Abstract
The objective of this study was to determine if there are common innate differences in gene expression or gene pathways in the ventral tegmental area (VTA) among 5 different pairs of rat lines selectively bred for high (HEC) or low (LEC) ethanol consumption: (a) alcohol-preferring (P) vs. alcohol-non-preferring (NP) rats; (b) high-alcohol-drinking (HAD) vs. low-alcohol-drinking (LAD) rats (replicate line pairs 1 and 2); (c) ALKO alcohol (AA) vs. nonalcohol (ANA) rats; and (d) Sardinian alcohol-preferring (sP) vs. alcohol-nonpreferring (sNP) rats. Microarray analysis revealed between 370 and 1340 unique named genes that significantly differed in expression between the individual line-pairs. Analysis using Gene Ontology (GO) and Ingenuity Pathways information indicated significant categories and networks in common for up to 3 line-pairs, but not for all 5 line-pairs; moreover, there were few genes in common in these categories and networks. ANOVA of the combined data for the 5 line-pairs indicated 1,295 significant (p < 0.01) differences in expression of named genes. Although no individual named gene was significant across all 5 line-pairs, there were 22 genes that overlapped in the same direction in 3 or 4 of the line-pairs. Overall, the findings suggest that (a) some biological categories or networks may be in common for subsets of line-pairs; and (b) regulation of different genes and/or combinations of multiple biological systems (e.g., transcription, synaptic function, intracellular signaling and protection against oxidative stress) within the VTA (possibly involving dopamine and glutamate) may be contributing to the disparate alcohol drinking behaviors of these line-pairs.
Keywords: gene expression, ventral tegmental area, selectively-bred rat lines, alcohol-preferring, Alko alcohol, high-alcohol-drinking, Sardinian alcohol-preferring
1. Introduction
Studies from animals (Li and McBride 1995) and humans (Cloninger et al., 1985, 1989; Heath 1995; Pickens et al., 1991; Sigvardsson et al., 1996) indicate that genetic factors have a significant impact on alcohol drinking behavior. Gene expression studies can contribute to the identification of genes associated with high alcohol preference and help elucidate mechanisms underlying high alcohol drinking behavior. Gene expression studies with human tissue, however, have been conducted using autopsy samples from individuals who have had a previous history of alcohol abuse (Flatscher-Bader et al., 2005; 2008; Lewohl et al., 2000; Mayfield et al., 2002). To disentangle genetic differences related to susceptibility from those resulting from long-term excess consumption, it is important to determine levels of gene expression in the central nervous system (CNS) of subjects who are genetically susceptible to high alcohol drinking but have not had any previous exposure to ethanol. Since this is not possible in humans, animal studies offer an alternative. To that end, multiple rat and mouse lines, selected on the basis of various criteria for high (HEC) or low (LEC) ethanol-consumption, have been studied.
Kerns et al. (2005) studied the effects of ethanol on gene expression in the nucleus accumbens, prefrontal cortex and VTA of DBA/2J and C57BL/6J mice, and reported region-specific changes in ethanol-responsive genes. Tabakoff et al., (2009) reported results of gene expression levels in HXB/BXH recombinant inbred rats that linked candidate genes involved in GABA release, activation of dopamine neurons, and postsynaptic GABA receptor trafficking in the hypothalamus, ventral tegmentum and amygdala to alcohol consumption. These animal studies are supported by human gene expression studies on the effects of alcohol abuse in the VTA (Flatscher-Bader et al., 2008, 2010), which reported that changes in neurotransmission and signal transduction in the VTA were associated with alcohol abuse.
The fact that both susceptibility to excess ethanol consumption and ethanol consumption itself influence gene expression in the VTA is not surprising because the VTA is a critical part of the brain reward system and has been implicated in mediating the rewarding actions of ethanol (McBride et al., 1999; Gatto et al., 1994; Rodd-Henricks et al., 2000) and in regulating alcohol drinking (Hodge et al., 1993; Nowak et al., 2000). Moreover, there is evidence that the VTA of P rats is more sensitive to the reinforcing effects of ethanol than is the VTA of either NP or stock Wistar rats (Gatto et al., 1994; Rodd et al., 2004), suggesting there may be innate differences in the VTA of P rats that makes ethanol more reinforcing and contributes to their high alcohol drinking characteristics.
Tabakoff et al., (2008) performed a meta-analysis across three types of mouse populations (high-alcohol-preference, low-alcohol-preference, BXD recombinant inbred, and 22 inbred strains) to correlate innate whole brain gene expression with previously reported levels of alcohol intake to identify candidate genes associated with the variance in alcohol preference in mice. These authors reported that the 8 candidate genes identified play important roles in neuronal migration, differentiation and synaptic remodeling. However, this study was conducted using whole brain and not all of the mice were selected for alcohol preference or non-preference. The present study was undertaken to better delineate innate differences in gene expression between 5 pairs of independent, divergently selected HEC and LEC rat line-pairs: (a) the alcohol-preferring (P) and alcohol-non-preferring (NP) rats (Murphy et al., 2002); (b) two replicate lines of high-alcohol-drinking (HAD) and low-alcohol-drinking (LAD) rats (Murphy et al., 2002); (c) ALKO alcohol (AA) and nonalcohol (ANA) rats (Eriksson, 1968); and (d) Sardinian alcohol-preferring (sP) and –non-preferring (sNP) rats (Colombo, 1997). Undoubtedly, genes unrelated to alcohol preference were fixed during the selection process of these lines. By analyzing differential gene expression both within and across line-pairs, it is hoped that any gene expression differences present by chance would be minimized and gene expression differences present because of phenotype would be maximized. It is also possible that different genes may contribute to alcohol preference via the same underlying mechanism if they are in the same biologic pathway. For this reason, it is important to also analyze pathways that may be affected within line-pairs.
The hypothesis to be tested is there will be common differences across the 5 pairs of selectively bred lines of rats in gene expression or within biological systems in the VTA that could influence neuronal function, contribute to the reinforcing actions of ethanol, and promote high alcohol consumption.
2. Method
2.1 Animals
Animals used for this study were ethanol-naïve, male adult selectively-bred rats (n = 9, 10/line). The alcohol-preferring (P), alcohol-non-preferring (NP), high-alcohol-drinking lines 1 & 2 (HAD1, HAD2) and low-alcohol-drinking lines 1 & 2 (LAD1, LAD2) were bred at the Indiana University School of Medicine. Animals were pair-housed in normal 12-hr light/dark cycle rooms (lights on at 0800 hr). The ALKO alcohol (AA) and ALKO nonalcohol (ANA) were housed in a similar manner in animal facilities in Helsinki, Finland. The Sardinian alcohol-preferring (sP) and Sardinian alcohol-nonpreferring (sNP) were also similarly housed in animal facilities in Cagliari, Italy.
The P and NP lines of rats were derived by selective breeding from an outbred stock of Wistar rats maintained at Walter Reed Army Institute of Research (Lumeng et al., 1977). The HAD and LAD replicate lines were derived from the N/Nih heterogeneous stock rats (Hansen and Spuhler, 1984; Li et al., 1993). The sP and sNP rats were selectively bred from a base population of Wistar rats obtained from a vendor in Italy (Fadda et al., 1989). The AA and ANA lines were originally derived from a foundation stock that included Wistar and Sprague-Dawley strains; these lines were subsequently crossed with F1 hybrids from Lewis and Brown Norway rats (Erikson 1981; Sinclair et al., 1989; Kiianmaa et al., 1992; Sommer et al., 2006). The selection criteria for the HEC lines were ethanol intakes of > 5 g/kg/day and a 10% ethanol to water preference ratio equal to or greater than 2:1, using a 24-hr free-choice drinking procedure. The LEC lines had ethanol intakes less than 1 g/kg/day.
The P, NP, HAD1,2 and LAD1,2 animals used in these experiments were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All research protocols were approved by the institutional animal care and use committee and are in accordance with the guidelines of (a) the Institutional Care and Use Committee of the National Institute on Drug Abuse, National Institutes of Health, and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council 1996), and (b) the European Communities Council Directive (86/609/EEC) on the “Protection of animals used for experimental and other scientific reasons”.
2.2. Sample preparation
Rats were killed by decapitation; the brains were quickly removed and frozen in isopentane in dry ice. Brains were stored at −80° C until sectioned. Whole brains from AA, ANA, sP and sNP rats were shipped overnight to Indianapolis in dry ice. On the day of preparation of micro-punch samples, brains were transferred, at least 2 hr prior to sectioning, to a cryostat set at −6 to −10° C. Sections (300 μm) were obtained and transferred to glass slides that had been pre-cooled in the cryostat. Micro-punch sampling was done on a frozen stage (−25 to −35° C) with an anatomic microscope equipped with a cool microscope lamp. The stereotaxic atlas of Paxinos and Watson (1998) was used to identify the VTA. Micro-dissection needles (Fisher Scientific) with an inner diameter of 0.77 mm were used to obtain samples. This inner diameter fits within the entire region and minimizes contamination from adjacent tissue. Punches were taken bi-laterally from 2–3 sections. A different fresh sterile micro-punch needle was used for each animal. After withdrawing the micro-punch sample, a distinct demarcated hole remained; this hole was used to validate the micro-dissection method. All equipment used to obtain tissue was treated with RNAse Zap (Ambion, Inc. Austin, TX) to prevent RNA degradation. To minimize batch and order effects within each line-pair, micro-punch samples were obtained from each line-pair of HEC and LEC rats in a counterbalanced design over a 2–3 day period, such that equal numbers within each line-pair were obtained on the same day. However, micropunch samples from the 5 line-pairs were processed at different times with intervals up to several months between line-pairs.
