Abstract
The biophysical basis of passive membrane permeability is well understood, but most methods for predicting membrane permeability in the context of drug design are based on statistical relationships that indirectly capture the key physical aspects. Here, we investigate molecular mechanics-based models of passive membrane permeability and evaluate their performance against different types of experimental data, including parallel artificial membrane permeability assays (PAMPA), cell-based assays, in vivo measurements, and other in silico predictions. The experimental data sets we use in these tests are diverse, including peptidomimetics, congeneric series, and diverse FDA approved drugs. The physical models are not specifically trained for any of these data sets; rather, input parameters are based on standard molecular mechanics force fields, such as partial charges, and an implicit solvent model. A systematic approach is taken to analyze the contribution from each component in the physics-based permeability model. A primary factor in determining rates of passive membrane permeation is the conformation-dependent free energy of desolvating the molecule, and this measure alone provides good agreement with experimental permeability measurements in many cases. Other factors that improve agreement with experimental data include deionization and estimates of entropy losses of the ligand and the membrane, which lead to size-dependence of the permeation rate.
INTRODUCTION
Permeability assessment is a crucial component in the drug development process for selecting and optimizing leads with favorable absorption, distribution, metabolism and excretion (ADME) properties. In order to avoid compounds with poor permeability or other ADME properties, a commonly adopted strategy is to comply with rules-of-thumb for ‘drug-likeness’ such as Lipinski’s Rules of Five.1 These rules qualitatively outline the physiochemical space defined by the majority of well-absorbed drugs, which include (1) molecular weight (MW) <500, (2) number of hydrogen bond donors ≤5, (3) number of hydrogen bond acceptors ≤10, and (4) octanol-water partition coefficient (LogPo/w) <5. Exceptions to these rules are known, and such rules do not provide quantitative measures.
Predictive quantitative models in use for drug design are primarily quantitative structure-permeability relationship (QSPR) methods that employ statistical relationships derived empirically from experimental permeability measurements and various physiochemical properties of a training set of compounds.2–14 The development of these quantitative models has previously been reviewed elsewhere.15,16 The QSPR approach has been widely applied in modeling oral bioavailability,17 intestinal absorption,18 skin permeation,19,20 and brain permeability.9,21,22 Some of the common QSPR descriptors include MW, polar surface area (PSA), partition coefficients, and hydrogen bond counts, all of which are known to correlate with rates of permeation. However, the main limitation of such an approach, like other empirical scoring models, is transferability. The performance of the model depends on the chemical similarity between the molecules of interest and the training set.12 Empirical statistical relationship can also be difficult to interpret, for example, what modifications to a compound would most successfully improve permeability?
An alternate approach is to develop models that are more directly based on the physics of the underlying processes. In the case of intestinal permeability, for example, there are (at least) three primary processes: passive diffusion across the cellular membrane, paracellular diffusion through the cell junctions, and active transport that includes chemical influx and efflux facilitated by transporter proteins. Increasing structural knowledge of key transport proteins, such as the efflux transporter P-glycoprotein (PGP),23,24 will undoubtedly advance predictive models of active transport mechanisms. Paracellular diffusion, which is primarily pertinent for very small and polar molecules, has been treated by others using hindered diffusion models.25–27
Here, we consider transmembrane passive diffusion which, for many molecules, is a dominant factor in intestinal absorption, rates of crossing the blood-brain barrier, and simply entering cells, as in in vitro cell-based experiments. In the extreme, it is possible to study passive membrane permeability in all-atom detail, i.e., using molecular dynamics simulations with explicit permeant and lipid molecules.28–36 Such simulations provided many important insights into the process and were shown to be capable of predicting relative permeability coefficients, at least for very small molecules. However, it remains difficult to predict absolute permeability by this underutilized approach, which is as yet computationally too expensive to be practically applied in drug design.29,35 A less computationally intensive alternative is to augment QSPR models with membrane-interaction descriptors obtained from MD simulations of permeants in lipid layers, such as the membrane-interaction QSAR method developed by Hopfinger and coworkers.37–41 Another approach is to use coarse-grained or implicit membrane models,42–49 which in principle provide an adequate representation of the membrane environment with lower computational expense. While implicit membrane models have been employed in studies of transmembrane proteins, their application to membrane permeation remains limited.46–48 One such example is the implicit model recently developed by Parisio and Ferrarini that accounts for the anisotropic and nonuniform membrane environment.49
Alternatively, solubility-diffusion theory has been used as the basis for a predictive framework, in which the permeability is assessed in terms of three components: the partitioning between water and the membrane, the diffusion across the membrane, and the width of the membrane.50,51 This theory was originated from the general permeability model for nonelectrolytes derived by Diamond et al.50 The statistical mechanical basis of this model was developed by Xiang and Anderson, who implemented methods to describe crucial aspects of the membrane physics, such as the size selectivity and chain ordering of lipids.52–52 A similar solubility-diffusion model was also utilized to study blood-brain barrier permeation by Seelig and coworkers, using parameters including the air/water partition coefficient and molecular shape of the permeant.59,60 The conceptual and theoretical underpinnings of these models are described in more detail in the following section.
While these solubility-diffusion approaches are typically more computationally expensive in comparison to QSPR models and remain less commonly employed, they do offer several advantages. Such methods capture the underlying physics of the permeation process and do not require training with permeability data, and thus are expected to be more general and transferrable. By the same token, the quality of a physical model, instead of being governed by the training set data, is determined by the theoretical representation of the process. Hence, in principle, physics-based models can be systematically improved by elevating the level of theory to describe the permeation process and to calculate each of the physical contributions. Since permeability is evaluated in physically-meaningful terms, the physics-based approach also offers a mechanistic understanding of the permeation process that allows for rational modulation.
Our own contributions in this area have emphasized the importance of performing extensive conformational sampling of the permeant, particularly for larger, conformationally complex molecules such as cyclic and linear peptides. 61–65 The free energy of desolvating the molecule, upon entering the hydrophobic interior of the membrane, is a critical parameter for accurate determination of the permeation rate. Conformationally flexible molecules can reduce this free energy cost by adopting conformations that, for example, maximize internal hydrogen bonding. Applications of this model to predict permeability of cyclic peptides and drug-like molecules resulted in good agreement with experimental data, including in prospective tests, warranting further development.61–65
In the present work, this simple physical model was expanded to provide a more sophisticated representation of the permeation process based on solubility-diffusion theory, building on prior work. 53,54,60 Additional attributes were incorporated in the model to account for contributions from both the permeant and the membrane that influence permeability, especially those related to the size- and shape-dependence of permeation. Along with combining these physical aspects with extensive conformational sampling, our major contribution is the systematic evaluation of different aspects of the model through a large and diverse group of experimental data sets as summarized in Table 1. The examined data ranges from in vitro measurements, which include parallel artificial membrane permeability assays (PAMPA), Caco-2 cell-based assays, and Madin-Darby canine kidney (MDCK) cell-based assays, to in vivo data, which includes percentage absorption in human (%Abs) and rat in situ perfusion assays.66–74
Table 1.
The examined data sets and their data types.
| Set | Compounds | Congeneric? | Data type (N) | Reference |
|---|---|---|---|---|
| Avdeef | FDA drugs | No | PAMPA (11) | 66 |
| Caco-2 (11) | ||||
| Balamine | FDA drugs | No | PAMPA (22) | 67 |
| Caco-2 (21) | ||||
| %Abs (22) | ||||
| Bermejo | Fluoroquinolones | Yes | PAMPA (16) | 68 |
| Caco2 (14) | ||||
| Perfusion assay (16) | ||||
| Di | FDA drugs | No | PAMPA (27) | 69 |
| Fujikawa | FDA drugs & small chemicals | No | PAMPA (70) | 70 |
| Caco-2 (29) | ||||
| Goodwin | Peptidomimetics | Yes | Caco-2 (12) | 71 |
| Irvine | FDA drugs | No | Caco-2 (50) | 72 |
| MDCK (51) | ||||
| %Abs (53) | ||||
| Li | Peptidomimetics | Yes | PAMPA (25) | 73 |
| Caco-2 (25) | ||||
| Whitlock | 1-(2-Phenoxyphenyl) methanamines | Yes | PAMPA (8) | 74 |
With the exception of PAMPA, all measurements involve biological membranes. The artificial membrane in PAMPA is typically composed of lipids and organic solvent, thus the assay measures strictly passive diffusion across an organic layer. The cell-based assays measure permeability across a monocellular layer. The Caco-2 and MDCK cell lines are well established models for biological processes, such as intestinal absorption or penetration across the blood brain barrier. The cellular membranes possess active transport facilitated by transport proteins and paracellular transport through cell junctions, in addition to transcellular passive permeation. Common approaches to evaluating passive permeation in these assays include using inhibiting substrate for transporter proteins, identifying possible active efflux or influx by measuring permeations in both apical-to-basolateral (A→B) and basolateral-to-apical (B→A) directions,71 measuring permeability of paracellular permeants as a control,72 and genetically knocking out known transporters.75
The in vivo data examined in this study are absorption measurements in human or rat. The percentage human absorption (%Abs) refers to intestinal absorption in human. The in situ perfusion assay allows assessment of gut absorption in rat by perfusing the substance of interest through the cannulated gut of an anesthetized rat and monitoring the change in substance concentration.76 Although the extent of drug absorption involves many properties, including solubility, passive membrane permeability is critical to the absorption process. Therefore, in vitro permeability measurements and calculated “hydrophobicity” measures, such as LogPo/w and PSA, are often utilized to predict absorption.
By evaluating the physics-based permeability model with respect to the different data types and conventional regression-based in silico models, the results highlight the generality of the approach and the ability to systematically improve the model.
THEORY
The physical models of passive membrane permeability considered here are based upon solubility-diffusion theory. The conceptual underpinnings and applications of this model have been described and reviewed in detail previously.50,53,54,55,60 A key concept of the theory is that membrane permeability is inversely related to the resistances presented in the permeation process, including the internal resistance of the membrane and the resistance at the water-membrane interface. If the aqueous unstirred layers and interfacial resistances are assumed to be negligible, then conceptually, the rate limiting step involves traversing a barrier region, i.e. the region of the membrane where the free energy of the molecule reaches a maximum. Molecule dynamics simulations suggest that this region, for molecules with polar groups, is located close to the middle of the membrane, where most permeants become nearly or completely desolvated.32,54
Using these assumption, solubility-diffusion theory estimates the permeation rate based on membrane/water partition, membrane diffusion and the permeation distance. Considering the membrane as a planar barrier in the xy-plane that varies only in the z direction, i.e. any plane parallel to the membrane is homogenous, the partition coefficient between water and the membrane, Km/w, and the diffusion coefficient in the membrane, Dm, can be expressed in terms of the membrane depth along the z-axis, z. In the steady state, the diffusional resistance of such membrane with thickness of d can be written as the reciprocal of the membrane permeability coefficient, Pm:
| (1) |
If the rate of permeation is limited by a homogenous “barrier region”, which the partition and the diffusion coefficients can be considered to be independent to the membrane depth, then the permeation rate across the membrane depends on the thickness of the barrier region, and the integral of Eq. 1 can be simplified as Eq. 2:
| (2) |
where Kbarrier is the permeant’s barrier/water partition coefficient of the permeant, Dbarrier is the diffusion coefficient within the barrier, and δbarrier is the effective thickness of the rate-limiting barrier. Conceptually, this model implies that rates of permeation are dominated by the free energy cost of desolvating the molecules in the hydrophobic interior of the membrane, as well as other free energy costs of inserting a molecule in the membrane, such as the free energy cost of creating a cavity of the appropriate size and shape; the rate of diffusion in the semi-ordered membrane environment is the other major contributor.
