Abstract
CD4+ helper T cells play critical roles for host defense and immune-mediated disease by their ability to differentiate into specialized subsets. These subsets attain restricted patterns of cytokine secretion and specific expression of master transcription factors in response to microbial pathogens. Classically, the various helper CD4+ T cell subsets have been viewed as terminally differentiated lineages with limited flexibility. However, following the recognition of new subsets, there is increased recognition of plasticity. In this review, we highlight recent advances that pertain to this topic and the mechanisms that contribute to helper CD4+ T cell differentiation and plasticity.
Introduction
Differentiation of CD4+ T cells into functionally distinct helper T subsets is critical for proper host defense and normal immunoregulation [1,2]. These subsets are specified by extrinsic and intrinsic cues, and the resultant cell populations acquire seemingly stable phenotypes, which are reinforced by epigenetic modifications [3,4]. Consequently, these subsets have been viewed as lineages, defined by expression of selective signature cytokines and “master regulator” transcription factors [5]. Originally, CD4+ T cells were viewed as having two major fates – T helper1 (Th1) cells, which express T-bet and selectively produce interferon (IFN)-γ and Th2 cells, which express Gata3 and produce interleukin (IL)-4 [1,2,6]. This represented a simple and tractable model system for understanding basic principals in cellular specification and gene regulation. However, the Th1/Th2 paradigm failed to explain a good deal about immunity and autoimmunity [7]. Regulatory T (Treg) cells are another CD4+ lineage with essential immunosuppressive functions that express the master transcription factor FoxP3 [8,9]. Comprising both thymic derived natural, nTreg cells and peripherally induced iTreg cells, the identification of Treg cells was a key discovery in refining our understanding of mechanisms of autoimmunity.
The recognition of cells that selectively produce IL-17 and the transcription factor RORγt (Th17 cells) led to renewed interest in the topic of helper T cell differentiation and even more refined views of the genesis of autoimmune disease [10,11]. Newer fates for helper T cells continue to be identified, with nomenclature based on production of their signature cytokines: Th9 and Th22 cells [12].
The newest “lineage” of CD4 T cells refers to cells that reside in proximity to B cells in germinal centers of lymphoid tissues. These follicular helper T (Tfh) cells that are critical for providing B cell help by promoting class switching of B cells and are defined by expression of master regulator Bcl6 and effector cytokine IL-21, along with key surface molecules (PD-1, CXCR5 and ICOS) [13,14]. However, as will be discussed below, the distinction between Tfh cells and cytokine-secreting effector subsets is a topic of intense ongoing investigation.
Although CD4+ T cell subsets have elements of stability and have been referred to as distinct lineages, there are increasing evidences pointing to substantial phenotypic flexibility of the “newer” helper T cells and indeed, previously identified subsets also appear to be more plastic than originally recognized [12,15,16].
Flexibility of helper T cell phenotypes
First, it is important to remember that although some cytokines are selectively produced by different subsets, many are broadly expressed. Initially thought to be a Th2 cytokine, IL-10 is now known to be made by Th1, Th2, Treg cells and a variety of innate immune cells [17]. IL-21 is the “signature cytokine” of Tfh cells but can also be made by Th17 and Th1 cells [18,19].
Second, it is now clear that helper cells can change their phenotype (Figure 1). Although IL-17-secreting helper T cells were initially suggested to represent a new lineage because they do not make the other lineage-defining cytokines, IFN-γ and IL-4, it is now appreciated that Th17 cells often become IFN-γ producers [20-22]. Although the subject is far from resolved, Treg cells can lose of FoxP3 expression and acquire the capacity to produce proinflammatory cytokines [23,24]. Perhaps the most dramatic example is that Gata3+Th2 cells can be reprogrammed to express T-bet+ and IFN-γ+ in the setting of viral infection. Interestingly, type I interferons are important drivers of reprogramming [25].