The micro-punched samples were immediately homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and processed according to the manufacturer’s protocol, but with twice the suggested ratio of Trizol to tissue (Edenberg et al., 2005). Ethanol precipitated RNA was further purified through RNeasy® columns (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The yield, concentration and purity of the RNA were determined by running a spectrum from 210 to 350 nm, and analyzing the ratio of large and small ribosomal RNA bands using an Agilent Bioanalyzer. Yields, purity and quality of the RNA were excellent; RNA integrity numbers (RIN) averaged 8.5 for the samples, showing little or no degradation.
2.3. Microarray procedures
Separate preparations of total RNA were made for each animal. Samples were not pooled at any stage. Samples from each line-pair were processed as one batch for labeling. Samples from the two HAD/LAD line-pairs were labeled using the Affymetrix 2-cycle protocol using 25 ng of total RNA. Due to the discontinuation of that kit, the remaining line pairs were labeled using the 3′ IVT Express Protocol using 100 ng of total RNA; the 3′ IVT express protocol uses only one round of IVT amplification. Samples from each line-pair were divided into two evenly distributed batches across the HEC and LEC animals for hybridization and scanning. The fragmented, biotinylated cRNA from each independent sample was mixed into 300 μl of hybridization cocktail, of which 200 μl were used for hybridization to Affymetrix Rat Genome 230 2.0 GeneChips. Hybridization was carried out for 17 hr at 42°C. Washing and scanning of the GeneChips were carried out according to standard protocols, as previously described (Edenberg et al., 2005; McClintick et al., 2003). Each GeneChip® was scanned using an Affymetrix Model 3000 scanner and underwent image analysis using Affymetrix GCOS software. Microarray data are available from the National Center for Biotechnology Information’s Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ under accession GSE31709.
2.4. Statistical and neuroinformatics analysis of microarray data
Raw CEL files were imported into the statistical programming environment R (R: A language and environment for statistical computing Ver 2.13.0; R Foundation for Statistical Computing, 2011) for further analysis with tools available from the Bioconductor Project (Gentleman et al., 2004). Expression data from the arrays of each experiment was normalized and converted to log(2) values using the Robust Multi-chip Average (RMA) method (Irizarry et al., 2003). As a standardization step to facilitate later comparisons with other experiments, expression levels were scaled such that the mean expression of all arrays was log2(1000). After the initial statistical analysis (Table 1), all probe sets currently annotated by Affymetrix as “expressed sequence tags” or whose gene names contain the words “riken”, “predicted”, or “similar to” were not included in any further analyses; only the remaining probe sets deemed “named genes” were further analyzed. Probe sets that were not detectable above background were filtered out; this has been shown to reduce noise in microarray experiments (McClintick and Edenberg, 2006). Probe sets that did not have a median expression across experiments of at least log2(100) were therefore excluded. To obtain lists of significant genes within-line-pair, a simple t-test was performed and the resultant p-values were used to calculate the FDR q value for each gene according to the method of Storey et al. (2004). Genes with a q value equal to or less than 0.10 were deemed significant. To test for significance of genes across line-pairs, 2 factor linear modeling including interaction term with ANOVA (p < 0.01) was conducted. One factor was line-pair (5 levels) and the other factor was ethanol consumption phenotype (2 levels). FDR was calculated using the same method as for the within-line t-tests and significance was again set at FDR equal to or less than 0.10.
TABLE 1.
Summary of probe sets that significantly differed (FDR = 0.1) in expression in the VTA between each of the selectively bred pairs of high (HEC) and low (LEC) ethanol-consuming lines of rats
| P-NP | HAD1-LAD1 | HAD2-LAD2 | AA-ANA | sP-sNP | |
|---|---|---|---|---|---|
| Total significant probe-sets | 2510 | 1238 | 1816 | 769 | 1961 |
| Total significant ESTs | 987 | 593 | 813 | 360 | 906 |
| Total significant probe-sets, named genes* | 1523 | 645 | 1003 | 409 | 1055 |
| Significant unique named genes* | 1344 | 578 | 894 | 370 | 947 |
Total probe-sets remaining after removing those not detectably expressed was 20,064.
Omits any annotated as “riken”, “predicted” or “similar to”.
Testing for over-representation of GO (Harris et al., 2004; Ashburner et al., 2000) biological processes (BP) and KEGG categories was performed using the Bioconductor package GOstats (Gentleman, 2004). Briefly, for each set of significant genes tested, a list of unique Entrez-Gene identifiers was constructed. This list was then compared to the list of all Entrez-Gene identifiers for the named genes that were called “present” using the filtering methods described above. Identification of over-represented categories was then accomplished using the hypergeometric distribution. Categories were called significant at p < 0.05.
Genes differentially expressed at FDR 10% for each individual line pair and for those significant at FDR 10% in the ANOVA analysis were uploaded separately into Ingenuity Pathway Analysis ® (Ingenuity® Systems, www.ingenuity.com). Genes were identified by their Affymetrix probe set id and then mapped to their corresponding objects in the Ingenuity® Knowledge Base. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity.
Present probe sets were analyzed for enrichment in regions of established Rat QTLs using the methodology of Gene Set Enrichment Analysis (GSEA) (Subramanian, et al., 2005) accompanied by linear modeling (Oron et al., 2008) with the Bioconductor packages GSEABase (R package version 1.17.1) and GSEAlm (R package version 1.15.0). For the across line-pairs analysis, the same linear model was used, as described above for the ANOVA analysis, and GSEA was conducted using the statistic for the ethanol consumption phenotype factor. For the within line-pair analyses, ethanol consumption phenotype was the only factor in the model. The aggregate gene set statistic used was the J-G statistic described in Jiang and Gentleman (2007).
3. Results
3.1. Individual line-pairs analyses
There were over 2,500 probe-sets that differed between the P and NP lines, over 1,200 probe-sets that differed between the HAD1 and LAD 1 lines, over 1,800 probe-sets that differed between the HAD2 and LAD2 lines, over 700 probe-sets that differed between the AA and ANA lines, and over 1,900 probe-sets that were different between the sP and sNP lines (Table 1). Approximately 40–50% of the significant probe-sets were Expressed Sequence Tags (ESTs). There were 370 (AA-ANA lines) to 1,344 (P-NP lines) significant differences in unique named genes between the individual line-pairs (Table 1). See Supplemental Tables A–E for list of significant differences in unique named genes for each line-pair. Supplemental Tables G - K have lists of unnamed probe sets that significantly differed within each line-pair.
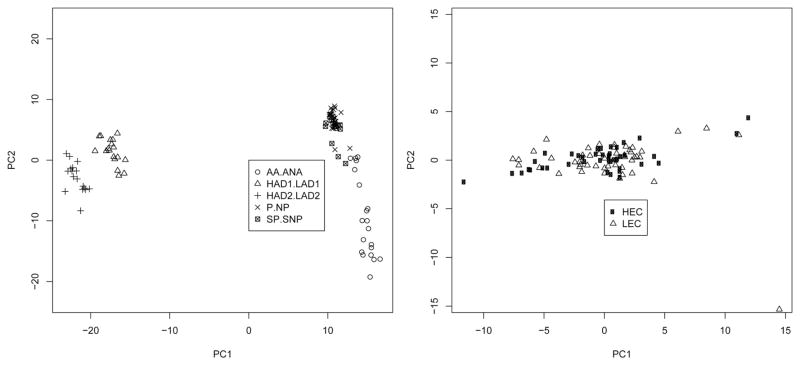
Principal Components Analysis (PCA) by line pair (Fig. 1) indicated that (a) the HAD-LAD replicate lines clustered together and separately from the other line-pairs; (b) the AA-ANA lines clustered separately from all other lines; and (c) the P-NP and sP-sNP lines clustered together. A second PCA indicated that genes for the HEC lines did not cluster separately from the genes for the LEC lines (Fig. 1). Heat map (clustergram; not shown) of genes that were significant (FDR = 0.10) between individual line-pairs did not reveal any clear pattern of genes associated with high or low ethanol-consumption. The patterns of gene expression in the heat map reinforced the findings from the PCA. These results suggest that differences in the genetic background of the strains (and potential batch effects in housing, sacrificing and hybridizing the different line pairs) predominate over any potential clusters of genes associated with high or low ethanol consumption.
Fig. 1.
Principal components (PCA) plot from the genes found significant (FDR < 0.10) in the ANOVA of the 5 line-pairs. Plot on the left is of processed expression data with animals labeled by line-pair. Plot on the right is derived from the residuals after linear modeling taking into account line-pair only, which allows for a view of the data with only ethanol consumption as a factor.