The functional form of Pm in this work follows Eq. 2. The overall scheme of the model is outlined in Figure 1. The focus of this study is on estimating the membrane/water partition based upon an expanded physical model of interfacial transfer in which the permeant is postulated to assume a neutral “membranephilic” state from the “hydrophilic” conformational ensemble in order to partition into a size-sensitive low-dielectric barrier region. The transmembrane diffusivity is estimated by diffusion coefficient across the low dielectric barrier. The effective thickness of the barrier is treated as an intrinsic characteristic of the membrane which is, in turn, assumed to be consistent for each assay of the same study and unaffected by the permeant. Since predictions were not cross-compared between different data sets or data types, the value of δbarrier was defined as 18 Å for a general membrane in all reported calculations.54
Figure 1.
The proposed physical model of passive membrane permeability.
Partition coefficient estimation
Following the barrier domain model proposed by Xiang and Anderson from experimental and theoretical studies,53,54 the lipid/water partition coefficient was estimated from the partition between water and a bulk organic solvent that resembles the membrane environment. In order to reproduce the chemical environment in the membrane, organic solvents with comparable hydrophobicity as that of the lipid membrane are employed as the model medium, such as 1,9-decadiene and 1-hexadecene.54 As such a reference medium is isotropic in nature, the estimation is augmented by a size-dependent factor to incorporate the anisotropic property of size/shape selectivity in membrane permeation. In essence, the correction factor relates the permeant’s size to the spatial constraints in the membrane, where are different from those in a disordered organic liquid; the correction factor is formulated as the reversible work of creating a cavity to accommodate the permeant under the pressure exerted by the lipid molecules. In the present implementation, the lipid barrier/water partition coefficient, Kbarrier, is computed from the partition coefficient between water and chloroform, Kc/w, and the size selectivity factor, ξ V, as Eq. 3 below.
| (3) |
Based upon properties of both the permeant (volume) and the membrane (interior pressure within the membrane), ξ V is defined as:
| (4) |
where kB is the Boltzmann constant, T is the temperature, Vp is the volume of the permeant, P‖ is the normal pressure, and P⊥ is the lateral pressure exerted by the lipid molecules. In the current model, P‖ and P⊥ are set to 1 bar and 300 bar, respectively.54 This factor lowers the partition coefficient exponentially as the volume of the permeant increases, thus incorporating at least a part of the observed size-dependence of membrane permeability. As shown elsewhere, differences in membrane composition, such as the addition of cholesterol, can be approximated as differences in lateral pressure.77
In our previous model, we effectively ignored the size-dependence of permeation, a limitation that we remedy here, along with other improvements. The focus of our previous work was explicit treatment of conformational flexibility of permeants, a strength that we retain in this work. Permeability was evaluated as the conformation-dependent partition free energy between a high dielectric environment (water) and a low dielectric environment (chloroform) using a conformational search approach.61–65 A key concept of the model was to link permeability with the structural flexibility of the permeant by allowing conformational change in response to the change of the surroundings from water to membrane. The predicted relative permeabilities were based on the lowest energy conformation in the low dielectric continuum, which we termed the low dielectric conformation (LDC). With the assumption that the desolvation penalty is proportional to the free energy cost of the partitioning, the partition free energy was calculated as the free energy of transfer of the LDC from water to chloroform, ΔGtransfer, as Eq. 5.
| (5) |
where ECHCl3 and EH2O are, respectively, the energies of the LDC in chloroform and water. Although this simple model clearly ignores many aspects of the actual permeation process, the predicted ΔGtransfer values, namely the desolvation costs, correlated well with experimental permeability measurements for cyclic peptides and drug-like molecules.61–65 Those promising results indicate that conformational sampling of the permeant can be important for flexible molecules, and the approximation of the lipid/water interface with implicit solvent models is reasonable in the context of the model.
Building upon this simple model and the work of others, this work presents a more sophisticated model with improved estimation of the Kbarrier. Here, the permeant is assumed to retain a neutral (deionized, if possible) state, as stated by the pH partition theory,78 and to adopt one particular “membranephilic conformation” when diffusing across the membrane. Conceptually, we utilize a thermodynamic cycle in which all the additional contributions to Kbarrier concern the permeant in the water phase prior to membrane permeation, including (1) the deionization penalty, (2) the tautomerization penalty of maintaining a certain tautomeric state, and (3) the conformational change from the lowest energy conformer in water to the membranephilic conformer (Fig. 1).
Charged molecules permeate through membranes much more slowly than neutral compounds, and our model assumes that for drug-like molecules, the permeation rate is dominated by the neutral species, even if it is present at a relatively low concentration in water. The free energy cost of charge neutralization of the permeant can be assessed by first predicting the pKa values for possible ionizable groups, followed by quantifying the free energy required to maintain a zero net charge with all ionization groups deionized, if possible, at a desired pH. Most in vitro assays are performed with the aqueous phase at pH 7.4, and so we adopted this value for the results represented here; it is straightforward to generate results for other pH’s if desired. Similarly, a tautomerization penalty is derived from possible tautomer states and their estimated relative populations. These two processes are combined as a state penalty, ΔGstate, that represents the free energy cost for the permeant to adopt a particular neutral, tautomeric form for membrane permeation.
In previous work, we emphasized the role of internal hydrogen bonds in minimizing the free energy cost of partitioning into the membrane. However, adopting such a LDC with favorable hydrogen bonds may result in internal strain in the compound as well as entropy loss relative to the ensemble of states present in water, both of which would reduce permeation rates. To estimate these effects, we assumed that each permeant has only one probable membranephilic conformation, which we identify by conformational sampling. The free energy cost corresponding to this conformational change is quantified as the conformer-focusing free energy, ΔGcf, which reflects the cost of retaining a specific conformation. Such an approach for quantifying the conformational penalty as ΔGcf was originally employed in the context of protein-ligand binding with the purpose of estimating the free energy cost required for the unbound ligand to adopt the bound conformation while giving up other conformational states.79 In light of the similar scenario in our model of passive permeation, this two-state model is applied to estimate a conformational penalty of adopting the membranephilic conformation. The conformational shift to the membranephilic state is assumed to occur in water, so the calculation of ΔGcf requires knowledge of the conformational ensemble and the corresponding energetics in water. In the simple case where the permeant has i conformational states in the high dielectric environment but only one state in the low dielectric environment, i.e. the membranephilic state, the ΔGcf can be estimated as the following:
| (6) |
where ELDC(HD) is the energy of the sole low dielectric state in the high dielectric environment, EHDC is the energy of the lowest energy conformer in the high dielectric environment, namely the high dielectric conformation (HDC), EHD,i is the energy of state i in the high dielectric environment, and ni is the degeneracy of state i. The first term in Eq. 6 is the reorganization cost obtained from the relative energy difference between the membranephilic state and the HDC in the high dielectric environment. The second term accounts for the loss of conformational states, which is quantified by the modified partition function that includes the relative energy difference between the HDC and all other conformation states in the high dielectric environment. While this approach is highly approximate, it is more rigorous than estimating the effect by rotatable bond count, which can be misleading without considering the conformational ensemble.80 As the calculation of ΔGcf is relatively straightforward, we tested whether this simple approximation would contribute positively to the ability to reproduce experimental permeation rates.
Combining ΔGtransfer with the state and conformational penalties, the partition free energy between chloroform and water, ΔGc/w, and the corresponding Kc/w can be obtained from Eq. 7 and Eq. 8, respectively.
| (7) |
| (8) |
Finally, Kbarrier can subsequently be computed from Kc/w and ξV as defined in Eq. 3.
An alternative method reported by Seelig et al. to estimate Kbarrier from the air/water partition is examined as well.60 The formulation of Kbarrier in this approach is very similar to that of Eq. 3 from which the air/water partition is scaled by a shape-dependent insertion factor. This approach to calculating Kbarrier is investigated with our physics-based model by which the air/water partition is derived from the solvation free energy and the shape is measured as the cross-sectional area of the permeant. However, relative to the barrier domain model using the chloroform/water partition as the reference state, application of this method does not result in any significant improvement in accuracy. A more detailed discussion of this approach and its performance against various experimental data is included in the Supporting Information.
Diffusion coefficient
We use a very simple assumption that the rate-limiting barrier is similar to that of the bulk solvent, and the Stokes-Einstein relation is applied to describe the diffusion across the barrier.53 The size-dependent diffusion coefficient of the permeant in the barrier, Dbarrier, is computed using the Stokes-Einstein equation for spherical particle:
| (9) |
where ηm is the membrane viscosity that is assumed to be 1 poise and rp is the radius of the permeant. Following the spherical approximation of Eq. 9, rp is estimate from volume of the permeant, Vp.
| (10) |
In this work, we do not consider the rate of diffusion across the unstirred water layer, because the width of this barrier will vary significantly between assays and is difficult to estimate.
COMPUTATIONAL PROCEDURES
The overall workflow of the permeability calculation is outlined in Figure 2. The coordinates of compounds were downloaded from the PubChem database if available or prepared in Maestro.81 All downloaded 2-dimensional structures were converted to 3-dimensional structures by the Ligprep module (Schrödinger). Prior to the permeability calculations, all compounds were minimized using MacroModel (Schrödinger) with the OPLS2005 force field and the implicit water model.82–84 Molecular descriptors and QSPR regression-based predictions were computed by QikProp (Schrödinger) for all optimized input structures.85
Figure 2.
Computational protocol of the physics-based permeability model.
Using EPIK, the probable ionization states and tautomeric forms of the permeant were first generated along with their pKa values, followed by the estimation of the free energy cost of neutralizing the compound at pH 7.4.86,87 The next step was to generate conformers for each of these neutral states using the PLOP program.88 The OPLS2005 force field parameters of the permeant were generated by the hetgrp_ffgen program (Schrödinger). A semi-exhaustive sampling in the torsional space was carried out to identify sterically allowed conformations, as described elsewhere.88 The subsequent clustering based on Cartesian atomic coordinates identified a maximum of 400 representative conformations. A major difference between the current protocol and previously published work is that, rather than focusing only on one LDC, all low energy conformations of the permeant were processed and evaluated. Energy evaluations in chloroform and water were performed for all unique conformations with the use of the implicit solvent model.89–91 For computational efficiency, only conformers that are within a 5 kcal/mol threshold from the lowest energy conformation in either water or chloroform were included in the final conformer ‘ensemble’. The solvent-accessible volume in water of each of the selected conformations was computed by QikProp to calculate the size-selectivity factor and diffusion coefficient. Finally, the conformer with the highest predicted permeability was selected as the membranephilic conformation. The calculation on average takes about 2–4 minutes per molecule on a single-core CPU using PRIME (Schrödinger) for conformational search and energy calculations.92 The Li set, which consists of flexible peptidomimetics with 11–18 rotatable bonds, is the only exception, with average CPU time of 38.6 minutes per molecule.
RESULTS
To benchmark and evaluate the computational models, we collected permeability data from the literature, which is dominated by PAMPA and cell-based assays. In many cases, the permeability studies were performed to evaluate or parameterize other methods. The general criteria we followed in selecting data sets include 1) permeability data reported from the same source (for at least one data type), 2) the dynamic range of the data spans at least 1 log unit, and 3) broad chemical diversity, including both diverse and congeneric series of drug-like (FDA approved drugs or other small molecules) and non-drug-like compounds (peptidomimetics that do not follow Lipinski’s Rules). The 9 examined data sets include 188 compounds in total from which 36 compounds have data reported in multiple sources.