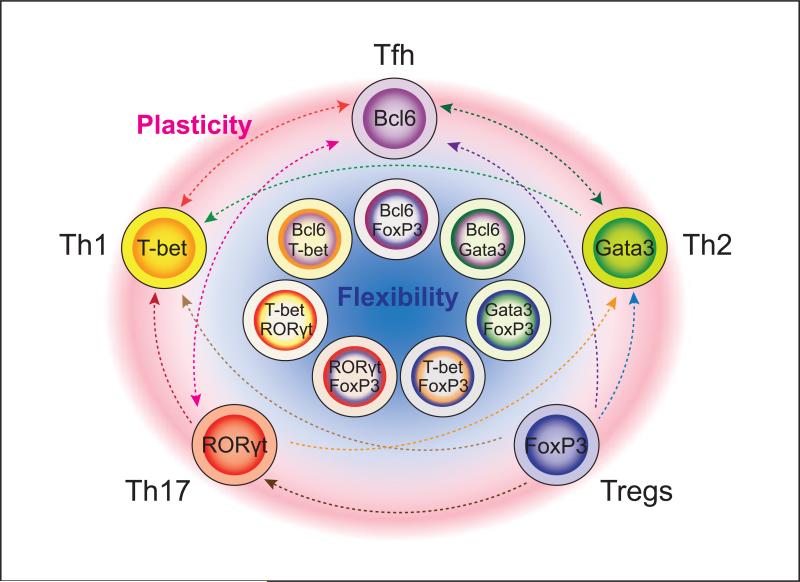
Figure 1.
Flexibility and plasticity of helper T cells. Recent studies continue to reveal surprising flexibility in expression of “master regulator” transcription factors. In addition, there are now many examples in which helper T cell phenotypes can change their pattern of expression of signature cytokines and gene expression. Striking examples exist in which apparently fully committed “lineages” readily switch their phenotype, and there are now many circumstances where helper T cells have been shown to express more than one master regulator. This may be advantageous in terms of host defense, but needs to be borne in mind in thinking about effective therapies for immune-mediated disease and vaccine development.
Tfh cells appear to be the most fluid subset. In vitro, Tfh cells can become Th1, Th2, and Th17 cells and conversely, Th1, Th2, and Th17 can acquire attributes of Tfh cells [26,27]. In vivo isolated Tfh cells can also express cytokines characteristic of other helper T cells [14,27,28]. In Peyer's patches, FoxP3+ T cells can convert to become Tfh cells [29]. Thus, it remains debatable whether Tfh cells should be designated a distinct lineage analogous to other subsets of helper T cells, or simply a state of differentiation that can be superimposed upon the phenotype of Th1, Th2, Th17 or Treg cells.
Collectively, these newer findings argue that although helper T cells may appear to be phenotypically distinct, their capacity for plasticity should not be ignored. This then raises a mechanistic question. What factors underlie the cell biology of commitment versus plasticity and how are they regulated?
Helper T cells can express more than one “master regulator”
A tacit assumption of a pure lineage commitment model is that expression of a single lineage-defining master regulator equates with the distinctive phenotype. However, it has become increasingly clear that CD4+ T cells can express more than one master regulator [15] (Figure 1). For instance, FoxP3+ cells can express T-bet, Gata3, RORγt, STAT3 and Bcl6 – all of which are functionally relevant. In response to IFN-γ, some Treg cells can express T-bet, which drives CXCR3 expression and promotes trafficking of the cells to sites of Th1-associated inflammation [30]. The Th2 master regulator, Gata3, is expressed in Treg cells that reside in barrier sites such as the gastrointestinal tract and the skin, but is also induced by TCR and IL-2 stimulation [31,32]. Although not required for basal Treg cell homeostasis and function, Treg cell expression of Gata3 is critical during inflammatory responses where it maintains FoxP3 expression and limits Treg cell conversion to an effector T cell phenotype. Considerable numbers of human CD4+ T cells coexpress FoxP3 and RORγt. These T cells express CCR6 and produce IL-17 upon activation, but also have suppressive activity [33]. Similarly, Treg cell-specific deletion of STAT3, a factor that drives Th17 and Tfh differentiation, also leads to autoimmunity [34]. Finally, Tregs present in the germinal center express Bcl6 and function to constrain B cell responses [35,36]. Together, these evidences suggest that Treg cells elicit effector response-specific suppression by using components of the transcriptional machinery important for the “target” effector lineage. In this respect, Tregs can be viewed as a heterogenous and potentially dynamic population.
Likewise, inflammatory Th17 cells can be generated that express RORγt together with T-bet or Gata3 [37,38]. In experimental autoimmune encephalomyelitis, T-bet+ RORγt+ cells are highly pathogenic and in this model expression of T-bet is associated with pathogenicity, even when IFN-γ is not.