Intersection of lists of named unique genes (see Supplemental Tables A-E) with a FDR < 0.10 indicated that there were no overlapping common genes across all 5 line-pairs. Even with a more relaxed criteria (either FDR < 0.20 or p < 0.05), there were no overlapping common genes across all 5 line-pairs. Furthermore, with FDR < 0.10 or p < 0.05, there were only 6 genes in common across 4 line-pairs, but only the 6 genes obtained with FDR < 0.10 were significant and did not occur by chance alone. Overall, the results do not support common differences in gene expression within the VTA as being associated with the disparate alcohol drinking characteristics across these 5 line-pairs of selectively bred rats.
GSEA was used to determine enrichment of probe sets in established rat QTLs. No enrichment was found for any of the QTLs within a line-pair or across the line-pairs.
3.1.1. Gene Ontology (GO) analyses of individual line-pairs
Although each line pair had significant categories, there were no biological categories that were in common across more than 3 line-pairs (Table 2). The ‘circadian rhythm’ category was significant for the P-NP, HAD1-LAD1 and sP-sNP lines. Among the 9–14 genes in this category for the 3 line-pairs, there was only one gene that was significant in all 3 line-pairs, i.e., Crem, however, its direction of change was not consistent across the line-pairs. There were several genes in common in 2 of the 3 line-pairs, although the differences were not always in the same direction. Comparison of the differences between the P-NP vs. the HAD1-LAD1 lines yielded 2 common genes: Bhlehe41 (both higher in P & HAD1) and Dpyd (both lower in P & HAD1). Comparison of the differences between the P-NP vs. the sP-sNP lines indicated the following common genes: Cdk4 (both lower in P & sP), and Per2, Per3 and Kcnma, which all changed in opposite directions. Comparison of the HAD1-LAD1 vs. sP-sNP rats indicated that Phlpp was higher in HAD1 and sP lines, whereas Homer1 and Ncor1 changed in opposite directions in the 2 line-pairs.
Table 2.
Significant Gene Ontology (GO) Biological Categories in the VTA of the Individual Line-Pairs
| Lines | Category | Significant Genes | P-value | Count/size |
|---|---|---|---|---|
| P-NP | Circadian rhythm | Alb, Atp7b, Bhlhe41, Crem, Cry1, Cry2, Cdk4, Cst3, Dpyd, Npas2, Per1, Per2, Per3, Kcnma1 | 0.004 | 14/48 |
| HAD1-LAD1 | Circadian rhythm | Bhlhe41, Crem*, Dpyd, Ddc, Hmgb1, Homer1, Ncor1, Opn4, Phlpp1 | 0.004 | 9/48 |
| sP-sNP | Circadian rhythm | Crem, Csk1e, Cdk4, Egfr, Homer1, Ncor1, Per2, Per3, Phlpp1, Kcnma1, Pmch | 0.011 | 11/48 |
| HAD2-LAD2 | Response to estrogen stimulus | Arsa, Brca2, Casp9, Cd24, Cd38, F3, Cnnd1, Gstm5, Ifi27, Mmp15, Mapk1, Ncor2, Ramp2, Sstr3, Tek, Txnip, Timp3, Tgfb3, Tnfrsf11b | 0.017 | 19/114 |
| AA-ANA | Response to estrogen stimulus | Bdh1, Arsb, Ash2l, Bmp4, Car2, F3, Gpx1, Ifi27, Mmp2, Pik3r1, Serpinb6, Tbl1x, Tgfb3 | 0.021 | 13/40 |
| P-NP | Tetrahydrobiopterin biosynthetic process | Pts, Qdpr, Spr, LOC685729 | 0.006 | 4/6 |
| HAD2-LAD2 | Nitric oxide mediated signal transduction | Gucy1a3, Mt1a, Rln1 | 0.026 | 3/7 |
| HAD2-LAD2 | Cytokine-mediated signaling pathway | Adipor1, Cd24, Cd74, Cx3cl1, Cx3cr1, Fkbp1a, Il6st, Irak3, Pf4, Traf6, Zc3h15 | 0.030 | 11/59 |
| AA-ANA | Glutathione metabolic process | Cth, G6pd, Gclc, Gpx1 | 0.029 | 4/29 |
| sP-sNP | Neuropeptide signaling pathway | Celsr2, Calca, Eltd1, Gpr126, Lphn3, Nmb, Npy5r, Ntsr2, Pmch, Pcsk1n | 0.024 | 10/47 |
| sP-sNP | Cell redox homeostasis | Dld, Erp44, Grxcr1, Qsox2, Sco1, Txn1, Txndc12, Txndc16, Txnrd3, Txnl1 | 0.046 | 10/52 |
Underlined genes had lower expression in the P, HAD, AA or sP compared to their respective low alcohol consuming counterparts. Asterisk indicates 2 probe sets for same gene that changed in opposite directions.
The ‘response to estrogen stimulus’ category was significant in the AA-ANA and HAD2-LAD2 line-pairs (Table 2). Among the 13–19 genes listed within this category, there were only 3 common genes, two (F3 & Ifi27) of which changed in opposite directions. The third gene (Tgfb3) was lower in the AA and HAD2 lines compared to ANA and LAD2 lines, respectively.
Other significant GO categories (Table 2) included tetrahydro-biopterin biosynthetic process (P-NP lines), NO-mediated signal transduction (HAD2-LAD2 lines), cytokine-mediated signaling (HAD2-LAD2 lines), glutathione metabolism (AA-ANA lines), neuropeptide signaling (sP-sNP lines) and cell redox homeostasis (sP-sNP rats).
3.1.2. Ingenuity single line-pair analyses
The top 5 Ingenuity pathways for each line pair were selected for further analysis. All networks had a score > 25. There were no common networks across all 5 line-pairs, although there were networks common to 3 line-pairs (Table 3). Glucocorticoid receptor signaling networks were found in the VTA of the P-NP, HAD2-LAD2 and sP-sNP line-pairs. NF-κB signaling networks were common among the P-NP, HAD2-LAD2 and AA-ANA line-pairs. Ephrin receptor signaling networks were common between the P-NP and HAD1-LAD1 line pairs, and glutathione metabolism networks were common for HAD1-LAD1 and HAD2-LAD2 line-pairs. However, except for Gst and Gsta4 in the glutathione metabolism network, there were no common genes in these networks between any line-pairs, suggesting that the mechanisms underlying these cellular processes may be different for each line-pair.
TABLE 3.