For the cell-based assay data, a major concern is that the permeability measurements were polarized by facilitated transport mechanisms, which are beyond the scope of the current model. It is important to reinforce the idea that the observed permeability from these assays is a combined effect of passive permeation, facilitated transport and paracellular transport. The cellular permeation rate in the A→B direction can be considered as an additive rate of different processes:
| (11) |
where Pcell,A→B is the cellular permeability rate in the A→B direction, Ptranscellular is the passive transcellular permeability, Pparacellular is the paracellular permeability, Pactive,A→B is the active “influx” rate in A→B direction, and Pactive,B→A is the active “efflux” rate in the B→A direction. Assessing passive permeability solely based on unidirectional cell-based permeability data may not be straightforward. For example, a non-PGP substrate with low passive permeability and a strong PGP substrate with high passive permeability may have similar Pcell,A→B values. In the present study, we invoke the assumption that the transmembrane or transcellular permeability is the dominant component of the overall permeation process. We identified 37 compounds in the data sets that have reported active transport with PGP or other transporters. As listed in the Supporting Information, six cell-based assay data sets, including the Avdeef, Balimane, Bermejo, Fujikawa and the two Irvine sets, have compounds with reported active transport mechanisms. Hence, in addition to the analysis with all compounds, a passive permeation subset with active transport compounds excluded was also examined for each of these sets. The following analyses and discussions for these cell-based data sets are based on the results from the passive permeation subsets. The analyses with all compounds including those with reported active transport are reported in the Supporting Information. The Caco-2 permeability measurements for the Goodwin set were obtained in the presence of a PGP substrate, verapamil, which was used to minimize the effect of facilitated transport. The reported efflux ratio also provided evidence that active transport did not take place, so we did not exclude any compounds from this data set. For the Li set, information from the original article is insufficient to identify actively transported compounds, and we did not eliminate any from this set either.
The metrics for assessing the performance of the computational predictions include 1) the correlation coefficient (r2) and the slope (m) from the linear regression model with the experimental data, and 2) the Spearman’s rank correlation coefficient (ρ), which compares the rankings by predictions and experimental data. We do not combine data sets from different papers, because there are frequently significant differences in the assay conditions and other experimental details. Some compounds appear in multiple data sets and indeed the reported permeation rates vary significantly in some cases. As a result, we focus our analysis on the ability to reproduce relative, not absolute, permeation rates. Also, few predictions with statistical insignificant ρ (p-value >0.05) are noted in the analyses. These predictions mostly show low correlation with the experimental data, or are for data sets that have either a small sample size (<10) or a relatively small dynamic range (Whitlock set).
Results for in vitro measurements
The correlation coefficients (r2) from linear regression analyses and the Spearman’s rank correlation coefficients (ρ) between the in vitro permeability data and our physics-based predicted permeabilities are summarized in Tables 2 and 3. Following the relationship stated in Eq. 8, the calculated free energy values were converted to LogK for comparisons with LogPm and experimental data. The linear regression models for the original (LogK(ΔGtransfer(LDC))) and the present (LogPm) physics-based predictions are shown in Figures 3 (PAMPA) and 4 (cell-based assays). Overall, the physics-based predictions correlate well with experimental data, producing r2 >0.55 and ρ >0.70 for most data sets.
Table 2.
The correlation coefficients between PAMPA data and computational predictions. The key component(s) of each physical model is/are marked with “X”. Physics-based predictions with correlation coefficient differences (Δr2 or Δρ) relative to LogK(ΔGtransfer(LDC)) ≥0.05 and ≤-0.05 are highlighted in green and red, respectively. QikProp predictions with correlation coefficient differences relative to LogPm ≥0.05 (Δ|r2| or Δ|ρ|) are highlighted in light green.
| Model’s component | Correlation coefficient | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictiona | Conf. search |
Conf. ensemble |
State penalty |
Conf. penalty |
Size selectivity |
Membrane diffusion |
Avdeef | Balimane | Bermejo | Di | Fujikawa | Li | Whitlock | |||||||
| r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | |||||||
| LogK(ΔGtransfer(LDC)) | × | 0.73 | 0.82 | 0.57 | 0.73 | 0.77 | 0.94 | 0.47 | 0.67 | 0.43 | 0.65 | 0.57 | 0.74 | 0.69 | 0.97 | |||||
| LogK(ΔGtransfer) | × | × | 0.73 | 0.83 | 0.57 | 0.75 | 0.80 | 0.95 | 0.49 | 0.68 | 0.43 | 0.63 | 0.52 | 0.71 | 0.62 | 0.89 | ||||
| LogK(ΔGtransfer+ΔGstate) | × | × | × | 0.75 | 0.90 | 0.65 | 0.85 | 0.90 | 0.98 | 0.58 | 0.71 | 0.54 | 0.71 | 0.79 | 0.78 | 0.74 | 0.85 | |||
| LogK(ΔGtransfer+ΔGcf) | × | × | × | 0.77 | 0.88 | 0.57 | 0.77 | 0.82 | 0.94 | 0.50 | 0.69 | 0.47 | 0.67 | 0.65 | 0.76 | 0.59 | 0.86 | |||
| LogK(ΔGc/w) | × | × | × | × | 0.78 | 0.88 | 0.64 | 0.83 | 0.88 | 0.95 | 0.57 | 0.72 | 0.56 | 0.73 | 0.82 | 0.76 | 0.74 | 0.85 | ||
| LogKbarrier | × | × | × | × | × | 0.80 | 0.91 | 0.62 | 0.83 | 0.82 | 0.95 | 0.56 | 0.70 | 0.58 | 0.73 | 0.83 | 0.81 | 0.78 | 0.87 | |
| LogPm | × | × | × | × | × | × | 0.80 | 0.91 | 0.61 | 0.83 | 0.81 | 0.95 | 0.56 | 0.70 | 0.58 | 0.74 | 0.83 | 0.81 | 0.78 | 0.87 |
| MW | Physiochemical descriptor | 0.11 | 0.33b | 0.03 | 0.13b | 0.12 | 0.62 | 0.00 | −0.31b | 0.19 | −0.47 | 0.13 | 0.33b | 0.41 | −0.36b | |||||
| Vol | Physiochemical descriptor | 0.21 | 0.55b | 0.10 | 0.23b | 0.22 | 0.67 | 0.02 | −0.17b | 0.13 | −0.41 | 0.15 | 0.32b | 0.07 | 0.13b | |||||
| PSA | Physiochemical descriptor | 0.61 | −0.76 | 0.45 | −0.68 | 0.16 | −0.72 | 0.45 | −0.66 | 0.39 | −0.60 | 0.12 | −0.47 | 0.17 | −0.66b | |||||
| QPlogPo/w | QSPR regression-based model | 0.86 | 0.92 | 0.52 | 0.72 | 0.65 | 0.81 | 0.53 | 0.75 | 0.18 | 0.33 | 0.12 | 0.36b | 0.16 | 0.68b | |||||
| LogQPPCaco | QSPR regression-based model | 0.67 | 0.75 | 0.71 | 0.87 | 0.29 | 0.80 | 0.59 | 0.76 | 0.47 | 0.60 | 0.63 | 0.84 | 0.16 | 0.68b | |||||
| LogQPPMDCK | QSPR regression-based model | 0.74 | 0.76 | 0.73 | 0.89 | 0.28 | 0.77 | 0.57 | 0.76 | 0.41 | 0.58 | 0.70 | 0.85 | 0.22 | 0.73b | |||||
| N(Total)c | 11 | 22 | 16 | 27 | 70 | 25 | 8 | |||||||||||||
| N(QikProp-TS)d | 10 | 15 | 0 | 14 | 37 | 0 | 0 | |||||||||||||
Optimal values were determined from the refined conformational ensemble for each physics-based prediction, except for ΔGtransfer(LDC), which were computed using the LDC, and for LogKbarrier, which was computed using the conformation with the optimal LogPm value.
The p-value of ρ is greater than 0.05.
Number of compounds included in the analysis.
Number of compounds in QikProp training sets (QPlogPo/w, QPPCaco, and QPPMDCK). The physics-based model does not use a training set.
Table 3.
The correlation coefficients between cell-based assay data and computational predictions. The key component(s) of each physical model is/are marked with “X”. Physics-based predictions with correlation coefficient differences (Δr2 or Δρ) relative to LogK(ΔGtransfer(LDC)) ≥0.05 and ≤-0.05 are highlighted in green and red, respectively. QikProp predictions with correlation coefficient differences relative to LogPm (Δ|r2| or Δ|ρ|) ≥0.05 are highlighted in light green.
| Model’s component | Correlation coefficient | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictiona | Conf. search |
Conf. ensemble |
State penalty |
Conf. penalty |
Size selectivity |
Membrane diffusion |
Avdeef (Caco-2) |
Balimane (Caco-2) |
Bermejo (Caco-2) |
Fujikawa (Caco-2) |
Goodwin (Caco-2) |
Irvine (Caco-2) |
Irvine (MDCK) |
Li (Caco-2) |
||||||||
| r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | r2 | ρ | |||||||
| LogK(ΔGtransfer(LDC)) | × | 0.67 | 0.89 | 0.56 | 0.44b | 0.39 | 0.50b | 0.32 | 0.35b | 0.83 | 0.89 | 0.59 | 0.77 | 0.68 | 0.83 | 0.49 | 0.61 | |||||
| LogK(ΔGtransfer) | × | × | 0.70 | 0.89 | 0.55 | 0.44b | 0.37 | 0.45b | 0.33 | 0.35b | 0.88 | 0.96 | 0.57 | 0.75 | 0.67 | 0.83 | 0.55 | 0.69 | ||||
| LogK(ΔGtransfer+cGstate) | × | × | × | 0.80 | 0.89 | 0.55 | 0.56b | 0.58 | 0.52b | 0.35 | 0.43b | 0.88 | 0.96 | 0.68 | 0.80 | 0.74 | 0.84 | 0.49 | 0.66 | |||
| LogK(ΔGtransfer+cGcf) | × | × | × | 0.75 | 0.89 | 0.53 | 0.54b | 0.44 | 0.47b | 0.30 | 0.30b | 0.82 | 0.92 | 0.61 | 0.78 | 0.70 | 0.84 | 0.54 | 0.65 | |||
| LogK(ΔGc/w) | × | × | × | × | 0.83 | 0.89 | 0.50 | 0.56b | 0.61 | 0.55b | 0.32 | 0.41b | 0.82 | 0.92 | 0.69 | 0.82 | 0.75 | 0.84 | 0.45 | 0.64 | ||
| LogKbarrier | × | × | × | × | × | 0.84 | 0.89 | 0.55 | 0.56b | 0.59 | 0.55b | 0.34 | 0.37b | 0.86 | 0.95 | 0.70 | 0.81 | 0.77 | 0.84 | 0.45 | 0.66 | |
| LogPm | × | × | × | × | × | × | 0.84 | 0.89 | 0.56 | 0.56b | 0.59 | 0.55b | 0.34 | 0.37b | 0.86 | 0.95 | 0.70 | 0.81 | 0.77 | 0.84 | 0.45 | 0.66 |
| MW | Physiochemical descriptor | 0.00 | 0.14b | 0.31 | −0.33b | 0.00 | −0.01b | 0.08 | −0.01b | 0.05 | −0.23b | 0.12 | −0.43 | 0.19 | −0.43 | 0.04 | 0.18b | |||||
| Vol | Physiochemical descriptor | 0.06 | 0.37b | 0.13 | −0.51b | 0.01 | −0.02b | 0.00 | 0.02b | 0.01 | 0.01b | 0.02 | −0.21b | 0.05 | −0.23b | 0.06 | 0.23b | |||||
| PSA | Physiochemical descriptor | 0.41 | −0.71b | 0.59 | −0.59b | 0.07 | −0.17b | 0.34 | −0.20b | 0.83 | −0.89 | 0.54 | −0.79 | 0.63 | −0.78 | 0.30 | −0.66 | |||||
| QPlogPo/w | QSPR regression-based model | 0.74 | 0.71b | 0.50 | 0.71 | 0.39 | 0.38b | 0.32 | 0.55 | 0.53 | 0.76 | 0.27 | 0.48 | 0.31 | 0.54 | 0.15 | 0.45 | |||||
| LogQPPCaco | QSPR regression-based model | 0.91 | 0.94 | 0.47 | 0.66b | 0.42 | 0.62 | 0.81 | 0.81 | 0.84 | 0.90 | 0.74 | 0.85 | 0.81 | 0.88 | 0.55 | 0.72 | |||||
| LogQPPMDCK | QSPR regression-based model | 0.91 | 0.94 | 0.51 | 0.66b | 0.32 | 0.47b | 0.68 | 0.74 | 0.76 | 0.83 | 0.73 | 0.83 | 0.81 | 0.87 | 0.56 | 0.68 | |||||
| N(Total)c | 6 | 9 | 12 | 20 | 12 | 31 | 32 | 25 | ||||||||||||||
| N(QikProp-TS)d | 6 | 7 | 0 | 12 | 0 | 31 | 32 | 0 | ||||||||||||||
Optimal values were determined from the refined conformational ensemble for each physics-based prediction, except for ΔGtransfer(LDC), which were computed using the LDC, and for LogKbarrier, which was computed using the conformation with the optimal LogPm value.