Gata3 and Bcl6 are co-expressed in the setting of helminth infestation [28]; in fact, Bcl6 is broadly expressed in CD4+ T cells and not just confined in bona fide Tfh cells. Moreover, even though IL-6 and IL-21 acting via STAT3 drive Tfh cell generation, Tfh cell differentiation can occur in the absence of these factors [39]. This can be explained by the fact that IL-12 via STAT4 is capable of driving IL-21 production and Bcl6 expression [18]. Bcl6 and T-bet can be simultaneously expressed in early Th1 cells. To compound matters, those factors not only antagonize each other's function but can also cooperate to repress gene expression [18,40].
Conversely, even if a “master regulator” is necessary for optimal generation of a subset, it may not be sufficient. Although FoxP3 is necessary for normal Treg cell differentiation, expression of FoxP3 is not sufficient to induce the full Treg phenotype [41,42]. Moreover, without FoxP3, some Treg functionalities remain. Similarly, absence of T-bet does not abrogate Th1 differentiation [43,44].
For all these reasons it should be clear that in helper T cells there is not a simple relationship between phenotype and master regulator expression. In this respect, the role of the “master regulators” is distinct from the classic cell biology (e.g. MyoD). In T cells, the master regulators are neither absolutely necessary nor are they sufficient. Moreover, expression of more than one master regulator is frequent event. Thus, rather than invoking simplistic models of distinct expression of a single master regulator in terminally differentiated cells, it may be more accurate to think of co-expressed master regulators. These factors can be dynamically regulated by extrinsic and intrinsic factors and can fine tune T cell capabilities as the situation requires [15].
Epigenetic views
Fully differentiated cells tend to maintain the established phenotype through multiple rounds of cell division without continued instructive signals. Though the notion of “epigenetics” preceded the era of modern molecular biology, it has come to describe hereditable changes in phenotype or gene expression without changes in DNA sequence. Mechanistically, this means that distinct cellular phenotypes and transcriptomes are not merely the result of expression of transcription factors, but also the consequence of chromatin architecture of the genes regulated.
The N-terminal tails of histone proteins can be covalently modified through acetylation, methylation, and phosphorylation [45]. Histone modifications are associated with gene expression by relaxing or condensing the chromatin structure to activate or repress transcription, respectively [46]. For example, trimethylation of H3K4 (H3K4me3) is associated with gene activation [47]. In contrast, polycomb complexes catalyze trimethylation of H3K27 (H3K27me3), which serves to repress gene expression.
Consistent with the standard “lineage commitment” model of helper cell differentiation, cytokine genes typically show permissive (H3K4me3) marks on lineage-appropriate cytokines in the corresponding lineages (e.g. Ifng in Th1 cells or Il4 in Th2 cells), and are accompanied by repressive (H3K27me3) marks in other subsets (Figure 2). However, unlike Th1 and Th2 cells, there is considerable intrinsic instability of Th17 cells [21]. Indeed, this instability is correlated with the rapid remodeling of the Ifng locus when Th17 cells are stimulated in the presence of IL-12 [48].
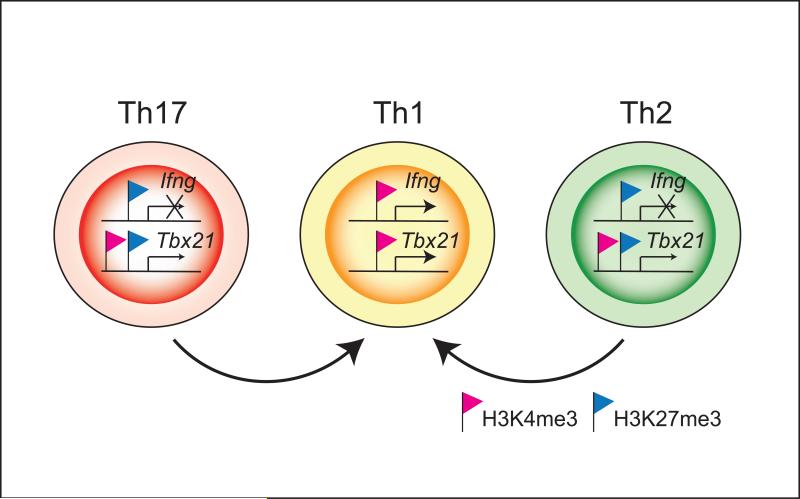
Figure 2.
