-I) like receptors (RLRs) are super family 2 (SF2) RNA helicase domain containing proteins [1, 2, 3]. Like all PRRs, RLRs are germ-line encoded and are constitutively expressed in most cells, including dendritic cells and macrophages. Pathogen associated molecular pattern (PAMP) recognition by PRRs results in the activation of type I interferons (IFNs) and leads to subsequent activation of IFN stimulated response elements (ISREs) that can ultimately control viral infections [4]. There are three members of the RLR family: retinoic acid inducible gene-I (RIG-I), melanoma differentiation associated factor gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) [3, 5, 6].
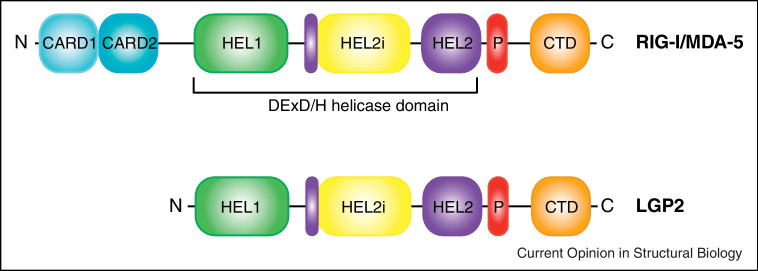
RIG-I, MDA5, and LGP2 all contain DEX/DH box RNA helicases (Figure 1 ). In addition to the common SF2 helicase domain, which contains a helicase insertion domain (HEL2i) along with helicase domain 1 (HEL1) and 2 (HEL2), all three proteins share an RNA binding domain known as C-terminal domains (CTD, also called repressor domain (RD)) [6, 7••]. The tandem caspase activation and recruitment domains (CARDs) are present at the N-terminus of RIG-I and MDA5. By contrast, LGP2 lacks the N-terminal CARD domains. The N-terminal CARD domains engage in protein-protein interactions with other CARD domain containing proteins, most notably with mitochondrial associated antiviral signaling molecule (MAVS, also known as IPS-1, VISA, CARDIF) [8]. In the context of RLR signaling, only MDA5 and RIG-I can interact with downstream effector molecule MAVS, while both agonist and antagonist roles have been described for LGP2 in the literature. CARD-CARD interactions between RLRs and MAVS lead to activation of interferon kinases, such as Tank binding kinase-1 (TBK-1) and Interferon kB kinase ɛ (IKKɛ), that can phosphorylate interferon regulator factors 3 (IRF3) and 7 (IRF7) [9]. Phosphorylation and nuclear localization of IRF3/7, as well as nuclear factor κB (NFκB), result in type I interferon (IFN-1) production [1, 4, 9]. IFN-α/β produced as a result of these signaling events can function in an autocrine and paracrine manner, leading to the induction of a large number of antiviral molecules [1, 10]. Expression of IFN stimulated genes (ISGs) establishes an antiviral state, which limits viral replication and spread, and often leads to viral clearance.
Figure 1.
Domain architecture of RIG-I-like receptors. RIG-I and MDA-5 have similar domain organization. The domains are: CARD1 (cyan), CARD2 (blue), helicase HEL1 (green), helicase insertion domain HEL2i (yellow), helicase HEL2 (purple), the regulatory pincer motif P (red), and C-terminal domain CTD (orange). LGP2 lacks the N-terminal CARDs.
Although the role of RLRs as key molecules orchestrating antiviral signaling has been recognized for some time, our knowledge of the complex regulatory mechanisms that control signaling through these key molecules is incomplete. Recent structural analysis by four independent groups of RIG-I proteins, including the first structure of an RLR protein containing the N-terminal CARDs, provide key snap shots that reveal important aspects of RNA recognition and activation mechanisms (see Table 1 Table 1) [11••, 12••, 13••, 14••]. Most significantly, the current structures reveal how the helicase and CTD regions interact with dsRNA. Therefore, these studies shed new light into the structural basis for dsRNA recognition and support an activation mechanism, which includes conformational changes to multiple contacts between double stranded RNA (dsRNA) and RIG-I domains as well as interactions with dsRNA. Together with previous structural studies on the C-terminal repressor domains, which was key to understanding the ligand recognition mechanisms (structures listed in Table 1), these studies greatly expands our understanding and paints an exciting picture of how RLRs are tightly regulated to protect host cells from invading pathogens (Table 1) [15•, 16•, 17, 18, 19, 20]. In this review, we will present an overview of structural and biochemical studies that define the structural basis for RNA recognition and activation of RLRs and discuss areas that require further investigation, including differences between RIG-I and MDA5, in order to fully understand the regulatory mechanisms that control RLRs.