The p-value of ρ is greater than 0.05.
Number of compounds included in the analysis.
Number of compounds in QikProp training sets (QPlogPo/w, QPPCaco, and QPPMDCK). The physics-based model does not use a training set.
Figure 3.
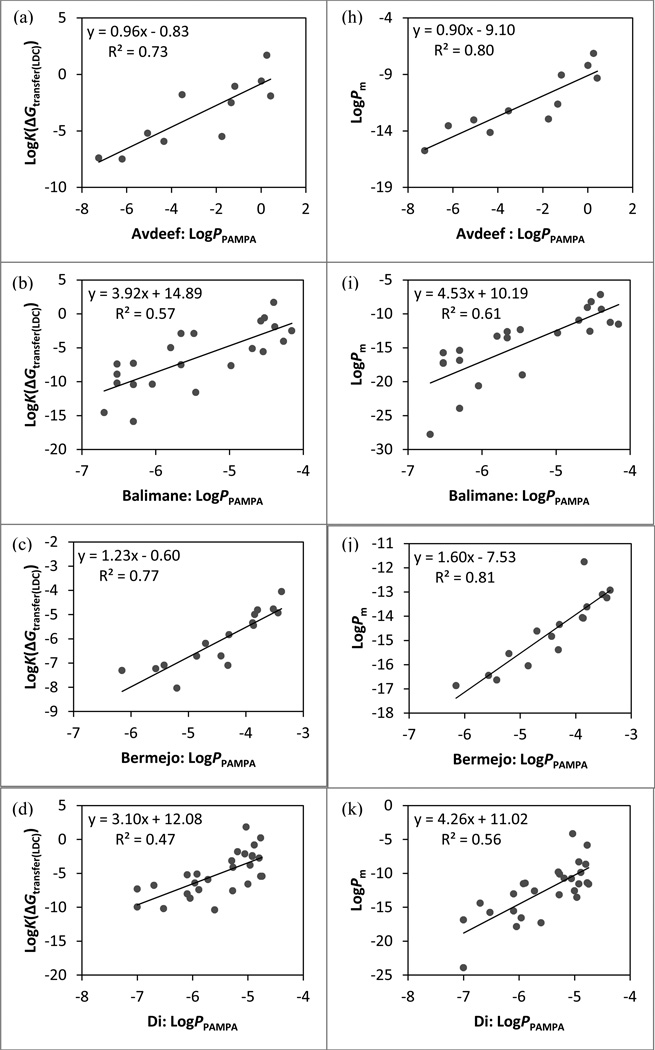
The linear regression models between PAMPA data and physics-based permeability predictions. Predictions based on LogK(cGtransfer(LDC)): (a) Avdeef set, (b) Balimane set, (c) Bermejo set, (d) Di set, (e) Fujikawa set, (f) Li set, and (g) Whitlock set. Predictions based on LogPm: (h) Avdeef set, (i) Balimane set, (j) Bermejo set, (k) Di set, (l) Fujikawa set, (m) Li set, and (n) Whitlock set.
Figure 4.
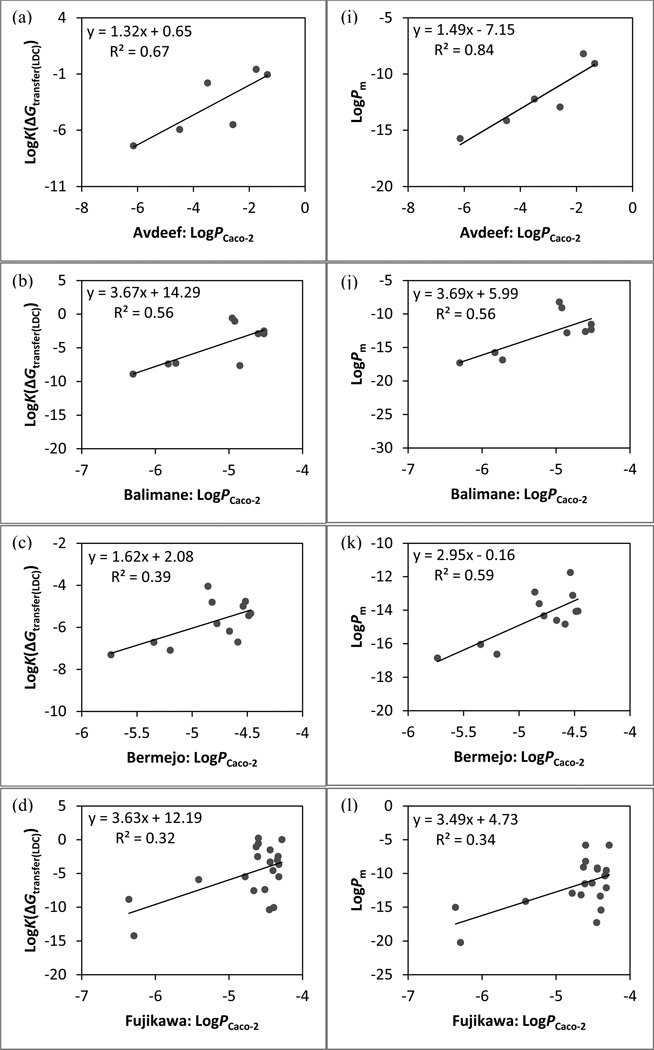
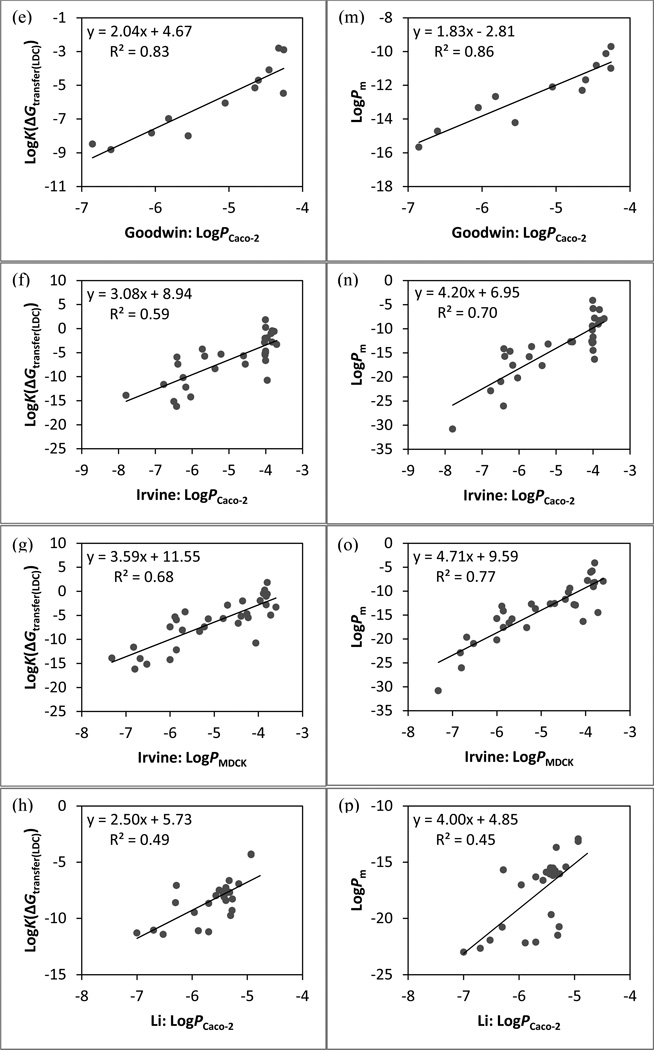
The linear regression models between cell-based assay data and physics-based permeability. Predictions based on LogK(cGtransfer(LDC)): (a) Avdeef set (Caco-2), (b) Balimane set (Caco-2), (c) Bermejo set (Caco-2), (d) Fujikawa set (Caco-2), (e) Goodwin set (Caco-2), (f) Irvine set (Caco-2), (g) Irvine set (MDCK), and (h) Li set (Caco-2). Predictions based on LogPm: (i) Avdeef set (Caco-2), (j) Balimane set (Caco-2), (k) Bermejo set (Caco-2), (l) Fujikawa set (Caco-2), (m) Goodwin set (Caco-2), (n) Irvine set (Caco-2), (o) Irvine set (MDCK), and (p) Li set (Caco-2).
In order to evaluate the performance of the updated method in calculating the partition free energy systematically, new components were added to the model progressively. Obtaining the free energy of transfer from the refined ensemble of low energy conformations (ΔGtransfer) instead of from the single lowest-energy conformation in low dielectric (ΔGtransfer(LDC)) has no significant effect on the correlation between calculations and experiments for most sets. The incorporation of either the state penalty (ΔGtransfer+ΔGstate) or the conformational penalty (ΔGtransfer+ΔGcf) is shown to be favorable for modeling both types of in vitro permeability data. Including the ΔGstate term leads to significant improvements in r2 and ρ for many data sets, reflecting the importance of accounting for neutralization of ionizable groups and tautomerization. While imposing the conformational penalty is constructive as well, the beneficial effects are noticeably smaller than that of the state penalty. We speculate that this may be due to the fact that the examined data sets contain many relatively rigid drug-like molecules for which the conformational effect is likely to be small.93 As anticipated, the biggest improvement in r2 by introducing the ΔGcf term alone is seen with the PAMPA data of flexible peptidomimetics with 11–18 rotatable bonds from the Li set (Δr2 = 0.13 between ΔGtransfer and ΔGtransfer+ΔGcf). Computing the partitioning free energy with both the state and conformational penalties (ΔGc/w) yields similar correlation coefficients as those by including ΔGstate alone for both PAMPA and cell-based data, suggesting that ΔGstate is the dominant term.
An important aspect of the physical model considered here, building on prior work, is the dependence of membrane permeation and diffusion on the size of the permeant. The results from LogKbarrier values of conformations with the highest LogPm values illustrate that the size-selectivity term does improve r2 over predictions by ΔGc/w for more than half of the in vitro data sets. However, the resulting improvements in accuracy are relatively small for either PAMPA or cell-based data as most changes in either correlation coefficient are less than 0.05. The addition of the size-dependent diffusion coefficient is also shown to have no significant effect. Given that the diffusion coefficient is inversely proportional to the radius of the permeant and the molecular sizes within each set are not very different, the diffusion terms of any pair of molecules are thus expected to be similar.