Networks identified by Ingenuity Pathway Analysis of significant differences in genes with higher or lower expression in the VTA of the P, HAD1, HAD2, AA and sP lines compared to their respective low ethanol consuming lines
| Lines | Network | Gene expression highera | Gene expression lowerb |
|---|---|---|---|
| P-NP | Glucocorticoid receptor signaling | 16 genes: Afg3l2, Atp13a5, Kif1b, Atl1, Afg3l1, Slit1, Mag12, Camlg, Zmynd8, Atn1, Zmynd11, Mag11, Fkbp15, Ppif, Fkbp2, Timm22 | 16 genes: Atp11b, Tmed9, Tjap1, Atpase, Spast, Dlg2, Rad51, Syn1, Lgtn, Fkbp11, Ppib, Ppil3, Pfkl, Timm9, Tomm7, Timm10 |
| HAD2-LAD2 | Glucocorticoid receptor signaling | 16 genes: Lama4, Fbln2, Mta3, Pf4, Mmp15, Appl1, Rbbp4, Ext2, Haus1, Mt1f, Phf21a, Hifd, Mcm4, Tagln, Hey2, Abcb1 | 10 genes: Pik3c3, Becn1, Vcan, Igfbp5, Adamts1, Wwc1, Hyou1, Brca2, Orc5l, Rrm2 |
| sP-sNP | RAR activation; Glucocorticoid receptor signaling; PPAR signaling | 18 genes: Usp47, Mllt4, Usp9x, Dub, Usp46, Ncoa7, Casp4, Usp33, Trim24, Nrip1, Sra1, Tbl1x, N-cor, Lrp2, Atn1, C20orf191, Pfkl, Megf8 | 13 genes: Scand1, Epha6, Trim33, Ncoa, Mvp, Scn3a, Rnf31, Anapc10, Zmynd8, Rere, Plekha5, Rbp1, Ltbp4 |
| P-NP | Ephrin receptor signaling | 12 genes: Creb3l3, Nr2c1, Foxn3, Pgk1, Hdac4, Ank3, Znf24, Rlim, Ino80c, Tex10, Kpna1, Clic4 | 18 genes: Creb5, Crem, Ankra2, Vegfb, Bcor, Creb, Lmo4, Vegr, Elk3, Ctbp1, Ddx42, Lmo2, Mml, Hhex, Lyl1, Eng, Rpl23a, Ran, Arpc3, Il33, Lta4h |
| HAD1-LAD1 | Ephrin receptor signaling; Integrin signaling | 12 genes: Csrp1, Tpm3, Vcl, Actr2, Wipf1, Elavl2, Ccnd2, Cd83, Smn1, C190rf2, Pbx3, Med9 | 17 genes: Pls1, Gas8, Arhgef18, Tmod1, Tmod3, Arp2/3, Arpc4, Lagls3bp, Smyd3, Actr3, Tfcp2, Hbb, Ccnc, Polr2, Dhfr, Med21, Rnmt |
| P-NP | NFkB signaling | 11 genes: Nlk, Rnf138, Atic, Ube2k, Rnf125, Setd2, Rnf103, Azi2, Ccnl2, Ube2g1, Ercc8 | 16 genes: Vta1, Rabac1, Pdia6, Mavs, Lsm2, Magoh, Pbx3, Ube2l6, Smyd3, Ube2n, Cbr1, Ube2, Jkamp, Znf268, Ube2a, Uqcrq |
| HAD2-LAD2 | NfkB signaling; p38 Mapk signaling | 15 genes: Armc6, Telo2, Pinx1, Tagn2, Anapc10, Nap1l1, Traf3ip2, Marcks, Map4k2, Stom, Cyb5a, Sqstm1, Eabp5, Grb14, Lss | 17 genes: Abcd3, Psmd6, Atpase, Dcxr, Psmc6, E3ring, Irak3, Cul2, Traf6, Myh9, Irak, Limd1, Acaa2, Klhl24, Srebf2, Tek, Sqle |
| AA-ANA | NFkB signaling | 15 genes: Lrp6, Rab22a, Appl2, Akt, App, Krt10, Hsp70, Hist1h2bb, Appbp2, Hspa9, Pcbp1, Csnk1a1, Traf6, Tsn, Bub3 | 11 genes: Lrp4, Appl1, Csnk2a1, Rufy1, Tars, Tnfrsf1a, Limd1, Nap1l1, Nhp2, Atp2a2, Il17ea |
| HAD1-LAD1 | Glutathione metabolism; RAR activation | 15 genes: Mgst, Ccnd1, Pdia4, Hsp90b1, Ncoa1, Agt, Cxcl12, C14orf1, Vcan, Hif1a, Pdk1, Sephs1, Bhlhe41, Cpne3, Pggt1b | 14 genes: Gst, Gstt2, Eno3, Rrm2, Gsta4, Eif2b1, Eif2b5, Abcb, Mtus1, Aldh2, Mgmt, Adamts1, Pf4, Ccs |
| HAD2-LAD2 | Glutathione metabolism | 7 genes: Nupl1, Hla-e, Nup133, Hla-c, Gstm3, Ctps, Mpg | 8 genes: Gmps, Gsta4, Mgst2, Gst, Gsta1, Nup98, Cbr1, Gsto1 |
| HAD1-LAD1 | cAMP-mediated signaling; protein kinase A signaling | 14 genes: Crem, Sstr2, Adam9, Arfgap1, Ints7, Lrg1, Hspa9, Elavl1, Cdkn1b, Mmp16, Ybx1, Tgfb3, Ogfod1, Tgfbl | 13 genes: Atp2a2, Sox17, Itga9, Tgfb2, Rgs2, Nfyb, Mmp2, Fgf1, Tgfb, Itgb2, Akap2, Prkar1a, Akap7 |
| HAD1-LAD1 | Ox40 signaling pathway | 17 genes: Glrx3, Tfdp2, Hifo, Vip, Dbt, Nfkb1, Mhc1, Nfkb1, Exosc7, Sgca, Hlac, Hlae, Bcl11b, Enpp2, B2m, H2t24, Mhca | 7 genes: Tmem126b, Prkd2, Utrn, Abcb1b, Commd7, Hladmb, Gmps |
| HAD2-LAD2 | Clathrin-mediated endocytosis signaling | 12 genes: Ssr3, Bat2, Napa, Dynamin, Myo1e, Shank2, Src, Gsn, Rpp38, Tom1l1, Ddr2, Isg20l2 | 11 genes: Sh3gl2, Amph, Bin1, Prmt2, Epb41l3, P2ry2, Cnn3, Gpr143, Trpv1, Gpsm3, Polr3e |
| HAD2-LAD2 | RAR activation; PPARα/RXRα activation; Wnt/β-catenin signaling | 19 genes: Slc27a1, Ilf3, Ets, Cpt1a, Hnrnpd, Elk3, Lmo4, Lrg1, Sox17, Vegr, Fgfb, Gabbr1, Ercc2, Fubp1, Ints7, Sox4, Ddx42, Smyd3, Tex10 | 13 genes: Acaa1, Ltb, Chi3l1, Acadl, Pkp1, Tgfb3, Hk2, Elavl1, Tgfb, Gabbr2, Arhgef9, Mll, Slc27a2 |
| AA-ANA | CREB signaling in neurons | 8 genes: Rac1, Flnb, N-cor, G6pd, Dhfr, Chrna5, H1fo, Gsta4 | 17 genes: Ptprj, Pik3r, Lcp2, Kif13a, Pi3k, Pik3r1, Pnp, Axl, Plor1b, Eml4, Dag1, Tbl1x, Actl6a, Gstm4, Nfx1, Bmp4, Nup153 |
| sP-sNP | Dopamine receptor signaling; Production of NO and reactive O2 species | 12 genes: Bar1, Aatk, Entpd2, Ppp2r4, Sorl1, Vps4b, Cdadc1, Vps35, Mipep, Vps29, Tm7sf2, Nipsnap1 | 8 genes: Stk39, Atpase, Pp2a, Chmp1b, Afg3l1, Abcd3, Cnnm3 |
Genes with higher expression in high vs. low ethanol consuming lines.
Genes with lower expression in high vs. low ethanol consuming lines.
Several networks were detected in only one of the line-pairs (Table 3). Comparison of gene expression between HAD1-LAD1 rats (Table 3) indicated differences in cAMP-mediated and protein kinase A signaling and Ox40 signaling. For the HAD2-LAD2 lines, there were differences in clathrin-mediated endocytosis signaling; RAR and PPARα/RXRα activation and Wnt/β-catenin signaling. The AA-ANA lines had differences in CREB signaling. The sP-sNP lines had significant differences in gene expression in a network related to dopamine receptor signaling and production of NO.
3.2. Overall ANOVA for all line-pairs combined
ANOVA, using ethanol consumption phenotype (HEC and LEC) and line-pair (5 categories) as factors, was conducted for each probe set. The results of the ANOVA showed 1,295 individual named genes that were significantly (p < 0.01) different between the HEC and LEC lines (see Supplemental Table F for named genes, and Supplemental Table L for unnamed probe sets that significantly differed).
Ingenuity pathway analyses revealed 6 significant (score > 30) networks (Table 4). All networks had more genes with higher than lower expression in the HEC compared to the LEC lines, with the most pronounced differences found in networks 1 (1.6-fold), 3 (2.5-fold) and 4 (1.9-fold). Within network 1, there were 22 genes in the App (amyloid beta precursor protein) cluster, with 17 genes having higher expression, and 6 genes with lower expression in the HEC lines compared to the LEC lines (Table 4, genes in cluster designated by asterisks). Network 3 was associated with proliferation of cells and transcription of DNA; within this network, there were 13 genes in the Ccnd1 (cyclin D1) cluster, with 10 genes having higher expression in the HEC lines. Network 4 was associated with the growth and outgrowth of neurites and migration of neurons; several genes within this network interact with Itga2b (integrin alpha 2b). Among the 9 genes in the Itga2b cluster, there were 7 genes with higher expression in the HEC than LEC group, with only Fn1 and Vwf having lower expression in the HEC group. Network 5 had almost the same number of genes with higher and lower expression (Table 4). However, of the 13 genes in the Rxra (retinoid X receptor, alpha) cluster, there were 10 genes that had higher expression levels in the VTA of the HEC vs. LEC lines. Networks 2 and 6 had a few more genes with higher than lower expression in the VTA of the HEC vs. the LEC lines. Within network 2, there were 17 genes in the Egfr (epidermal growth factor receptor) cluster, with 10 genes having higher and 7 genes having lower expression in the HEC lines. Network 6 had 9 genes in the Vcl (vinculin) cluster, with 5 genes having higher expression in the HEC group.
Table 4.