Table 1.
Summary of RLR structural studies
| RLRa | Domainsb | ATPc | RNA | PDB | Reference |
|---|---|---|---|---|---|
| hRIG-I | T | 2QFB, 2QFD | [34] | ||
| hRIG-I | T | 2RMJ | [33•] | ||
| hMDA-5 | T | 3GA3 | |||
| hLGP2 | T | 2W4R | [43] | ||
| hLGP2 | T | 5′ OH dsRNA (8 mer) | 3EQT | [18] | |
| hLGP2 | T | 2RQA | [20] | ||
| hMDA-5 | T | 2RQB | [20] | ||
| hRIG-I | T | 5′ppp dsRNA (12 mer) | 3NCU | [16•] | |
| hRIG-I | T | 5′ppp dsRNA (14 mer) | 3LRN | [15•] | |
| hRIG-I | T | 5′ppp dsRNA (12 mer) | 3LRR | [15•] | |
| hRIG-I | T | 5′OH dsRNA (14 mer) | 3OG8 | [19] | |
| dRIG-I | T | 4A2V | [13••] | ||
| dRIG-I | T | 5′OH dsRNA (14 mer) | 4A2X | [13••] | |
| dRIG-I | H | Analog | 5′OH dsRNA (19 mer) | 4A36 | [13••] |
| dRIG-I | H | 4A2P | [13••] | ||
| mRIG-I | H | Analog | 3TBK | [11••] | |
| hMDA5 | HEL2i | 3TS9 | [39•] | ||
| hRIG-I | H, T | Analog | 5′ppp dsRNA (14 mer) | 3TMI | [12••] |
| hRIG-I | H, T | 5′OH dsRNA (10 mer) | 2YKG | [14••] | |
| dRIG-I | C, H | 4A2Q | [13••] | ||
| dRIG-I | C, H, T | 4A2 W | [13••] |
human (h), mouse (m), duck (d).
CARDS (C), Helicase (H), Helicase2 insert (HEL2i), C-terminal domain (T).
ATP or analog.
dsRNA containing ligands activate RLRs
Detection of a variety of RNA PAMPs by RLRs is critical for viral detection and activation of IFN-α/β [3, 21]. In their seminal study, Yoneyama et al. reported that the activity of RIG-I through CARD domains is under negative regulation [7••]. While RIG-I, MDA-5, and LGP2 all retain the C-terminal sequence homology, including the helicase domains, dsRNA binding and ATPase activity are both required for signal transduction by RIG-I [7••]. Subsequent studies have identified many different RNA ligands or moieties that are involved in RLR regulation, including ligands such as poly I:C [7••], 5′ triphosphate (5′ppp) [22, 23], double strandedness [7••, 24, 25, 26], and the panhandle structure formed by the 5′ and 3′ untranslated regions (UTRs) [27]. Together, these studies demonstrate that many of the RLR activators are either parts of viral genomes or products and byproducts of viral replication.
RIG-I and MDA5 are thought to recognize different ligands [28] and are implicated in the recognition of distinct viruses [29, 30]. For example, RIG-I receptors are critical for limiting infection by rhabdoviruses (vesicular stomatitis virus and rabies virus), paramyxoviruses (Sendai virus, respiratory syncytial virus, and Newcastle disease virus), orthomyxoviruses (influenza A and B) and filoviruses (Ebolavirus and Marburgvirus), whereas picornaviruses (EMCV, coronavirus, and murine hepatitis virus, and murine norovirus-1 type I) are detected by MDA5 predominantly [4, 29]. Interestingly, flaviviruses (Dengue virus and West Nile viruses) as well as reoviruses (rotavirus) can signal through both RIG-I and MDA5 [4, 29, 30, 31, 32]. These differences are probably owing to differences in dsRNA recognition as MDA5 can be activated by long dsRNA, whereas much shorter dsRNA and those that contain 5′ppp can activate RIG-I more efficiently [28]. Collectively these studies have shown that RIG-I can be activated by either dsRNA or DNA-RNA heteroduplexes, but not dsDNA.