The slopes of the linear regression models are reported in Tables 4 and 5 for the PAMPA and cell-based assays, respectively. These slopes are variable and generally greater than one. The slopes do not generally vary dramatically based on which aspects of the model are included. That is, as previously reported, the simplest model based on ΔGtransfer(LDC) values (conformation-dependent solvation free energy) produces linear regression models with slopes steeper than one.63 It is important to note that the slopes are sensitive to the dielectric constant used for the membrane; using chloroform as the reference state, with dielectric ~4, may underestimate the effective dielectric constant of the barrier region. We cannot fully account for the variability in slope observed for the different data sets. Differences in assay conditions, such as pH, the composition of the artificial membrane, cell layer, may contribute. We speculate that entropic losses, which we cannot account for in a rigorous manner, may contribute to this variability as well (i.e. some of the series involve larger, more flexible ligands, while others have predominantly small, rigid compounds). In addition, the current model neglects the aqueous boundary layer at the water/membrane interface, which can act as another rate limiting region for passive permeation. The volume-based size-selectivity factor in the present implementation is a relatively simple approximation. Exploring other approaches, such as free-surface-area theory used to model membrane order, may also be constructive.57,58
Table 4.
The slopes from linear regression analyses between PAMPA data and computational predictions.
| Slope of linear regression model (|m|) | |||||||
|---|---|---|---|---|---|---|---|
| Prediction | Avdeef | Balimane | Bermejo | Di | Fujikawa | Li | Whitlock |
| LogK(ΔGtransfer(LDC)) | 0.96 | 3.92 | 1.23 | 3.10 | 2.64 | 1.36 | 3.76 |
| LogK(ΔGc/w) | 0.99 | 4.73 | 1.77 | 4.35 | 3.29 | 2.83 | 5.08 |
| LogKbarrier | 0.91 | 4.54 | 1.61 | 4.26 | 3.51 | 2.74 | 5.19 |
| LogPm | 0.90 | 4.53 | 1.60 | 4.26 | 3.53 | 2.74 | 5.19 |
| QPlogPo/w | 0.45 | 1.41 | 0.86 | 2.11 | 0.64 | 0.29 | 0.66 |
| LogQPPCaco | 0.14 | 0.70 | 0.17 | 0.87 | 0.58 | 0.46 | 0.32 |
| LogQPPMDCK | 0.13 | 0.77 | 0.16 | 0.97 | 0.61 | 0.47 | 0.42 |
Table 5.
The slopes from linear regression analyses between cell-based assay data and computational predictions.
| Slope of linear regression model (|m|) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Prediction | Avdeef (Caco-2) |
Balimane (Caco-2) |
Bermejo (Caco-2) |
Fujikawa (Caco-2) |
Goodwin (Caco-2) |
Irvine (Caco-2) |
Irvine (MDCK) |
Li (Caco-2) |
| LogK(ΔGtransfer(LDC)) | 1.32 | 3.67 | 1.62 | 3.63 | 2.04 | 3.08 | 3.59 | 2.50 |
| LogK(ΔGc/w) | 1.52 | 3.47 | 3.10 | 3.49 | 1.79 | 4.16 | 4.65 | 4.15 |
| LogKbarrier | 1.49 | 3.67 | 2.96 | 3.49 | 1.83 | 4.20 | 4.71 | 4.00 |
| LogPm | 1.49 | 3.69 | 2.95 | 3.49 | 1.83 | 4.20 | 4.71 | 4.00 |
| QPlogPo/w | 0.51 | 2.18 | 1.42 | 1.57 | 0.71 | 0.67 | 0.70 | 0.64 |
| LogQPPCaco | 0.22 | 0.72 | 0.48 | 0.95 | 0.29 | 0.63 | 0.70 | 0.85 |
| LogQPPMDCK | 0.24 | 0.76 | 0.39 | 1.02 | 0.20 | 0.66 | 0.74 | 0.82 |
Overall, the results suggest that the conformation-dependent free energy cost of desolvating permeants (as quantified as ΔGtransfer) is a dominant factor affecting the rate of permeation. However, other factors expected to also be important, such as charge neutralization and size-dependent effects, do appear to also contribute, in that including these terms improves correlation with in vitro measures of permeability. The Pm prediction yields higher r2 and ρ values with experimental data than the previous basic model of ΔGtransfer for almost all cases, illustrating the ability to improve a physical model by incorporating description of the underlying physical processes. Further improvements may be possible in future work.
In contrast to the obvious improvements when comparing the PAMPA data, the favorable effects of the additional components to the model are less profound for the cell-based assay data but, as expected, the physical model performs better for compounds not known to be substantially affected by active transport (Supporting Information). These findings suggest that the model does capture key aspects of the permeation process across the biological membrane, albeit that many assumptions and approximations have been made to simplify the calculations required. On the other hand, many of the compounds with known active transport mechanisms are predicted as outliers by our model when compared to the cell-based data, such as occurs in the Irvine set (Supporting Information).
Results for in vivo measurements
Our focus in this study is comparison with in vitro permeability assays, but three of the studies whose data we employ also reported in vivo measurements, and we provide comparisons to those data as well. While the current permeability model does not include other cellular transport components, such as active transport and paracellular transport, the following comparisons with in vivo data are done under the simple assumption that passive transmembrane permeability is a major contributor to the absorption process. Despite being overly simplistic and approximate, such an assumption is implicit in other QSPR models of absorption and in the physiochemical principles underlying Lipinski’s Rules of Five for druglikeness, i.e. LogPo/w <5 and the hydrogen bond donor and acceptor counts.
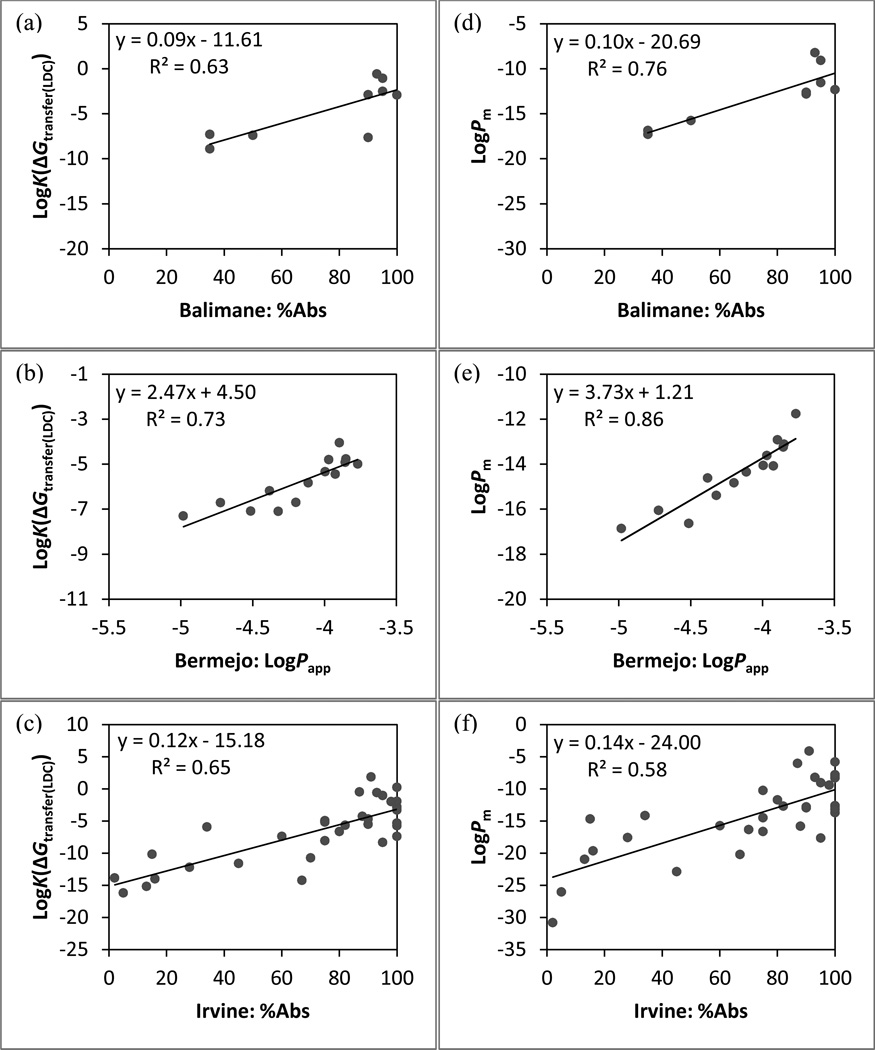
As with the cell-based data sets, compounds with known active transport were not included in the analyses below for the in vivo data sets and the analyses with all compounds included are reported in the Supporting Information. The correlation coefficients r2 and ρ are summarized in Table 6. The linear regression models of predictions, LogK(ΔGtransfer(LDC)) and LogPm, are shown in Figure 5, and the slopes of linear regression models are summarized in Table 7. Overall, these results are consistent with the previous findings from comparisons with the in vitro data. Excluding compounds with active transport yields higher r2 for both Balimane and Irvine sets, while the changes in r2 are minor for the Bermejo set as only two compounds are excluded (Supporting information). The physics-based predictions correlate well with the in vivo measurements, yielding r2 ≥0.7 and ρ ≥0.8 for most cases except for the %Abs data from the Irvine set.
Table 6.
The correlation coefficients between PAMPA data and computational predictions. The key component(s) of each physical model is/are marked with “X”. Physics-based predictions with correlation coefficient differences (Δr2 or Δρ) relative to LogK(ΔGtransfer(LDC)) ≥0.05 and ≤−0.05 are highlighted in green and red, respectively. The QikProp prediction with correlation coefficient difference relative to LogPm (Δ|r2|) ≥0.05 is highlighted in light green.
| Model’s component | Correlation coefficient | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictiona | Conf. search |
Conf. ensemble |
State penalty |
Conf. penalty |
Size selectivity |
Membrane diffusion |
Balimane (%Abs) |
Bermejo (Papp) |
Irvine (%Abs) |
|||
| r2 | ρ | r2 | ρ | r2 | ρ | |||||||
| LogK(ΔGtransfer(LDC)) | × | 0.63 | 0.66c | 0.73 | 0.87 | 0.65 | 0.74 | |||||
| LogK(ΔGtransfer) | × | × | 0.63 | 0.66c | 0.75 | 0.89 | 0.66 | 0.76 | ||||
| LogK(ΔG transfer+ΔGstate) | × | × | × | 0.72 | 0.84 | 0.88 | 0.94 | 0.62 | 0.74 | |||
| LogK(ΔGtransfer+ΔGcf) | × | × | × | 0.65 | 0.76 | 0.78 | 0.91 | 0.63 | 0.74 | |||
| LogK(ΔGc/w) | × | × | × | × | 0.69 | 0.84 | 0.88 | 0.96 | 0.59 | 0.72 | ||
| LogKbarrier | × | × | × | × | × | 0.76 | 0.84 | 0.86 | 0.96 | 0.58 | 0.73 | |
| LogPm | × | × | × | × | × | × | 0.76 | 0.84 | 0.86 | 0.96 | 0.58 | 0.73 |
| MW | Physiochemical descriptor | 0.36 | −0.30b | 0.01 | 0.34b | 0.03 | −0.17b | |||||
| Vol | Physiochemical descriptor | 0.10 | −0.23b | 0.07 | 0.41b | 0.01 | 0.03b | |||||
| PSA | Physiochemical descriptor | 0.65 | −0.84 | 0.16 | −0.70 | 0.54 | −0.59 | |||||
| QPlogPo/w | QSPR regression-based model | 0.52 | 0.76 | 0.62 | 0.77 | 0.40 | 0.56 | |||||
| LogQPPCaco | QSPR regression-based model | 0.68 | 0.86 | 0.38 | 0.80 | 0.47 | 0.55 | |||||
| LogQPPMDCK | QSPR regression-based model | 0.69 | 0.86 | 0.32 | 0.73 | 0.44 | 0.53 | |||||
| QP%HOA | QSPR regression-based model | 0.70 | 0.85 | 0.59 | 0.84 | 0.63 | 0.67 | |||||
| N(Total)c | 9 | 14 | 33 | |||||||||
| N(QikProp-TS)d | 7 | 0 | 33 | |||||||||
Optimal values were determined from the refined conformational ensemble for each physics-based prediction, except for ΔGtransfer(LDC), which were computed using the LDC, and for LogKbarrier, which was computed using the conformation with the optimal LogPm value.