Networks and gene clusters identified by Ingenuity Pathway Analysis of significant differences in higher or lower gene expression in the VTA of the HEC compared to the LEC lines
| Network/gene cluster | Higher Expressiona | Lower Expressionb |
|---|---|---|
| Network 1: neurological disorder; neuronal death; interactions clustered around App | 21 genes: Ank3, Appb1*, Sorl1*, Dnm1l*, Sparcl1*, Aplp2*, Taz*, Pgam1, Bcl2l2*, Apba2*, App*, Erp44*, Kif1a, Hmox2*, Ccdc92*, Git1, Nbl1*, Cfb*, Sncb*, Rtn3*, Lingo1* | 13 genes: Nfia, Rgnef, Gusb, Rp2, Arl4d, Vps33a*, Gas7, Hyou1*, Clic1*, C1qa*, Nptx1*, Ngb*, Fam160a2 |
| Network 2: migration of cells; proliferation of cells; interactions clustered around Egfr | 19 genes: Rab6b, Bmpr1a, Slc9a3r2, Smad7*, Gref1*, Adora1, Cdh2, Rhot2, Bcan*, Sh2b1*, Sorbs1, Rnf41*, Adam9*, Ptprs*, Grb14*, Capza2*, Tom1l1*, Surf4, Cops7a | 15 genes: Chn1, Adora2b, Sash3, Acvr1, Fancl, Col3a1*, Epb41l2, Col6a1*, Trak2*, Axl*, Egfr*, Cd2ap*, Itga9, F11r, Degs1* |
| Network 3: proliferation of cells; transcription of DNA; interactions clustered around Ccnd1 | 25 genes: Anapc2, Nedd9, Rfxap, Id2*, Hla-dqa1, Dnajb1, Cirbp*, Sox2*, Cdk9*, Suds3, Appl2*, Hexim1*, Zeb2*, Ccnk, Ccnd1*, Runx1t1, Krt10*, Supt5h, Tubb3, Ctnnbip1, Mcf2l*, Heafx, Luc7l3, Mdc1, Thoc4 | 10 genes: Anapc10, Hinfp, Mdfic, Hdac1, Csf1r*, Ccndbp1*, Rilpl2, Srpk2*, Cbfb, Phip |
| Network 4: growth & outgrowth of neurites; migration of neurons; interactions clustered around Itga2b | 23 genes: Cir1, Nt5e, Calcb, Vcan, Ncor2, Zbtb7a, Gpx1, Sec23b, Gmnn, Sparc, Hif1a*, Tesk1, Spred1, Pdia4, Itga2b*, Mta3, Mbd3*, Myl6b*, Pc*, L1cam*, Cntn2, Aup1*, Bat5 | 12 genes: Mfng, Jag1, Nov, Mgp, Fn1*, Adamts1, Blnk, Rragd, Vwf*, Crkl, Kidins220, Wdr77 |
| Network 5: interactions clustered around Rxra | 17 genes: Map4k2, Sox4, Nr2c1, Rxrb, Dgkz, Cpt1a*, Rxra*, Znhit3*, Ppargc1a*, Dlat, Tnrc6b, Hmgcs2*, Pcsk2*, Pkn1Ubqln4*, Ncoa4*, Zfpm2*, | 16 genes: Rhoc, Pnlip, Rock2, Leprot, Aprx, Lepr, Slco1a2*, Rbp1*, Cript, Acat1, Ubr7, Tfam, Jazf1*, Slit3, Pdk1, Pmch |
| Network 6: interactions clustered around Vcl | 19 genes: Cog5, Ppp1r12c, Cdc42ep1, Golga2, Letm2*, Tmed2, Adam19, Nrg1*, Vcl*, Ddx19a, Pdcd4, Pabpc1*, Psma2*, Vdac1, Csde1, Nudc, Pcbp1, Hla-c, Nfyc | 16 genes: Cog4, Lpcat3, Scfd1*, Dad1, Snrpf*, Bag4*, Psma4, Pik3r1*, Upf3b, Eif4g2, Eif4g1, Psmc4, Lig3, Psmc6, Psmd6, Tapbp |
Genes with higher expression in high vs. low ethanol consuming lines.
Genes with lower expression in high vs. low ethanol consuming lines.
Indicates each gene that is associated with the cluster within that particular network.
Two important biological pathways emerged from the analysis of KEGG pathways in the overall ANOVA (Table 5). These include 11 genes associated with the dopamine system and 13 genes associated with the glutamate system. Although there were subunits of the GABA-A receptor that differed in any one line pair, there were only 2 genes that were significant in the GABA system in the overall analysis.
Table 5.
Significant genes in the overall analysis of the 5 line-pairs associated with certain pathways identified by Ingenuity Pathway Analysis or KEGG
| Biological System | Genes with higher expressiona | Genes with lower expressionb |
|---|---|---|
| Dopamine | 8 genes: Adcy9, Comt, Ppp1r12c, Ppm1b, Ppp2r5b, Ppp2r5c, Ppp4r1, Qdpr | 3 genes: Prkar2a, Ppp2r1b, Ppp3r1 |
| Glutamatergic synapse | 9 genes: Adcy9, Glul, Gls, Gnas, Gng13, Gng3, Slc38a1, Slc38a2, Slc38a3 | 4 genes: Homer2, Pla2g12a, Pld2, Ppp3r1 |
| GABA | 1 gene: Abat | 1 gene: Aldh9a1 |
Genes with higher expression in high vs. low ethanol consuming lines.
Genes with lower expression in high vs. low ethanol consuming lines.
There were 22 genes differentially expressed (in the same direction) in at least 3 of the 5 line-pairs of HEC vs. LEC lines in the overall analysis (Table 6). There were 8 genes in common across 4 line-pairs, but all 8 genes were not in common across the same 4 line-pairs. There were 8 genes in common across the HAD1-LAD1, HAD2-LAD2 and P-NP lines (Table 6).
TABLE 6.
Genes that were significantly different in the ANOVAs of the 5 line-pairs and were significantly changed in the same direction in at least 3 line-pairs
| Symbol | Description | Higher or lower in 3 or 4 line-pairsa | H1b | H2 | P | AA | sP |
|---|---|---|---|---|---|---|---|
| Ankrd12 | Ankyrin repeat domain 12 | Higher | X | X | X | X | |
| Crebl2 | cAMP responsive element binding protein-like 2 | Lower | X | X | X | ||
| Fam102b | Family with sequence similarity 102, member B | Lower | X | X | X | ||
| Fam40a | Family with sequence similarity 40, member A | Higher | X | X | X | ||
| Gsta4 | Glutathione S-transferase alpha 4 | Lower | X | X | X | X | |
| H1f0 | H1 histone family, member 0 | Higher | X | X | X | ||
| Itga9 | Integrin alpha 9 | Lower | X | X | X | ||
| Itgb2 | Integrin beta 2 | Lower | X | X | X | ||
| Mx2 | Myxovirus resistance 2 | Higher | X | X | X | ||
| Ncaph | Non-SMC condensing l complex, subunit H | Higher | X | X | X | X | |
| Pls1 | Plastin 1 | Lower | X | X | X | ||
| Psd3 | Pleckstrin & Sec7 domain containing 3 | Lower | X | X | X | X | |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 | Higher | X | X | X | ||
| Rock2 | Rho-associated coiled-coil containing protein-kinase 2 | Lower | X | X | X | ||
| RT1-A2 | RT1 classIa, locus A2 | Higher | X | X | X | ||
| RT1-T24-4 | RT1 class I, locus T24, gene 4 | Higher | X | X | X | X | |
| RT1-DMb | RT1 claaa II, locus DMb | Lower | X | X | X | X | |
| Scpep1 | Serine carboxypeptidase 1 | Lower | X | X | X | ||
| Slc38a10 | Solute carrier family 38, member 10 | Lower | X | X | X | X | |
| Smtnl2 | Smoothelin-like 2 | Lower | X | X | X | ||
| Zfp212 | Zinc finger protein 212 | Lower | X | X | X | X | |
| Znf23 | Zinc finger protein 23 | Higher | X | X | X |
To qualify for this list, the gene needed to be significantly different in the overall analysis and (a) be significantly different in 4 of the line-pairs and have the same direction of change in all 4 line-pairs, or (b) be significantly different in 3 of the line- pairs, have the same direction of change in the 3 line-pairs, not have a significant change in the opposite direction in more than 1 of the remaining 2 line-pairs, and be listed in at least one network identified by Ingenuity Pathway Analysis. ‘Higher’ (or ‘lower’) indicates values were higher (or lower) in the high ethanol consuming rats compared to the low ethanol consuming rats.
Indicates which line-pair was significantly different; H1 = HAD1-LAD1; H2 = HAD2-LAD2; P = P-NP; AA = AA-ANA; sP = sP-sNP
Validation studies on key genes, using RT-PCR, were not conducted because there was not sufficient sample remaining for this measurement. Previous studies from our laboratory indicated good agreement between the data obtained with microarrays and the results found with RT-PCR (Kimpel et al., 2007; Rodd et al., 2008; Bell et al., 2009).
4. Discussion
The results of the analyses of the individual line-pairs indicate that, although there are biological categories or networks in common for up to 3 line-pairs, within these categories or networks, there were few genes in common. On the other hand, the overall ANOVAs of the 5 line-pairs indicated there were ~1,300 genes that were significantly different between the HEC and LEC rats, suggesting some common mechanisms may exist across all 5 line-pairs. This apparent disagreement between the individual line-pair comparisons vs. the overall ANOVAs may be a result of prominent differences in a subset of line-pairs contributing to the overall significance of a given gene in the ANOVA.
The finding that there did not appear to be similar differences in gene expression across all 5 line-pairs could be a result of a combination of factors including: (a) differences in the range of genetic variation in the foundation stocks of the different line-pairs; (b) multiple mechanisms may contribute to innate differences in responses to alcohol that contribute to high ethanol intakes; (c) other brain regions are involved in regulating alcohol drinking; (d) accumulated effects of several small differences may not be detected with the microarray procedure; and/or (e) common differences may be found in expression of genes that have not yet been identified and so were not analyzed. Selection acts on the existing genetic differences in the founder stock, and, if there are multiple pathways that can lead to the phenotype, an early divergence in one pathway is likely to be reinforced by continuing selection.