Epigenetic status of lineage specific cytokine and transcription factor gene loci in helper T cell subsets. H3K4me3 (red flag) and H3K27me3 (blue flag) modifications on Ifng and Tbx21 loci in Th1, Th2 and Th17 cells are shown. Consistent with the selective expression of IFN-γ in Th1 cells, the Ifng gene displays H3K4me3 (permissive) marks in Th1 cells accompanied by H3K27me3 (repressive) marks in Th2 and Th17 cells. The Tbx21 gene is associated with permissive H3K4me3 marks in the absence of H3K27me3 marks in Th1 cells. Although, repressive H3K27me3 marks are present in Th2 and Th17 cells, H3K4me3 marks are also present. The Bcl6 gene is another example where accessible marks are maintained. Such bivalent epigenetic modifications on master regulator may functionally contribute to helper T cell plasticity. Thus, the epigenetic modifications provide explanations for elements of stability (e.g. cytokine genes) that coexist with modes of plasticity (e.g. master regulators genes) within the same subset of helper cells.
Even more striking is that the histone methylation patterns of genes encoding several master regulators do not confirm to this simple “on or off” mode of regulation. They exhibit both repressive and accessible marks, even when these transcription factors are not expressed [24] (Figure 2). For example, the Tbx21 gene is associated with accessible H3K4me3 marks in Th1 cells. Although, repressive H3K27me3 marks are present in Th2 and Th17 cells, H3K4me3 marks are also present. Such regions, marked by both chromatin modifications, are termed “bivalent” domains and have been seen in genes poised for expression in stem cells [49]. This “poised” state of the T-bet locus can explain how T-bet might be induced in Treg, Th17 and even Th2 cells. Similarly, the Bcl6 locus has H3K4me3 marks in polarized Th1, Th2 and Th17 cells and conversely, the Tbx21, Rorc and Gata3 genes have accessible marks in Tfh cells [18,26]. This argues that this transcription factor may be readily induced – even in cells that appear to be firmly committed to being Th1, Th2, and Th17 cells and provides additional evidence to support plasticity of Tfh cells. It should be noted that the appearance of bivalent domains could be a reflection of heterogeneity of the cell populations; regardless though, it needs to be emphasized that cytokine genes do not exhibit this complexity.
MicroRNAs and phenotypic stability
MicroRNAs (miRNAs or miRs) are ~22-nucleotide non-coding small RNAs that bind to target messenger RNAs (mRNAs) and lead to inhibition of translation or mRNA degradation [50]. Drosha and Dicer are two key components of the machinery responsible for miRNA generation and loss of these factors is associated with autoimmune diseases and loss of Treg cell stability [51]. Our understanding of how miRNAs regulate helper cell differentiation is expanding rapidly.
Distinctive expression of miRNAs in each helper T cell subset was demonstrated in both mouse and human, suggesting that each miRNA seems to have some specific roles for the subsets [52,53]. It is clear that miR-146a is important for suppressive function of Treg [54], whereas miR-155 influences Th1, Th2 differentiation and Treg cell development [55-57]. By contrast, miR-326 enhances Th17 differentiation by targeting Ets-1, and miR-29 regulates Th1 differentiation by targeting T-bet, Eomes and IFN-γ [58-60]. Bcl6 represses expression of many miRNAs predicted to control the Tfh cell signature, including miR-17-92, which represses CXCR5 expression [61]. Yet another miR, miR-125b contributes to maintaining the naïve state of helper T cells and miR-182 promotes clonal expansion [53,62]. This is surely an area that will continue to provide new insights into control of CD4 phenotypic stability. Only small numbers of evidences that demonstrate critical roles of miRNA in T cell differentiation has been reported over recent five years even though miRNAs must be apparently involved in both T cell stability and plasticity and seem to contribute to fine-tuning of interplay among crucial transcription factors.
Conclusion
While there are aspects of CD4+ T helper cells in which they appear to behave as stable “lineages”, there is also no shortage of molecular mechanisms that permit flexibility in their repertoire of responses. In fact, global histone epigenetic analysis supported the idea that elements of both terminal differentiation (e.g. cytokine genes) and plasticity (e.g. master regulators genes) can coexist within the same subset. Recent studies have also shown that various types of miRNAs control phenotypic stability of CD4+ T cells. With the present terminology, there is no simple way of denoting stability versus plasticity. What is clear is that CD4+ T cells have the capacity for substantial flexibility. In fact, this may be advantageous in terms of host defense as well as pathogenic under certain circumstances. A better understanding of the extrinsic and intrinsic signals that control stability and plasticity of CD4+ T cells will have important therapeutic applications to combat infections and to control autoimmunity.