The p-value of ρ is greater than 0.05.
Number of compounds included in the analysis.
Number of compounds in QikProp training sets (QPlogPo/w, QPPCaco, QPPMDCK, and QP%HOA). The physics-based model does not use a training set.
Figure 5.
The linear regression models between in vivo data and physics-based permeability predictions for all compounds. Predictions based on LogK(ΔGtransfer(LDC)): (a) Balimane set, (b) Bermejo set, and (c) Irvine set. Predictions based on LogPm: (d) Balimane set, (e) Bermejo set, and (f) Irvine set.
Table 7.
The slopes from linear regression analyses between in vivo data and computational predictions.
| Slope of linear regression model (|m|) | |||
|---|---|---|---|
| Prediction | Balimane (%Abs) |
Bermejo (Papp) |
Irvine (%Abs) |
| LogK(ΔGtransfer(LDC)) | 0.09 | 2.46 | 0.12 |
| LogK(ΔGc/w) | 0.10 | 3.97 | 0.14 |
| LogKbarrier | 0.10 | 3.75 | 0.14 |
| LogPm | 0.10 | 3.73 | 0.14 |
| QPlogPo/w | 0.05 | 1.86 | 0.03 |
| LogQPPCaco | 0.02 | 0.47 | 0.02 |
| LogQPPMDCK | 0.02 | 0.40 | 0.02 |
| QP%HOA | 0.64 | 19.24 | 0.54 |
Relative to the basic desolvation model, the current model of Pm prediction show improved correlations with Balimane (%Abs) and Bermejo (perfusion assay) sets. No improvement is evident for the %Abs data from the Irvine set, although the physics-based predictions does give reasonable correlation with the data (r2 ~0.6 and ρ ~0.7 for the passive permeation subset). Given that in vivo absorption is a complicated biological process, being able to predict absorption data at this level of accuracy with physics-based passive permeation is encouraging, especially considering that 1) no prior training with existing data was performed, and 2) there is no reason to expect a simple linear relationship between our computed values and %Abs.
To compare the performance of different approaches in predicting human absorption data, the correlation coefficients with %Abs from both experiments and predictions for the Balimane and Irvine sets are summarized in Table 8. The LogPm model is shown to predict %Abs with accuracy comparable to PAMPA data (Balimane set), as well as cell-based assay data when excluding compounds that are actively transported (Irvine set). However, without discriminating compounds with active transport, the cell-based assays are demonstrated to have a stronger correlation with %Abs than the passive permeability prediction for both data sets, as expected.
Table 8.
The correlation coefficients between absorption data and experimental models.
Comparison to other computational methods
To further evaluate the utility of the physics-based permeability model, its predictive power is compared to that of common regression-based QSPR physiochemical descriptors and permeability predictions computed by QikProp, including molecular weight (MW), solvent-accessible volume (Vol), octanol/water partition coefficients (QPlogPo/w), polar surface area (PSA), predicted Caco-2 permeability (QPPCaco), predicted MDCK permeability (QPPMDCK), and predicted percentage human oral absorption (QP%HOA). The results of linear regression analyses and Spearman’s rank correlation coefficients between these descriptors and experimental data are summarized in Tables 2, 3 and 6.
For the in vitro data (Tables 2 and 3), MW and Vol show weak or no correlation with experimental data in most cases, whereas QPlogPo/w and PSA produce stronger correlations. Among all the regression-based predictions, LogQPPCaco, which was trained using many of the compounds in these data sets, yields the highest correlations for both the PAMPA and cell-based assay data, as anticipated. While QPPMDCK is optimized for MDCK data, its performance is similar to that of QPPCaco. In comparison, the physics-based LogPm values correlate better with the PAMPA data than those by QikProp predictions for most cases, generating higher r2 and ρ values. For cell-based assay data, predictions by LogPm values are more accurate than QPlogPo/w and comparable to the QPPCaco and QPPMDCK models.
These parameterized models are, as expected, most effective in predicting sets containing FDA-approved drugs and small molecules, such as for the Avdeef, Irvine and Fujikawa sets (r2 >0.7 for Caco-2 data and r2 >0.8 for MDCK data), which dominate the training sets, but are less optimal for sets with congeneric series and peptidomimetic compounds, like the Bermejo and Li sets (r2 <0.6 for Caco-2 data). An exception is the congeneric series of Goodwin set (LogQPPCaco: r2 = 0.84) from which the Caco-2 permeabilities correlate well with the hydrophobicity of the side-chain substitutions as supported by the strong correlation with PSA (r2 = 0.83). The physics-based model, on the other hand, performs considerably better in predicting relative permeabilities among congeneric series and peptidomimetic compounds. In addition, the quality of the physics-based predictions for drug-like molecules is frequently nearly as good as those given by the regression-based models that were trained specifically for this chemical space.
A notable difference between the two computational approaches is that the slopes of linear regression models with QikProp permeability predictions (QPlogPo/w, QPPCaco and QPPMDCK) are closer to 1 than those by physics-based predictions as shown in Tables 4, 5 and 7. However, the slopes of these QikProp models do vary across different data sets, and the range of slopes is similar to that of the physics-based predictions. This suggests that the slopes of the correlations between the experimental data and computational predictions may be related in part to the details of the experimental assays, and not solely due to limitations of the computational models.
DISCUSSION AND CONCLUSIONS
The performance of a physical model for passive membrane permeability was investigated by comparing its predictive power to experimental data of nine diverse compound sets, as well as regression-based in silico QSPR predictions. In general, the physics-based prediction of Pm is shown to consistently correlate well with the assorted permeability data that covers diverse chemical space. In many cases, the QSPR descriptors (PSA and QPlogPo/w) and parameterized permeability models (QPPCaco, QPPMDCK, and QP%HOA) also deliver comparable agreement with the experimental data. However, a “fair” comparison between the physics-based model and the regression-based model is not straightforward because many compounds in the examined data sets are part of the training sets for the parameterized models. Although QSPR prediction is still significantly faster than the physics-based method, the CPU time of the LogPm calculation is short enough to be practical for many drug design applications (~15 ligands per CPU-hour). As the physics-based predictions are fundamentally different from the QSPR method, performing both types of calculation can be of value as the agreement or disagreement of the results may provide further insight into the molecule.
We wish to emphasize several other potential advantages of this “physics-based” approach. First, the performance in predicting relative permeability differences for a wide-range of data types and compounds reflects the general applicability of the molecular-mechanics based approach. Second, systematically analyzing the contributions of different components of the model provides important physical insights into the contribution of each physical component, such as the effects of ionizable groups and conformational flexibility, elucidating the underlying basis of permeability. Third, as we have shown here, a physically grounded model is systematically improvable by escalating the level of theory and methods employed. The current model can be further improved: in particular, our attempts to estimate entropic losses and the size dependence of the diffusion rate are rather crude (largely for computational expediency), possibly accounting for their limited contribution to the accuracy of the model.
On the other hand, as a future direction, we envision that components of the physics-based model presented here may be used as QSPR descriptors in order to provide a better description of the physical process than many commonly used physiochemical descriptors. That is, the “physics-based” and empirical/statistical approaches may be complementary. For example, we posit that the permeant’s polarity or hydrophilicity, which is often measured by its PSA, is better described by the solvation free energy. Similarly, molecular size and shape are likely more appropriate descriptors than molecular weight. Therefore, we envision a regression-based model using physically-meaningful terms, such as ΔGtransfer, ΔGsolv, and ΔGcf; reweighing each component to better agree with experimental data would be possible while retaining the same functional form of the current model. One potential advantage of such a model would be the ability to estimate a confidence in the prediction that is related to the distance in chemical space between the compounds of interest and the training set.
The current physical model performs best at reproducing the PAMPA data. Considering that the artificial membrane of PAMPA is essentially two half-membrane layers sandwiching a middle organic/hydrophobic layer, the physics-based model and PAMPA both share the same approximation in mimicking the membrane with a low dielectric organic or lipophilic layer. On the other hand, passive permeation across the cell layer in the cell-based assays is more complicated. Efforts were made in the current study to filter the cell-based permeability data by excluding compounds with possible active transport mechanisms. However, factors like paracellular transport and variation in the cell monolayers are expected to also introduce noise in the comparisons. By the same token, intestinal absorption and perfusion assays include contributions from many biological events, in addition to passive permeation. The fact that the proposed model of passive permeability provides reasonable predictions that rival the accuracy of in vitro assays is promising.
Interestingly, the limitations of the physics-based permeability prediction parallel those of predicting protein/ligand interactions. While the permeability model presented in this work is demonstrated to be proficient in predicting relative permeability, calculating absolute membrane permeability would require further development and parameterization. Similarly, predicting relative binding affinity is technically less difficult, less expensive, and more accurate than computing absolute binding affinity. The slopes of the correlations between the computed values and the experimental values are also generally >1 in both cases. Another common feature between the two is that a congeneric series of similar compounds is easier to model than a diverse chemical set.
In summary, the performance of a physics-based permeability model is investigated and benchmarked against in vitro permeability data, in vivo data, and in silico predictions. Without any training against any existing data, the calculated LogPm values are shown to correlate well with different types of experimental data from various studies, suggesting that the model does provide a reasonable physical description of passive membrane permeation. Efforts to apply the model in the context of molecular design and to develop a physical model for gut absorption are underway.
Supplementary Material
ACKNOWLEDGMENT
Gratitude is expressed to the National Institutes of Health (GM086602) for financial support, to Katrina Lexa and Chakrapani Kalyanaraman for suggestions on the manuscript, and to other Jacobson group members, Tyler Day and Ramy Farid from Schrödinger, Inc. for helpful discussions. MPJ is a consultant to Schrödinger, Inc., Pfizer, and Third Rock Ventures.