4.1. Individual line-pair findings
Comparisons of the individual line-pair differences with GO (Table 2) and Ingenuity (Table 3) analyses indicated some common categories or networks for up to 3 line-pairs, but very few genes in common within these categories or networks, suggesting that different molecular mechanisms may be underlying the disparate alcohol drinking and other characteristics of each line-pair.
P-NP, HAD1-LAD1 and sP-sNP line-pairs had a significant GO ‘circadian rhythm’ biological category, suggesting that there may be a relationship between innate preference for alcohol and functions associated with disturbances in the circadian rhythm. Both preclinical and clinical studies have shown that the clock gene Per2, and variations of this gene, may influence high ethanol-consuming behavior in mice and humans (Comasco et al., 2010; Spanagel et al., 2005, 2010).
Ingenuity Pathway analysis revealed differences between the P-NP, HAD2-LAD2 and sP-sNP lines in glucocorticoid receptor (GR) signaling (Table 3), which is involved in regulating nuclear transcription and intracellular signaling in response to stress (Pariante & Miller 2001; Gutierrez-Mecinas et al., 2011). P-NP and HAD1-LAD1 lines had differences in ephrin receptor signaling, which is involved in clustering and modulating NMDA receptors (Antion et al., 2010; Murai & Pasquale 2004). Differences in NFkB signaling, which is a transcription factor involved in a wide variety of cellular functions, including regulation of neurite growth and neuro-protective mechanisms (Kimpinski et al., 1999; Mattson 1997; Tsatsanis et al., 2006), were observed between the P-NP, HAD2-LAD2 and AA-ANA lines. Differences between various high and low ethanol drinking lines were observed for glutathione metabolism, RAR activation and Ox40 signaling pathway (Table 3), which are all involved in protection against oxidative stress and other cellular insults (Li et al., 2011; Kostenko et al., 2011; van Neerven et al., 2008; Maher 2005; Shinozaki et al., 2007). Intracellular signaling and synaptic function can be altered by line differences in cAMP-mediated and protein kinase A signaling, clathrin-mediated endocytosis, PPAR and RXR signaling, CREB and dopamine receptor signaling (Table 3).
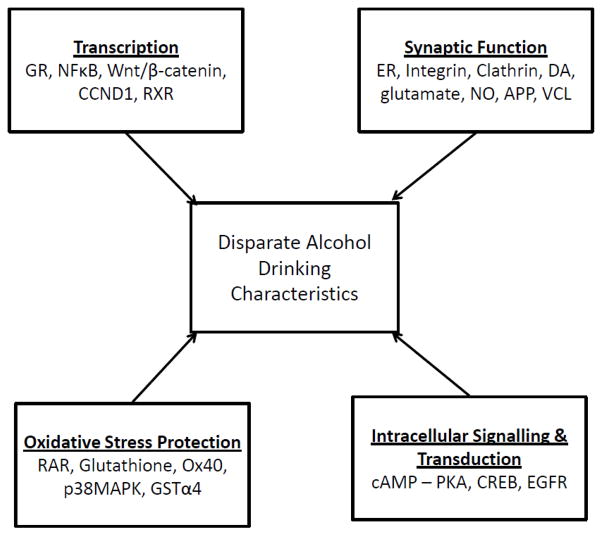
Overall, the Ingenuity results for the individual line-pairs suggest there are innate differences in the regulation of transcription in the VTA between each of the individual line-pairs. These differences in transcription could alter expression of genes involved in synaptic function and intracellular signaling pathways. In addition, differences in regulation of transcription could also alter expression of genes providing neuro-protection against oxidative and other cellular stress factors, which could influence the consequences of chronic alcohol drinking. Finally, although there were 4 common general themes (i.e., regulation of transcription, synaptic function, protection against oxidative stress and intracellular signaling), the molecular mechanisms underlying the differences between the line-pairs in each of these functions are not likely to be similar because there were very few genes in common between the line-pairs in any of the biological functions.
4.2. Combined line-pair findings
Examining expression across all 5 line-pairs can reveal differences that might be too small to detect in a single line-pair comparison. Almost 1,300 individual named genes were significantly different between the HEC and LEC lines. Ingenuity Pathway analysis of gene expression differences between the high and low alcohol consuming lines (Table 4) revealed significant differences in networks clustered around 6 genes: App, Egfr, Ccndl, Itga2b, Rxra and Vcl. Amyloid precursor protein (APP) is involved in normal neuronal functions, such as signal transduction, cell adhesion and motility, plasticity and memory (Turner et al. 2003). Three genes, or their isoform, (Aplp1, Hyou1, Vps11) from the APP pathway were identified by Mulligan and colleagues (2006) as candidate genes for high ethanol consumption in mice.
Epidermal growth factor receptor (EGFR) is a member of the receptor tyrosine kinase family, which is involved in signal transduction (Carpenter & Cohen, 1990), cell proliferation, migration, differentiation and apoptosis (Citri & Yarden, 2006). Cyclin D1 (CCND1) is a co-factor to cyclin-dependent kinases (CDKs), which can induce nuclear receptor activity and regulate transcription (Kim & Diehl, 2009). Retinoid X receptor (RXRa) is a nuclear receptor that regulates transcription of approximately 500 different genes, including enzymes, transcription factors and cytokines (Van Neerven et al., 2008). Brain RXRs expression was enhanced with chronic ethanol consumption and appeared to be involved in ethanol-induced memory impairment (Alfos et al., 2001).
ITGa2b (Table 4, Network 4) is a member of a family of extracellular matrix (ECM)-interacting membrane receptors, which are critical for cell adhesion and survival. Vinculin (VCL) is a cytoskeleton protein involved in cell adhesion and motility, as well as cell survival and proliferation (Carisey & Ballestrem, 2011).
Overall, the results suggest that differences in cell-to-cell interactions and transcription may be contributing factors to the disparate alcohol drinking characteristics of the HEC and LEC lines.
4.2.1. Genes associated with the DA system
The VTA is a major source of DA neurons that project to several limbic regions and are involved in regulating the rewarding actions of ethanol and alcohol drinking behavior (Gatto et al., 1994; Hodge et al., 1993; Nowak et al., 2000; Rodd-Henricks et al., 2000). In the dopamine system, differences were found in expression of genes involved in presynaptic dopamine homeostasis, i.e., Qdpr and Comt, as well as postsynaptic signal transduction, i.e., Adcy9, Prkar2a and several protein phosphatase genes (Table 5). Quinoid dihydropteridine reductase (QDPR) is an enzyme involved in the generation of BH4, the co-factor for tyrosine hydroxylase, which is the rate-limiting enzyme in the synthesis of dopamine. Catechol-O-methyltransferase (COMT) is an enzyme involved in dopamine degradation. COMT plays an important role in cognitive functions and may be involved in various psychiatric diseases including schizophrenia (Tunbridge et al., 2005). Adenylate cyclase 9 (Adcy9) is a downstream effector protein to dopamine receptor activation, which has been shown to be associated with bipolar disorder (Toyota et al., 2002). The activity of protein kinase A associated protein (Prkar2a) in the mesolimbic system appears to regulate ethanol consumption in rats (Misra & Pandey, 2006). Protein phosphatases (Ppm1b, Ppp2r5b, Ppp2r5c, Ppp2r1b) modulate the activity of various protein kinases and are involved in the regulation of cell growth, cytoskeleton dynamics and apoptosis, among their diverse multiple cellular functions (Janssens & Goris, 2001). Overall, the results suggest differences in dopamine neuronal function within the VTA may be contributing to a predisposition for high ethanol consumption.
4.2.2. Genes associated with the glutamate system
The VTA receives excitatory glutamatergic inputs from several sources, e.g., pedunculopontine nucleus, medial prefrontal cortex, insular cortex, etc, and may also contain neurons that use glutamate as a transmitter or co-transmitter (Stuber et al., 2010). Within the glutamate system, higher expression of genes involved in glutamate metabolism was found (Table 5). Glutamate-ammonia ligase (Glul), a member of the glutamine synthetase family, is involved in neuroprotection against excessive glutamate and ammonia, as well as in astrocyte differentiation (Suarez et al., 2002). There were differences in several sodium-coupled neutral amino acid transporters (Slc38a1, 2, & 3), which preferentially transfer glutamine from glial cells to neurons. Also, there was higher expression of the gene for glutaminase (Gls), an enzyme that converts glutamine to glutamate in neurons. The latter two groups of genes play important roles in the glutamate/GABA-glutamine homeostasis in the brain (Mackenzie & Erickson, 2004; Mates et al., 2009).