RLR C-terminal domains recognize blunt end dsRNA
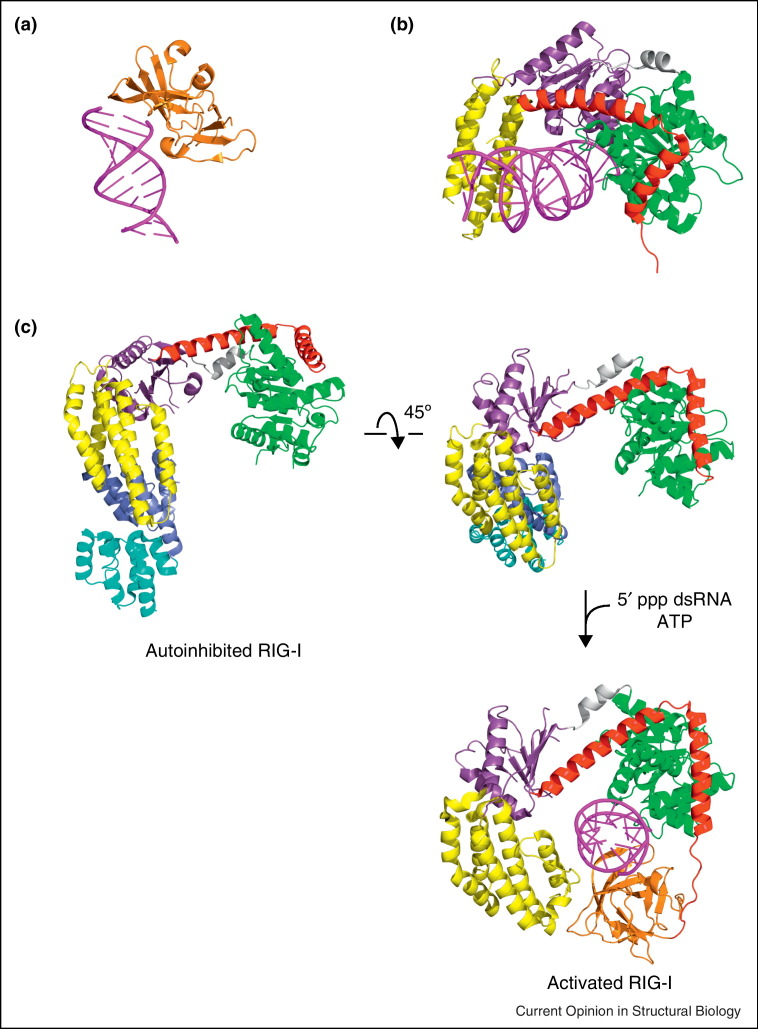
The RIG-I C-terminal RNA binding domain (CTD) was the first experimentally determined structural domain of RLRs. Takahasi et al. [33•] and Cui et al. [34•] reported structures of the RIG-I CTD that was identified through a series of biochemical and structural mapping studies. Guided by the structures, subsequent mutagenesis experiments identified key basic residues that formed the dsRNA interaction surface [33•, 34•]. The binding site for the 5′-ppp was predicted on the basis of these studies. The following structures of RIG-I CTD bound to 5′-ppp containing dsRNA, as well as MDA5 and LGP2 bound to blunt ended dsRNA, confirmed previous mutagenesis and binding interactions (Figure 2 and Table 1) and also identified the structural determinants required for 5′-ppp binding [15•, 16•]. Comparison of RIG-I CTD bound to RNA with structures of RIG-I helicase–CTD (Figure 2c) show that interactions between the CTD and the dsRNA remain largely unchanged. These include contacts with both strands of the dsRNA as well as base stacking by the conserved phenylalanine residue (Figure 2c, in human RIG-I). Interestingly, comparison of RIG-I CTD–dsRNA complex structures with helicase–CTD–dsRNA complexes suggests that the RNA in the 5′-ppp containing dsRNA–CTD complex structure align best with the 5′OH dsRNA from the RIG-I helicase–dsRNA complex structure. These observations suggest that 5′-ppp binding aligns the dsRNA in the most energetically favorable orientation. Such binding may correspond to potent RIG-I activation, as judged by ATPase activities that are seen when 5′-ppp ligands are used.
Figure 2.
Structural basis for dsRNA recognition and activation of RLRs. (a) C-terminal RNA binding domain in the presence of RNA (PDB: 3NCU). (b) Helicase domain in the presence of RNA (PDB: 4A36). (c) In the autoinhibited conformation, the N-terminal CARDs are sequestered from signaling and the pincer maintains RIG-I in an autoinhibited state (PDB: 4A2W). Binding of dsRNA to the CTD brings HEL2i in contact with dsRNA (PDB: 2YKG). The change in conformation upon dsRNA binding presumably releases the CARD domains for signaling.