Footnotes
SUPPORTING INFORMATION
Experimental data of examined data sets, in silico predictions by the physical models and QikProp, compound classifications, chemical structures from congeneric series, analyses with complete sets of compounds for cell-based assay and in vivo data, and the investigation of an alternative approach to estimating lipid barrier/water partition. This material is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 2.Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. Journal of Medicinal Chemistry. 2000;43(21):3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 3.Ekins S, Durst GL, Stratford RE, Thorner DA, Lewis R, Loncharich RJ, Wikel JH. Three-dimensional quantitative structure-permeability relationship analysis for a series of inhibitors of rhinovirus replication. Journal of Chemical Information and Computer Sciences. 2001;41(6):1578–1586. doi: 10.1021/ci010330i. [DOI] [PubMed] [Google Scholar]
- 4.Ekins S, Waller CL, Swaan PW, Cruciani G, Wrighton SA, Wikel JH. Progress in predicting human ADME parameters in silico. Journal of Pharmacological and Toxicological Methods. 2000;44(1):251–272. doi: 10.1016/s1056-8719(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 5.Ekins S, Rose J. In silico ADME/Tox: The state of the art. Journal of Molecular Graphics and Modelling. 2001;20(4):305–309. doi: 10.1016/s1093-3263(01)00127-9. [DOI] [PubMed] [Google Scholar]
- 6.Fujikawa M, Ano R, Nakao K, Shimizu R, Akamatsu M. Relationships between structure and high-throughput screening permeability of diverse drugs with artificial membranes: Application to prediction of Caco-2 cell permeability. Bioorganic & Medicinal Chemistry. 2005;13(15):4721–4732. doi: 10.1016/j.bmc.2005.04.076. [DOI] [PubMed] [Google Scholar]
- 7.Fujiwara S, Yamashita F, Hashida M. Prediction of Caco-2 cell permeability using a combination of MO-calculation and neural network. International Journal of Pharmaceutics. 2002;237(1–2):95–105. doi: 10.1016/s0378-5173(02)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Krarup LH, Christensen IT, Hovgaard L, Frokjaer S. Predicting drug absorption from molecular surface properties based on molecular dynamics simulations. Pharmaceutical Research. 1998;15(7):972–978. doi: 10.1023/a:1011905522110. [DOI] [PubMed] [Google Scholar]
- 9.Liu RF, Sun HM, So SS. Development of quantitative structure-property relationship models for early ADME evaluation in drug discovery. 2. Blood-brain barrier penetration. Journal of Chemical Information and Computer Sciences. 2001;41(6):1623–1632. doi: 10.1021/ci010290i. [DOI] [PubMed] [Google Scholar]
- 10.Malkia A, Murtomaki L, Urtti A, Kontturi K. Drug permeation in biomembranes in vitro and in silico prediction and influence of physicochemical properties. European Journal of Pharmaceutical Sciences. 2004;23(1):13–47. doi: 10.1016/j.ejps.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Refsgaard HHF, Jensen BF, Brockhoff PB, Padkjaer SB, Guldbrandt M, Christensen MS. In silico prediction of membrane permeability from calculated molecular parameters. Journal of Medicinal Chemistry. 2005;48(3):805–811. doi: 10.1021/jm049661n. [DOI] [PubMed] [Google Scholar]
- 12.Stouch TR, Kenyon JR, Johnson SR, Chen XQ, Doweyko A, Li Y. In silico ADME/Tox: why models fail. Journal of Computer-Aided Molecular Design. 2003;17(2):83–92. doi: 10.1023/a:1025358319677. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita F, Wanchana S, Hashida M. Quantitative structure/property relationship analysis of Caco-2 permeability using a genetic algorithm-based partial least squares method. Journal of Pharmaceutical Sciences. 2002;91(10):2230–2239. doi: 10.1002/jps.10214. [DOI] [PubMed] [Google Scholar]
- 14.van de Waterbeemd H, Gifford E. ADMET in silico modelling: Towards prediction paradise? Nature Reviews Drug Discovery. 2003;2(3):192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 15.Dudek AZ, Arodz T, Galvez J. Computational methods in developing quantitative structure-activity relationships (QSAR): A review. Combinatorial Chemistry & High Throughput Screening. 2006;9(3):213–228. doi: 10.2174/138620706776055539. [DOI] [PubMed] [Google Scholar]
- 16.Katritzky AR, Kuanar M, Slavov S, Hall CD, Karelson M, Kahn I, Dobchev DA. Quantitative Correlation of Physical and Chemical Properties with Chemical Structure: Utility for Prediction. Chemical Reviews. 2010;110(10):5714–5789. doi: 10.1021/cr900238d. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida F, Topliss JG. QSAR model for drug human oral bioavailability (vol 43, pg 2578, 2000) Journal of Medicinal Chemistry. 2000;43(24):4723–4723. doi: 10.1021/jm0000564. [DOI] [PubMed] [Google Scholar]
- 18.Suenderhauf C, Hammann F, Maunz A, Helma C, Huwyler J. Combinatorial QSAR Modeling of Human Intestinal Absorption. Molecular Pharmaceutics. 2011;8(1):213–224. doi: 10.1021/mp100279d. [DOI] [PubMed] [Google Scholar]
- 19.Potts RO, Guy RH. A Predictive Algorithm for Skin Permeability – The Effects of Molecular-Size and Hydrogen-Bond Activity. Pharmaceutical Research. 1995;12(11):1628–1633. doi: 10.1023/a:1016236932339. [DOI] [PubMed] [Google Scholar]
- 20.Ottaviani G, Martel S, Carrupt PA. In silico and in vitro filters for the fast estimation of skin permeation and distribution of new chemical entities. Journal of Medicinal Chemistry. 2007;50(4):742–748. doi: 10.1021/jm0611105. [DOI] [PubMed] [Google Scholar]
- 21.Zhao YH, Abraham MH, Ibrahim A, Fish PV, Cole S, Lewis ML, de Groot MJ, Reynolds DP. Predicting penetration across the blood-brain barrier from simple descriptors and fragmentation schemes. Journal of Chemical Information and Modeling. 2007;47(1):170–175. doi: 10.1021/ci600312d. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Zhu H, Oprea TI, Golbraikh A, Tropsha A. QSAR modeling of the Blood-Brain Barrier permeability for diverse organic compounds. Pharmaceutical Research. 2008;25(8):1902–1914. doi: 10.1007/s11095-008-9609-0. [DOI] [PubMed] [Google Scholar]
- 23.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo RP, Harrell PM, Trinh YT, Zhang QH, Urbatsch IL, Chang G. Structure of P-Glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding. Science. 2009;323(5922):1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolghih E, Bryant C, Renslo AR, Jacobson MP. Predicting Binding to P-Glycoprotein by Flexible Receptor Docking. PloS Computational Biology. 2011;7(6):11. doi: 10.1371/journal.pcbi.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adson A, Raub TJ, Burton PS, Barsuhn CL, Hilgers AR, Audus KL, Ho NFH. Quantitative approaches to delineate paracellular diffusion in cultured epithelial-cell monolayers. Journal of Pharmaceutical Sciences. 1994;83(11):1529–1536. doi: 10.1002/jps.2600831103. [DOI] [PubMed] [Google Scholar]
- 26.Knipp GT, Ho NFH, Barsuhn CL, Borchardt RT. Paracellular diffusion in Caco-2 cell monolayers: Effect of perturbation on the transport of hydrophilic compounds that vary in charge and size. Journal of Pharmaceutical Sciences. 1997;86(10):1105–1110. doi: 10.1021/js9700309. [DOI] [PubMed] [Google Scholar]
- 27.Matsson P, Bergstrom CAS, Nagahara N, Tavelin S, Norinder U, Artursson P. Exploring the role of different drug transport routes in permeability screening. Journal of Medicinal Chemistry. 2005;48(2):604–613. doi: 10.1021/jm049711o. [DOI] [PubMed] [Google Scholar]
- 28.Alper HE, Stouch TR. Orientation and diffusion of a drug analog in biomembranes: molecular dynamic simulations. Journal of Physical Chemistry. 1995;99(15):5724–5731. [Google Scholar]
- 29.Bemporad D, Essex JW, Luttmann C. Permeation of small molecules through a lipid bilayer: A computer simulation study. Journal of Physical Chemistry B. 2004;108(15):4875–4884. [Google Scholar]
- 30.Bemporad D, Luttmann C, Essex JW. Computer simulation of small molecule permeation across a lipid bilayer: Dependence on bilayer properties and solute volume, size, and cross-sectional area. Biophysical Journal. 2004;87(1):1–13. doi: 10.1529/biophysj.103.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bemporad D, Luttmann C, Essex JW. Behaviour of small solutes and large drugs in a lipid bilayer from computer simulations. Biochimica Et Biophysica Acta-Biomembranes. 2005;1718(1–2):1–21. doi: 10.1016/j.bbamem.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Marrink SJ, Berendsen HJC. Permeation process of small molecules across lipid membranes studied by molecular dynamics simulations. Journal of Physical Chemistry. 1996;100(41):16729–16738. [Google Scholar]
- 33.Wilson MA, Pohorille A. Mechanism of unassisted ion transport across membrane bilayers. Journal of the American Chemical Society. 1996;118(28):6580–6587. doi: 10.1021/ja9540381. [DOI] [PubMed] [Google Scholar]
- 34.Xiang TX, Anderson BD. A computer simulation of functional group contributions to free energy in water and a DPPC lipid bilayer. Biophysical Journal. 2002;82(4):2052–2066. doi: 10.1016/S0006-3495(02)75553-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksson ESE, dos Santos D, Guedes RC, Eriksson LA. Properties and Permeability of Hypericin and Brominated Hypericin in Lipid Membranes. Journal of Chemical Theory and Computation. 2009;5(12):3139–3149. doi: 10.1021/ct9002702. [DOI] [PubMed] [Google Scholar]
- 36.Eriksson ESE, Eriksson LA. The Influence of Cholesterol on the Properties and Permeability of Hypericin Derivatives in Lipid Membranes. Journal of Chemical Theory and Computation. 2011;7(3):560–574. doi: 10.1021/ct100528u. [DOI] [PubMed] [Google Scholar]
- 37.Iyer M, Mishra R, Han Y, Hopfinger AJ. Predicting blood-brain barrier partitioning of organic molecules using membrane-interaction QSAR analysis. Pharmaceutical Research. 2002;19(11):1611–1621. doi: 10.1023/a:1020792909928. [DOI] [PubMed] [Google Scholar]
- 38.Iyer M, Tseng YJ, Senese CL, Liu JZ, Hopfinger AJ. Prediction and mechanistic interpretation of human oral drug absorption using MI-QSAR analysis. Molecular Pharmaceutics. 2007;4(2):218–231. doi: 10.1021/mp0600900. [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni A, Han Y, Hopfinger AJ. Predicting caco-2 cell permeation coefficients of organic molecules using membrane-interaction QSAR analysis. Journal of Chemical Information and Computer Sciences. 2002;42(2):331–342. doi: 10.1021/ci010108d. [DOI] [PubMed] [Google Scholar]
- 40.Liu JZ, Li Y, Pan DH, Hopfinger AJ. Predicting permeability coefficient in ADMET evaluation by using different membranes-interaction QSAR. International Journal of Pharmaceutics. 2005;304(1–2):115–123. doi: 10.1016/j.ijpharm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Santos OA, Hopfinger AJ. Combined 4D-Fingerprint and clustering based membrane-interaction QSAR analyses for constructing consensus Caco-2 cell permeation virtual screens. Journal of Pharmaceutical Sciences. 2008;97(1):566–583. doi: 10.1002/jps.21086. [DOI] [PubMed] [Google Scholar]
- 42.Bennett WFD, Tieleman DP. Water Defect and Pore Formation in Atomistic and Coarse-Grained Lipid Membranes: Pushing the Limits of Coarse Graining. Journal of Chemical Theory and Computation. 2011;7(9):2981–2988. doi: 10.1021/ct200291v. [DOI] [PubMed] [Google Scholar]
- 43.De Nicola A, Zhao Y, Kawakatsu T, Roccatano D, Milano G. Hybrid Particle-Field Coarse-Grained Models for Biological Phospholipids. Journal of Chemical Theory and Computation. 2011;7(9):2947–2962. doi: 10.1021/ct200132n. [DOI] [PubMed] [Google Scholar]
- 44.Orsi M, Michel J, Essex JW. Coarse-grain modelling of DMPC and DOPC lipid bilayers. Journal of Physics-Condensed Matter. 2010;22(15):15. doi: 10.1088/0953-8984/22/15/155106. [DOI] [PubMed] [Google Scholar]