Differences were also found in genes involved in postsynaptic glutamate signal transduction, including: (1) G-protein subunits coupled to metabotropic glutamate receptors (mGluRs); (2) phopholipase D2 (Pld2), a downstream effector enzyme to the activation of mGluRs, important for synaptic plasticity (Niswender & Conn, 2010); and (3) a member of homer protein family (Homer2), which function as scaffolding proteins and regulate receptor trafficking and clustering (Foa & Gasperini 2009). Several studies indicated an association between alcohol drinking and the mesolimbic mGluR-NMDAR-Homer2 function (Cozzoli et al., 2009; Goulding et al., 2011; Obara et al., 2009; Szumlinski et al., 2006). Overall, the results suggest that alterations in glutamate homeostasis and synaptic function may contribute to the disparate alcohol drinking characteristics of the 5 line-pairs.
4.3. Genes identified in both the individual line-pair and combined line-pair analyses
Several individual genes were significantly different in the ANOVA and also in 3 or 4 of the line-pair comparisons with the same direction of change (Table 6). Four of the 22 genes in Table 6 are located in an alcohol preference QTL. Fam102b and Fam40a are both located in Alc15, Psd3 is located in Alc11, and Zfp212 is located in Alc18. Little is known about Fam40a and Fam102b other then their location on chromosome 2 (Gregory et al., 2006; Nagase et al., 2000). Fam40a is higher and Fam102b is lower in all of the 4 significant HEC lines, suggesting this is not the case of a coding region being open or closed for transcription.
There are 42 genes within the region flanked by Fam40a and Fam102b (Entrez Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/, build RGSC v3.4); 11 of these genes encode for glutathione S-transferases, but only 4 are represented on Affymetrix rat chip 230.2, and, of these, only Gst4 is significant in any line (higher in the AA than ANA line). Gsta4 expression in several brain regions may have a role in alcohol preference and longevity in AA vs. ANA rats (Bjork et al., 2006). Seventeen other genes in this interval are represented on the chip and 6 of these are significant, but all are significant in only 1 line. Taken together, it seems that there may be something important about this region or nearby regions.
Psd3 contains 2 domains of interest. Although the exact functions of Psd3 are unknown, the 2 domains can be found in many other genes with better characterized functions, e.g., cell signaling and cytoskeletal regulation (Rebecchi & Scarlata, 1998). The final gene found in an alcohol QTL, Zfp212, is a zinc-finger protein with presumed transcriptional regulatory ability (Strausberg et al., 2002), but its specific function is unknown.
Several other genes in Table 6 are of interest. Crelbl2 shares 41% identity to Creb (Hoornaert et al., 1998) and the function of CREB in alcohol preference is well documented (Constantinescu et al., 1999; Li et al., 2003; Yang et al., 1998). Gsta4 is a glutathione S-transferase, which functions primarily to detoxify lipid peroxidation products, but has been implicated in ethanol preference (Bjork et al., 2006; Liang et al., 2004). There are 2 integrins in the table, Itga9 and Itgb2, which are located on different chromosomes. In the nervous system, integrins have been shown to interact with extra-cellular matrix proteins during regeneration after injury (Gardiner, 2011), suggesting a role in neuroplasticity. Several genes in Table 6 are involved with cytoskeletal architecture. Rock2 is a serine/threonine kinase that regulates both cytokinesis and focal adhesions and activates c-fos (Ivanov et al., 2009; Olsen et al., 2006). Pls1 is a member of the plastin family, which are actin-binding proteins (Lin et al., 1993). Smtnl2 encodes a structural protein that associates with actin fibers (Gerhard et al., 2004).
Finally, Slc38a10 is a putative neutral amino-acid transporter (Ota et al., 2004). The neutral amino acids tryptophan and tyrosine are precursors to the biogenic amines. This gene has reduced expression in 4 of the line-pairs, which could eventually lead to decreased synthesis of dopamine and serotonin in the VTA in the HEC compared to the LEC rats.
Some notable genes in Table 6 include cAMP responsive element binding protein-like 2, a glutathione S-transferase, intergrins, a protein kinase and a transporter. The products of these genes could have important effects on transcription, oxidative stress and metabolic aspects of cellular function that could result in an altered response to the actions of alcohol within the VTA.
Significant differences in the expression of genes related to glutamate neurotransmission in the VTA have been reported between alcoholics and non-alcoholics; these included two glutamate transporter genes, i.e., Slc17a and Slc1a2 (Flatscher-Bader et al., 2008). Multiple differences in the glutamate system were also observed in the present study (Table 5), including neutral amino acid transporters, which are involved in transferring glutamine from glial cells to neurons (Mackenzie & Erickson 2004; Mates et al., 2009). Increased expression of Sparc was reported in the VTA of alcoholics compared to non-alcoholics (Flatscher-Bader et al., 2010) and in the present study (Table 4, network 4). SPARC is a glycoprotein involved in regulating cell-matrix interactions (Brekken and Sage, 2001). Differences in expression of Fn1, which encodes for an extracellular matrix protein (Potts and Campbell, 1996), were also observed in the present study (Table 4, network 4) and in the VTA of alcoholics (Flatscher-Bader et al., 2010), although the changes were in opposite directions. These parallel a number of differentially expressed genes observed in the present study that are associated with cell adhesion molecules, proteins and integrins. Combined, these findings suggest that there may be differences in cell-cell interactions in the VTA between alcoholics vs. controls, as well as between selectively bred high vs. low ethanol-consuming rats.
The relationship between specific genes and associated pathways, identified herein, and ethanol-associated effects and consumption have also been reported in published meta-analyses and comprehensive genomic studies. In this regard, multiple reports have implicated the Nfkb, Il6 and Mapk pathways in alcohol abuse (Table 3, c.f., Aroor & Shukula, 2004; Crews et al., 2006; Mulligan et al., 2006; Zhai et al., 2008; Zou & Crews, 2005, 2010). Similarly, multiple reports have associated members of the glutathione S-transferase (Gst) family and its metabolic pathway with excessive ethanol intake (Tables 2 & 6; Bjork et al., 2006; Grisel et al., 2002; Hashimoto et al., 2011; Liang et al., 2004; Mulligan et al., 2006; Thibault et al., 2000). Both the AA-ANA and HAD2-LAD2 line-pairs had ‘Response to estrogen stimulus’ as a significant GO category (Table 2), which has also been reported recently in a comparison of ethanol-treated withdrawal seizure prone (WSP) and resistant (WSR) mice (Hashimoto et al., 2011).
It is noteworthy that a number of genes and/or biological systems identified in this study have also been identified as candidate genes for the development and/or expression of alcoholism through investigations of polymorphisms detected in alcoholic vs. control populations. Some of the genes identified in the present study and previous clinical literature include Gabra2 (Bierut et al., 2010; Enoch et al., 2009), Nfkb1 (a subunit of the transcription factor NFkB: Edenberg et al., 2008) and Per2 (Spanagel et al., 2005, 2010), as well as others, e.g., Npy5r, Grm8 and Comt.
4.4. Conclusions
In summary, no single pathway appears to account for the disparate alcohol drinking characteristics of all 5 line-pairs of HEC and LEC rats. Instead, the interactions of different combinations of multiple biological systems mediating transcription, oxidative stress protection, synaptic function and intracellular signaling, possibly involving dopamine and/or glutamate function within the VTA, may contribute to the disparate alcohol drinking characteristics of the 5 line-pairs (Fig. 2). Additional studies examining other CNS regions (and/or other genes not yet identified) will be needed to identify molecular mechanisms and biological pathways contributing to the alcohol drinking behaviors of the HEC and LEC rats.
Fig. 2.
Simplified block diagram showing the possible involvement and interactions of 4 general biological functions that could be associated with selective breeding for high or low ethanol consumption. Systems that could significantly alter transcription include glucocorticoid receptor (GR) signaling, NFkB signaling, Wnt/B-catenin signaling, cyclin D1 (CCND1), and retinoid X receptor (RXR) signaling. Protection against oxidative stress involves retinoic acid receptor (RAR) activation, glutathione, Ox40 signaling, p38 mitogen-activated protein kinase (p38MAPK), and glutathione-S-transferase (GST). Synaptic function could be altered by ephrin receptor (ER) signaling, integrin signaling, clathrin-mediated endocytosis, dopamine (DA), glutamate, nitric oxide (NO), amyloid precursor protein (APP) and vinculin (VCL). Different expression of genes for cyclic AMP-protein kinase A (PKA), CREB and epidermal growth factor receptor (EGFR) could significantly alter intracellular signaling pathways.
Supplementary Material
Highlights.
Gene expression 5 line-pairs of rats selectively bred for high or low alcohol consumption
Ventral tegmental area important for alcohol reward
Disparate alcohol drinking characteristics associated with differences in gene expression involved in transcription, synaptic function, oxidative stress protection, intracellular signaling & transduction
Acknowledgments
This project was supported in part by AA07611, INIA grants AA013522, AA016652 & AA016660, and INGEN (which is partially funded by Lilly Endowment, Inc).
Footnotes
None of the authors has a conflict of interest associated with this research.