Autoinhibition and activation of RIG-I/RLRs require ATPase activity and dsRNA binding
Helicases in the SF2 family bind and/or remodel nucleic acids, which sometimes result in unwinding as well as translocation of the helicase containing protein on the nucleic acid strands [35]. RLR helicases are part of the Dicer-RIG-I clade in the family of SF2 helicases and contain two RecA like domains that are required for ATPase activity [35]. It was previously reported that RIG-I may also unwind dsRNA [33•], but most recent studies show that RIG-I and potentially RLRs are unlikely to participate in dsRNA unwinding. Comparison of the RIG-I helicase bound to dsRNA with other helicases, such as Hepatitis C virus (HCV) NS3, reveals that key structural elements such as the Phe-loop of HCV is absent in RLR helicases [12]. Consistent with an RNA recognition and binding role, the complex structure of RIG-I helicase–CTD dual domain bound to dsRNA reveals extensive protein-RNA contacts in excess of 1500 Å that cover about 8 base pairs (Figure 2c) [12]. dsRNA recognition by the helicase domain may facilitate translocation along the dsRNA, where the ATPase activity of the RLR helicases was shown to be tightly coupled to the ability to translocate along dsRNA in a length dependent manner [36].
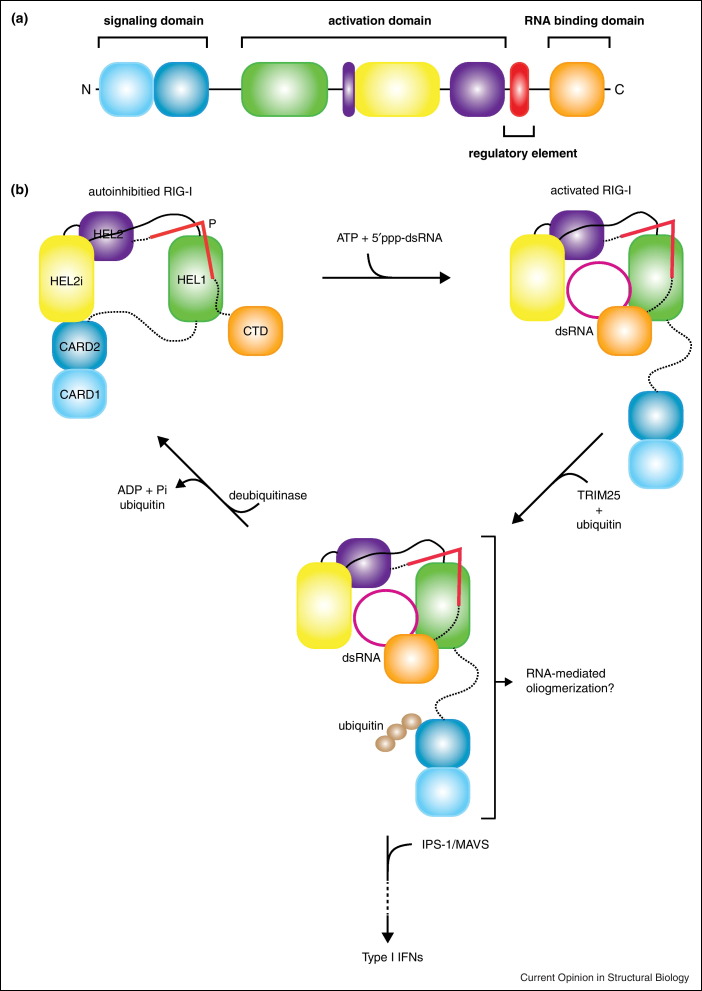
Since the CARD domains are required for signaling, it was proposed that interactions between the N-terminal CARDs and other domains of RLRs result in an autoinhibited state [7••, 33•, 34•]. Consistent with these findings, Kowalinski et al. showed that the two N-terminal CARD domains form a head to tail interaction, where the N-terminus of the CARD2 head interacts directly with the C-terminal region of CARD1 (Figure 2c). Interestingly, the C-terminus of CARD2 also makes extensive contacts with HEL2i, suggesting that the CARDs and HEL2i regions contribute to the formation of a rigid inactive conformation (Figure 2c). These CARD–helicase interactions may also prevent access to MAVS directly or by blocking ubiquitination by TRIM25 [37] or interactions with unanchored polyubiquitin chains [38], which are important for downstream signaling. Comparison of the dsRNA-bound structures of helicase and C-terminal domains with dsRNA free structures suggests that the pincer motif (also called the bridging domain or regulatory element) is also important for regulating RIG-I activation [13••]. The pincer motif interacts with both HEL1 and HEL2 domains and connects the helicase domain to the CTD, which binds RNA (Figure 2c). Consistent with these findings, the helicase domain alone binds dsRNA with low affinity, and the addition of the CTD markedly improves dsRNA binding [12]. While 5′-ppp containing dsRNA are more potent activators, these studies show that dsRNA longer that 8-10 base pairs are likely to bind and activate RLRs. Altogether, these results suggest that initial dsRNA binding to CTD enhances dsRNA binding to helicase, and that deletion (or mutation of key residues in CTD) result in significant reduction in dsRNA binding by the RLR helicases and subsequent blunting of innate immune signaling. A model consistent with these findings has been proposed (Figure 3 ), where the signaling CARD domains are sequestered through autoregulatory contacts within RIG-I [12••, 13••, 14••]. Interactions of dsRNA first with the CTD and subsequently with the helicase result in the reorientation of the pincer domain leading to ATP hydrolysis and release of the N-terminal CARDs for signal transduction. It is likely that ubiquitination of RLR signaling may result in more sustained signaling, with higher burst activity.