- 45.Orsi M, Essex JW. Permeability of drugs and hormones through a lipid bilayer: insights from dual-resolution molecular dynamics. Soft Matter. 2010;6(16):3797–3808. [Google Scholar]
- 46.Spassov VZ, Yan L, Szalma S. Introducing an implicit membrane in generalized Born/solvent accessibility continuum solvent models. Journal of Physical Chemistry B. 2002;106(34):8726–8738. [Google Scholar]
- 47.Tanizaki S, Feig M. A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes. Journal of Chemical Physics. 2005;122(12) doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 48.Ulmschneider MB, Ulmschneider JP, Sansom MSP, Di Nola A. A generalized Born implicit-membrane representation compared to experimental insertion free energies. Biophysical Journal. 2007;92:2338–2349. doi: 10.1529/biophysj.106.081810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parisio G, Ferrarini A. Solute Partitioning into Lipid Bilayers: An Implicit Model for Nonuniform and Ordered Environment. Journal of Chemical Theory and Computation. 2010:2267–2280. doi: 10.1021/ct100210u. [DOI] [PubMed] [Google Scholar]
- 50.Diamond JM, Katz Y. Interpretation of nonelectrolyte partition-coefficients between dimyristoyl lecithin and water. Journal of Membrane Biology. 1974;17(2):121–154. doi: 10.1007/BF01870176. [DOI] [PubMed] [Google Scholar]
- 51.Walter A, Gutknecht J. Permeability of small nonelectrolytes through lipid bilayer-membranes. Journal of Membrane Biology. 1986;90(3):207–217. doi: 10.1007/BF01870127. [DOI] [PubMed] [Google Scholar]
- 52.Xiang TX. A computer-simulation of free-volume distributions and related structural-properties in a model lipid bilayer. Biophysical Journal. 1993;65(3):1108–1120. doi: 10.1016/S0006-3495(93)81156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang TX, Anderson BD. Molecular distributions in interphases: statistical mechanical theory combined with molecular dynamics simulation of a model lipid bilayer. Biophysical Journal. 1994;66(3):561–572. doi: 10.1016/s0006-3495(94)80833-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang TX, Anderson BD. The relationship between permeant size and permeability in lipid bilayer-membranes. Journal of Membrane Biology. 1994;140(2):111–122. doi: 10.1007/BF00232899. [DOI] [PubMed] [Google Scholar]
- 55.Xiang TX, Anderson BD. Phospholipid surface-density determines the partitioning and permeability of acetic-acid in DMPC:cholesterol bilayers. Journal of Membrane Biology. 1995;148(2):157–167. doi: 10.1007/BF00207271. [DOI] [PubMed] [Google Scholar]
- 56.Xiang TX, Anderson BD. Permeability of acetic acid across gel and liquid-crystalline lipid bilayers conforms to free-surface-area theory. Biophysical Journal. 1997;72(1):223–237. doi: 10.1016/S0006-3495(97)78661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiang TX, Anderson BD. Influence of chain ordering on the selectivity of dipalmitoylphosphatidylcholine bilayer membranes for permeant size and shape. Biophysical Journal. 1998;75(6):2658–2671. doi: 10.1016/S0006-3495(98)77711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang TX, Chen J, Anderson BD. A quantitative model for the dependence of solute permeability on peptide and cholesterol content in biomembranes. Journal of Membrane Biology. 2000;177(2):137–148. doi: 10.1007/s002320001107. [DOI] [PubMed] [Google Scholar]
- 59.Gerebtzoff G, Seelig A. In silico prediction of blood - Brain barrier permeation using the calculated molecular cross-sectional area as main parameter. J. Chem. Inf. Model. 2006;46:2638–2650. doi: 10.1021/ci0600814. [DOI] [PubMed] [Google Scholar]
- 60.Seelig A. The role of size and charge for blood-brain barrier permeation of drugs and fatty acids. Journal of Molecular Neuroscience. 2007;33(1):32–41. doi: 10.1007/s12031-007-0055-y. [DOI] [PubMed] [Google Scholar]
- 61.Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. Journal of the American Chemical Society. 2006;128(8):2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 62.Rezai T, Bock JE, Zhou MV, Kalyanaraman C, Lokey RS, Jacobson MP. Conformational flexibility, internal hydrogen bonding, and passive membrane permeability: Successful in silico prediction of the relative permeabilities of cyclic peptides. Journal of the American Chemical Society. 2006;128(43):14073–14080. doi: 10.1021/ja063076p. [DOI] [PubMed] [Google Scholar]
- 63.Kalyanaraman C, Jacobson MP. An atomistic model of passive membrane permeability: application to a series of FDA approved drugs. Journal of Computer-Aided Molecular Design. 2007;21(12):675–679. doi: 10.1007/s10822-007-9141-z. [DOI] [PubMed] [Google Scholar]
- 64.White TR, Renzelman CM, Rand AC, Rezai T, McEwen CM, Gelev VM, Turner RA, Linington RG, Leung SSF, Kalgutkar AS, Bauman JN, Zhang Y, Liras S, Price DA, Mathiowetz AM, Jacobson MP, Lokey RS. On-resin N-methylation of cyclic peptides for discovery of orally bioavailable scaffolds. Nature Chemical Biology. 2011;7(11):810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rafi SB, Hearn BR, Vedantham P, Jacobson MP, Renslo AR. Predicting and improving the membrane permeability of peptide small molecules. Journal of Medicinal Chemistry. 2012;55(7):3163–3169. doi: 10.1021/jm201634q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avdeef A, Artursson P, Neuhoff S, Lazorova L, Grasjo J, Tavelin S. Caco-2 permeability of weakly basic drugs predicted with the Double-Sink PAMPA pK(a)(flux) method. European Journal of Pharmaceutical Sciences. 2005;24(4):333–349. doi: 10.1016/j.ejps.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 67.Balimane PV, Han YH, Chong SH. Current industrial practices of assessing permeability and P-glycoprotein interaction. Aaps Journal. 2006;8(1):E1–E13. doi: 10.1208/aapsj080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bermejo M, Avdeef A, Ruiz A, Nalda R, Ruell JA, Tsinman O, Gonzalez I, Fernandez C, Sanchez G, Garrigues TM, Merino V. PAMPA - a drug absorption in vitro model 7. Comparing rat in situ, Caco-2, and PAMPA permeability of fluoroquinolones. European Journal of Pharmaceutical Sciences. 2004;21(4):429–441. doi: 10.1016/j.ejps.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 69.Di L, Kerns EH, Fan K, McConnell OJ, Carter GT. High throughput artificial membrane permeability assay for blood-brain barrier. European Journal of Medicinal Chemistry. 2003;38(3):223–232. doi: 10.1016/s0223-5234(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 70.Fujikawa M, Nakao K, Shimizu R, Akamatsu M. QSAR study on permeability of hydrophobic compounds with artificial membranes. Bioorganic & Medicinal Chemistry. 2007;15(11):3756–3767. doi: 10.1016/j.bmc.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 71.Goodwin JT, Conradi RA, Ho NFH, Burton PS. Physicochemical determinants of passive membrane permeability: Role of solute hydrogen-bonding potential and volume. Journal of Medicinal Chemistry. 2001;44:3721–3729. doi: 10.1021/jm010253i. [DOI] [PubMed] [Google Scholar]
- 72.Irvine JD, Takahashi L, Lockhart K, Cheong J, Tolan JW, Selick HE, Grove JR. MDCK (Madin-Darby canine kidney) cells: A tool for membrane permeability screening. Journal of Pharmaceutical Sciences. 1999;88(1):28–33. doi: 10.1021/js9803205. [DOI] [PubMed] [Google Scholar]
- 73.Li C, Nair L, Liu TT, Li FB, Pichardo J, Agrawal S, Chase R, Tong X, Uss AS, Bogen S, Njoroge FG, Morrison RA, Cheng KC. Correlation between PAMPA permeability and cellular activities of hepatitis C virus protease inhibitors. Biochemical Pharmacology. 2008;75(5):1186–1197. doi: 10.1016/j.bcp.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 74.Whitlock GA, Blagg J, Fish PV. 1-(2-Phenoxyphenyl)methanamines: SAR for dual serotonin/noradrenaline reuptake inhibition, metabolic stability and hERG affinity. Bioorganic & Medicinal Chemistry Letters. 2008;18(2):596–599. doi: 10.1016/j.bmcl.2007.11.080. [DOI] [PubMed] [Google Scholar]
- 75.Di L, Whitney-Pickett C, Umland JP, Zhang H, Zhang X, Gebhard DF, Lai YR, Federico JJ, Davidson RE, Smith R, Reyner EL, Lee C, Feng B, Rotter C, Varma MV, Kempshall S, Fenner K, El-Kattan AF, Liston TE, Troutman MD. Development of a New Permeability Assay Using Low-Efflux MDCKII Cells. Journal of Pharmaceutical Sciences. 2011;100(11):4974–4985. doi: 10.1002/jps.22674. [DOI] [PubMed] [Google Scholar]
- 76.Martín-Villodre A, Plá-Delfina JM, Moreno J, Pérez-Buendía D, Miralles J, Collado EF, Sánchez-Moyano E, Del Pozo A. Studies on the reliability of a bihyperbolic functional absorption-model. I. Ring-substituted anilines. Journal of Pharmacokinetics and Biopharmaceutics. 1986;14(6):615–633. doi: 10.1007/BF01067967. [DOI] [PubMed] [Google Scholar]
- 77.Marqusee JA, Dill KA. Solute partitioning into chain molecule interphases – monolayers, bilayer-membranes, and micelles. Journal of Chemical Physics. 1986;85(1):434–444. [Google Scholar]
- 78.Shore PA, Brodie BB, Hogben CAM. The gastric secretion of drugs: a pH partition hypothesis. Journal of Pharmacology and Experimental Therapeutics. 1957;119(3):361–369. [PubMed] [Google Scholar]
- 79.Tirado-Rives J, Jorgensen WL. Contribution of conformer focusing to the uncertainty in predicting free energies for protein-ligand binding. Journal of Medicinal Chemistry. 2006;49(20):5880–5884. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 80.Guimarães CRW, Cardozo M. MM-GB/SA rescoring of docking poses in structure-based lead optimization. Journal of Chemical Information and Modeling. 2008;48(5):958–970. doi: 10.1021/ci800004w. [DOI] [PubMed] [Google Scholar]
- 81.Maestro, version 9.0. New York, NY: Schrödinger, LLC; 2009. [Google Scholar]
- 82.MacroModel, version 9.7. New York, NY: Schrödinger, LLC; 2009. [Google Scholar]
- 83.Jorgensen WL, Maxwell DS, TiradoRives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society. 1996;118(45):11225–11236. [Google Scholar]
- 84.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. Journal of Physical Chemistry B. 2001;105(28):6474–6487. [Google Scholar]
- 85.QikProp, version 3.2. New York, NY: Schrödinger, LLC; 2009. [Google Scholar]
- 86.Epik, version 2.0. New York, NY: Schrödinger, LLC; 2009. [Google Scholar]
- 87.Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. Epik: a software program for pK (a) prediction and protonation state generation for drug-like molecules. Journal of Computer-Aided Molecular Design. 2007;21(12):681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 88.Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins-Structure Function and Bioinformatics. 2004;55(2):351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 89.Gallicchio E, Zhang LY, Levy RM. The SGB/NP hydration free energy model based on the surface generalized born solvent reaction field and novel nonpolar hydration free energy estimators. Journal of Computational Chemistry. 2002;23(5):517–529. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- 90.Ghosh A, Rapp CS, Friesner RA. Generalized born model based on a surface integral formulation. Journal of Physical Chemistry B. 1998;102(52):10983–10990. [Google Scholar]
- 91.Luo R, Head MS, Given JA, Gilson MK. Nucleic acid base-pairing and N-methylacetamide self-association in chloroform: affinity and conformation. Biophysical Chemistry. 1999;78(1–2):183–193. doi: 10.1016/s0301-4622(98)00229-4. [DOI] [PubMed] [Google Scholar]
- 92.Prime, version 3.1. New York, NY: Schrödinger, LLC; 2012. [Google Scholar]
- 93.Swift RV, Amaro RE. Modeling the pharmacodynamics of passive membrane permeability. Journal of Computer-Aided Molecular Design. 2011;25(11):1007–1017. doi: 10.1007/s10822-011-9480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.