Highlights.
Increasing evidences point the phenotypic flexibility of the helper T cells.
Helper T cells can express more than one master regulator.
Epigenetic data help explain both stability and plasticity of helper T cells.
MicroRNAs may play a critical role in control of CD4 phenotypic stability.
Acknowledgements
This work was supported by the Intramural Research Program (IRP) of NIH/NIAMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 2.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 3.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 4.O'Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 6.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 7.Gor DO, Rose NR, Greenspan NS. TH1-TH2: a procrustean paradigm. Nat Immunol. 2003;4:503–505. doi: 10.1038/ni0603-503. [DOI] [PubMed] [Google Scholar]
- 8.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 11.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 14.Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T cells: lineage and location. Immunity. 2009;30:324–335. doi: 10.1016/j.immuni.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Plasticity of CD4(+) FoxP3(+) T cells. Curr Opin Immunol. 2009;21:281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 18•.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [This study described the phenotypic heterogeneity of Tfh cells and Th1 cells, demonstraing molecular interplay between T-bet and Bcl6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suto A, Kashiwakuma D, Kagami S, Hirose K, Watanabe N, Yokote K, Saito Y, Nakayama T, Grusby MJ, Iwamoto I, et al. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [See annotation to Ref. [21].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [Along with Ref. [20•], this study described plasticity of Th17 cells for conversion to IFN-γ producers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med. 2011;17:673–675. doi: 10.1038/nm.2389. [See annotation to Ref. [23].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, O'Brien S, Blank R, Lamb E, Natarajan S, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [Along with Ref. [22•], this study demonstrated instability of FoxP3 and capacity to produce proinflammatory cytokines in Treg cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [This study described the presence of bivalent epigenetic marks on transcription factor genes that helps to explain emerging views of helper T cell plasticity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Lohning M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [One of the first reports to show that Th2 can be redirected to Th1 cells.] [DOI] [PubMed] [Google Scholar]
- 26••.Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, Anderson SM, Wei L, Sun H, O'Shea JJ, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. [This study showed that Tfh cells can be reprogrammed to other helper T cell subsets and vice versa, supporting phenotypic plasticity of Tfh cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaretsky AG, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, Hori S, Fagarasan S. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [This study demonstrated that Treg cells can convert to become Tfh cells in Peyer's patches in the gut.] [DOI] [PubMed] [Google Scholar]
- 30••.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [This study clearly showed that T-bet regulates Treg-specific functions at sites of Th1-cell mediated inflammation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [See annotation to Ref. [32].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, et al. GATA3 controls Foxp3 regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [Along with Ref. [31••], this study demonstrated Gata3 is expressed in Treg cells and is important to maintain FoxP3 expression.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voo KS, Wang YH, Santori FR, Boggiano C, Arima K, Bover L, Hanabuchi S, Khalili J, Marinova E, Zheng B, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A. 2009;106:4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh WI, McWilliams IL, Harrington LE. Autoreactive Tbet-positive CD4 T cells develop independent of classic Th1 cytokine signaling during experimental autoimmune encephalomyelitis. J Immunol. 2011;187:4998–5006. doi: 10.4049/jimmunol.1100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A, Ocheltree EL, Greenberg PD, Ochs HD, Rudensky AY. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A. 2006;103:6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Furuta S, Kagami S, Tamachi T, Ikeda K, Fujiwara M, Suto A, Hirose K, Watanabe N, Saito Y, Iwamoto I, et al. Overlapping and distinct roles of STAT4 and T-bet in the regulation of T cell differentiation and allergic airway inflammation. J Immunol. 2008;180:6656–6662. doi: 10.4049/jimmunol.180.10.6656. [DOI] [PubMed] [Google Scholar]
- 44.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O'Shea JJ, Strober W. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 48.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y, Kim J, Yuan X, Braun T. Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circ Res. 2011;109:1067–1081. doi: 10.1161/CIRCRESAHA.111.243709. [DOI] [PubMed] [Google Scholar]
- 50.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 51.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [See annotation to Ref. [53].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [Along with Ref. [52•], this study demonstrated distinctive expression pattern of miRNAs in each helper T cell subset.] [DOI] [PubMed] [Google Scholar]
- 54.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 58.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 59.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 60.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]