Figure 3.
A structure based model dsRNA-mediated RLR regulation and signaling. (a) Functional elements in RLRs. The CARD domains comprise the signaling region, the helicase serves as the activation domain, the pincer is the regulatory element, and the C-terminal domain is the RNA binding domain. (b) Schematic model of the activation mechanism in RIG-I. In the absence of dsRNA, RIG-I exists in an autoinhibited conformation that is regulated by the pincer motif, which prevents the N-terminal CARDs from signaling. Binding of dsRNA to the CTD relieves repression by the pincer motif, initiates dsRNA binding to HEL2i, and releases the CARDs from HEL2i to become polyubiquinated and activate production of Type I IFNs.
How different is MDA5 from RIG-I?
Ligand length preferences between MDA5 and RIG-I and their ability to uniquely detect different viral families are well established. A recent study extends our understanding of the potential differences between MDA5 and RIG-I in their regulation and activity [39•]. Berke and Modis reveal that in contrast to RIG-I, the N-terminal CARDs are not likely to interact with the HEL2i or other domains within MDA5 [39•]. Moreover, ATP leads to differential binding affinities to RNA ligands by MDA5 in an ATP concentration dependent manner whereas ATPase mutations in RIG-I lead to dominant negative effects. Taken together, these new results on MDA5 suggest that despite high sequence conservation and structural similarity, large differences in MDA5 and RIG-I regulation may exist. Additional studies will be required to fully appreciate how these differences translate into functional consequences.
Concluding remarks and prospects
The recent structures of RIG-I proteins together with previous studies provide a wealth of information to understand how RLRs are structurally regulated and suggest a simplified model where the activity of the signaling module (CARDs) is repressed by the repression element (pincer motif) through interactions with the activation domain (helicase) (Figure 3a, b). RNA recognition by the CTD leads to a conformational change in the repression element, resulting in signaling. But, there are several aspects of RLR regulation that needs further clarification. For example, it is still not clear how the ATPase activity is linked to all RLR functions. A recent study showed that the pincer motif may function as a repressor element, where deletion of the pincer region resulted in constitutive activation of RIG-I signaling [40]. By contrast, the recent crystal structure of the MDA5 Hel2i domain revealed that while it is structurally similar to RIG-I proteins, the regulatory mechanism of MDA5 may be significantly different [39]. Moreover, MDA5 is able to bind both short and long dsRNA (and RNA with complex secondary structures) with varying affinities that are dependent on the ATP concentration [39•]. By contrast, an ATPase inactive mutant (K270A) was also functional in the context of a mutant RIG-I construct where key hydrophobic interactions within the pincer motif were mutated. These results raise the question whether RLRs with defective ATPases can also signal or if the ATPase activity is required to move the pincer motif in order to release CARDs from the HEL2i domain. Although we have several structures of dsRNA bound to the CTD and to the helicase domain, as well as helicase–CTD dual domains (Table 1), the relative structural orientations of the helicase and CTD in the absence of dsRNA is unknown. Moreover, oligomerization is thought to enhance translocation rates and provide a basis for cooperativity in ATPase hydrolysis and/or translocation [41, 42]. Is ubiquitination important for activation, sustained signaling, or both? Given the 8-18 base footprint of RLRs, what is the basis for length preference shown by MDA5 and RIG-I and the role of subsequent activation of signaling? Furthermore, what is the role of LGP2? These are some of the questions whose answers will bring us a step closer to uncovering a significant antiviral signaling mechanism with broad implications for normal function and response to pathogens.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Work in our laboratory is supported by NIH grants (1R56AI089547 to GKA (PI-Basler) and R01AI059536). We thank P. Ramanan and J. Binning for critical reading of the manuscript.
References
- 1.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki T., Kawai T., Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Ramos H.J., Gale M., Jr. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B.G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K.A. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M., Jr., Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 7••.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]; This work initially described the autorepression of RIG-I and laid the foundation for studies on the dsRNA-mediated regulatory mechanisms of RLRs.
- 8.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 10.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Civril F., Bennett M., Moldt M., Deimling T., Witte G., Schiesser S., Carell T., Hopfner K.P. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation for reference [13••].
- 12••.Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Jr., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation for reference [13••].
- 13••.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]; Together with references [11••, 12••], and [14••], this study provides structural insight into dsRNA recognition and activation of RLRs. These studies also reveal the structures of the helicase domains of Dicer-like SF2 family. In addition, this study is the first to report the structure of an RLR protein with intact CARDs and the helicase domain.
- 14••.Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation for reference [13••].
- 15•.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation for reference [16•].
- 16•.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]; Along with reference [15•], the current study provided the structural basis for dsRNA 5′ppp motif recognition. Of note, in the structure for reference 16, the γ-phosphate moeity was not visible, while the structure in reference 15 contained the full tri-phosphate moeity.
- 17.Li X., Lu C., Stewart M., Xu H., Strong R.K., Igumenova T., Li P. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Li X., Ranjith-Kumar C.T., Brooks M.T., Dharmaiah S., Herr A.B., Kao C., Li P. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahasi K., Kumeta H., Tsuduki N., Narita R., Shigemoto T., Hirai R., Yoneyama M., Horiuchi M., Ogura K., Fujita T., Inagaki F. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar H., Kawai T., Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 22.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 23.Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 24.Hausmann S., Marq J.B., Tapparel C., Kolakofsky D., Garcin D. RIG-I and dsRNA-induced IFNbeta activation. PLoS ONE. 2008;3:e3965. doi: 10.1371/journal.pone.0003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt A., Schwerd T., Hamm W., Hellmuth J.C., Cui S., Wenzel M., Hoffmann F.S., Michallet M.C., Besch R., Hopfner K.P. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habjan M., Andersson I., Klingstrom J., Schumann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Muhlberger E. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 30.Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., Garcia-Sastre A., Katze M.G., Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broquet A.H., Hirata Y., McAllister C.S., Kagnoff M.F. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 32.Leung D.W., Basler C.F., Amarasinghe G.K. RIG-I like receptors. Trends Microbiol. 2012;20:139–146. doi: 10.1016/j.tim.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]; See annotation for reference [34•].
- 34•.Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K., Fujita T., Conzelmann K.K., Krug A., Hopfner K.P. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]; References [33•] and [34•] were the first reported structures of RLR CTD structures. In addition to the first reports of the RLR-like dsRNA binding domain, these studies used structure-based methods to map the domain boundaries and binding site mapping.
- 35.Fairman-Williams M.E., Guenther U.P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myong S., Cui S., Cornish P.V., Kirchhofer A., Gack M.U., Jung J.U., Hopfner K.P., Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 38.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Berke I.C., Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study further highlights many differences between MDA5 and RIG-I, including differences in the potential role for ATP binding and oligomerization. In addition, this study suggests that unlike RIG-I, there may not be autoregulatory interactions between the N-terminal CARDs and other domains in MDA5.
- 40.Kageyama M., Takahasi K., Narita R., Hirai R., Yoneyama M., Kato H., Fujita T. 55 Amino acid linker between helicase and carboxyl terminal domains of RIG-I functions as a critical repression domain and determines inter-domain conformation. Biochem Biophys Res Commun. 2011;415:75–81. doi: 10.1016/j.bbrc.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Peisley A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T., Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Binder M., Eberle F., Seitz S., Mucke N., Huber C.M., Kiani N., Kaderali L., Lohmann V., Dalpke A., Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pippig D.A., Hellmuth J.C., Cui S., Kirchhofer A., Lammens K., Lammens A., Schmidt A., Rothenfusser S., Hopfner K.P. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]