The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIAAA or NIH.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfos S, Boucheron C, Pallet V, Hiqueret D, Enderlin V, Beracochea D, Jaffard R, Hiqueret P. A retinoid acid receptor antagonist suppresses brain retinoid acid receptor overexpression and reverses a working memory deficit induced by chronic ethanol consumption in mice. Alcohol Clin Exp Res. 2001;25:1506–1514. doi: 10.1097/00000374-200110000-00015. [DOI] [PubMed] [Google Scholar]
- Antion MD, Christie LA, Bond AM, Dalva MB, Contractor A. Ephrin-B3 regulates glutamate receptor signaling at hippocampal synapses. Mol Cell Neurosci. 2010;45:378–88. doi: 10.1016/j.mcn.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroor AR, Shukula SD. MAP kinase signaling in diverse effects of ethanol. Life Sci. 2004;74:2339–2364. doi: 10.1016/j.lfs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, Rodd ZA, Mayfield RD, Edenberg HJ, McBride WJ. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94:131–147. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork K, Srrtikoski ST, Arlinde C, Kovanen L, Osei-Hyiaman D, Ubaldi M, Reimers M, Hyytia P, Heilig M, Sommer WH. Glutathione-S-transferase expression in the brain: possible role in ethanol preference and longevity. FASEB J. 2006;20:1826–1835. doi: 10.1096/fj.06-5896com. [DOI] [PubMed] [Google Scholar]
- Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- Carpenter G, Cohen S. Epidermal growth factor. J Biol Chem. 1990;265:7709–7712. [PubMed] [Google Scholar]
- Carisey A, Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signaling. Eur J Cell Biol. 2011;90:157–163. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signaling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Bohman M, Sigvardsson S, Von Knorring AL. Psychopathology in adopted-out children of alcoholics. Recent Dev Alcoholism. 1985;3:37–51. doi: 10.1007/978-1-4615-7715-7_4. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Sigvardsson S, Gilligan SB, Von Knorring AL, Reich T, Bohman M. Genetic heterogeneity and the classification of alcoholism. Adv Alcohol Substance Abuse. 1989;7:3–16. doi: 10.1300/J251v07n03_02. [DOI] [PubMed] [Google Scholar]
- Colombo G. Ethanol drinking behaviour in Sardinian alcohol-preferring rats. Alcohol Alcohol. 1997;32:443–453. doi: 10.1093/oxfordjournals.alcalc.a008279. [DOI] [PubMed] [Google Scholar]
- Comasco E, Nordquist N, Gokturk C, Aslund C, Hallman J, Oreland L, Nilsson KW. The clock gene Per2 and sleep problems: association with alcohol consumption among Swedish adolescents. Upsala J Med Sci. 2010;115:41–48. doi: 10.3109/03009731003597127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu A, Diamond I, Gordon AS. Ethanol-induced translocation of cAMP-dependent protein kinase to the nucleus. Mechanism and functional consequences. J Biol Chem. 1999;274:26985–26991. doi: 10.1074/jbc.274.38.26985. [DOI] [PubMed] [Google Scholar]
- Cozzoli DK, Goulding SP, Zhang PW, Xiao B, Hu J-H, Ary AW, Obara I, Rahn A, Abou-Ziab H, Tyrrel B, Marini C, Yoneyama N, Metten P, Snelling C, Dehoff MH, Crabbe JC, Finn DA, Klugmann M, Worley PF, Szumlinski KK. Binge drinking upregulates accumbens mGluR5-Homer2-PI3K signaling: functional implications for alcoholism. J Neurosci. 2009;29:8655–8668. doi: 10.1523/JNEUROSCI.5900-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Bechara R, Brown LA, Guidot DM, Mandrekar P, Oak S, Qin L, Szabo G, Wheeler M, Zou J. Cytokines and alcohol. Alcohol Clin Exp Res. 2006;30:720–730. doi: 10.1111/j.1530-0277.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephans M, Jerome RE, Lumeng L, Li T-K, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and –non-preferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Wetherill LF, Bierut L, Bucholz K, Dick DM, Hesselbrock V, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Almasy LA, Nurnberger JI, Jr, Foroud T. Association of NFKB1, which encodes a subunit of the transcription factor NFkB, with alcohol dependence. Hum Mol Genet. 2008;17:963–970. doi: 10.1093/hmg/ddm368. [DOI] [PubMed] [Google Scholar]
- Enoch M-A, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34:1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K. Genetic selection for voluntary alcohol consumption in the albino rat. Science. 1968;159:739–741. doi: 10.1126/science.159.3816.739. [DOI] [PubMed] [Google Scholar]
- Eriksson CJ. Finnish selection studies on alcohol-related behaviors: factors regulating voluntary alcohol consumption. In: McClearn GE, Erwin G, editors. Development of Animal Models as Pharmacogenetic Tools. Rockville, MD: US Government Printing Office; 1981. pp. 119–145. [Google Scholar]
- Fadda F, Mosca E, Colombo G, Gessa G. Effect of spontaneous ingestion of ethanol on brain dopamine metabolism. Life Sci. 1989;44:281–287. doi: 10.1016/0024-3205(89)90186-0. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Zuvela N, Landis N, Wilce PA. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. Human Mol Genetics. 2008;17:38–51. doi: 10.1093/hmg/ddm283. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcohol Clin Exp Res. 2010;34:1291–1302. doi: 10.1111/j.1530-0277.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Foa L, Gasperini R. Developmental roles for Homer: more than just a pretty scaffold. J Neurochem. 2009;108:1–10. doi: 10.1111/j.1471-4159.2008.05726.x. [DOI] [PubMed] [Google Scholar]
- Gardiner NJ. Integrins and the extracellular matrix: Key mediators of development and regeneration of the sensory nervous system. Dev Neurobiol. 2011 doi: 10.1002/dneu.20950. [E pub ahead of print] [DOI] [PubMed] [Google Scholar]
- Gatto GJ, McBride WJ, Murphy JM, Lumeng L, Li T-K. Ethanol self-infusion into the ventral tegmental area by alcohol-preferring rats. Alcohol. 1994;11:557–564. doi: 10.1016/0741-8329(94)90083-3. [DOI] [PubMed] [Google Scholar]
- Gentleman RC. Using GO for statistical analysis. Proc COMPSTAT. 2004:171–180. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Malek J. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding SP, Obara I, Lominac KD, Gould AT, Miller BW, Klugmann M, Szumlinski KK. Accumbens Homer2-mediated signaling: a factor contributing to mouse strain differences in alcohol drinking? Genes Brain behav. 2011;10:111–126. doi: 10.1111/j.1601-183X.2010.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Barlow KF, McLay KE, Kaul R, Swarbreck D, Dunham A, Prigmore E. The DNA sequence and biological annotation of human chromosome 1. Nature. 2006;441:315–321. doi: 10.1038/nature04727. [DOI] [PubMed] [Google Scholar]
- Grisel JE, Metten P, Wenger CD, Merrill CM, Crabbe JC. Mapping of quantitative trait loci underlying ethanol metabolism in BXD recombinant inbred mouse strains. Alcohol Clin Exp Res. 2002;26:610–616. [PubMed] [Google Scholar]
- Gutierrez-Mecinas M, Trollope AF, Collins A, Morfett H, Hesketh SA, Kersante F, Reul JM. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2-MSK1-Elk-1 signaling. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1104383108. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C, Spuhler K. Development of the National Institute of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res. 1984;8:477–479. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Database issue. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto JG, Forquer MR, Tanchuck MA, Finn DA, Wiren KM. Importance of genetic background for risk of relapse shown in altered prefrontal cortex gene expression during abstinence following chronic alcohol intoxication. Neurosci. 2011;173:57–75. doi: 10.1016/j.neuroscience.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC. Genetic influences on alcoholism risk: A review of adoption and twin studies. Alcohol Health Res World. 1995;19:166–171. [PMC free article] [PubMed] [Google Scholar]
- Hodge CW, Haraguchi M, Erickson H, Samson HH. Ventral tegmental microinjections of quinpirole decrease ethanol and sucrose-reinforced responding. Alcohol Clin Exp Res. 1993;17:370–375. doi: 10.1111/j.1530-0277.1993.tb00778.x. [DOI] [PubMed] [Google Scholar]
- Hoornaert I, Marynen P, Baens M. CREBL2, a novel transcript from the chromosome 12 region flanked by ETV6 and CDKN1B. Genomics. 1998;51:154–157. doi: 10.1006/geno.1998.5366. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ivanov AI, Samarin SN, Bachar M, Parkos CA, Nusrat A. Protein kinase C activation disrupts epithelial apical junctions via ROCK-II dependent stimulation of actomyosin contractility. BMC Cell Biol. 2009;10:36–45. doi: 10.1186/1471-2121-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serin/threonine phophatases implicated in cell growth and signaling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Gentleman R. Extensions to gene set enrichment analysis. Bioinformatics. 2007;23:306–313. doi: 10.1093/bioinformatics/btl599. [DOI] [PubMed] [Google Scholar]
- Kerns DT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RB, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiianmaa K, Hyytia P, Sinclair JD. Development of an animal model of ethanol abuse: genetic approach. In: Boulton A, Baker G, Wu PH, editors. Neuromethods, Vol. 24; Animal Model of Drug Addiction. Totowa, NJ: The Humana Press; 1992. pp. 29–63. [Google Scholar]
- Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and –non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpinski K, Jelinski S, Mearow K. The anti-p75 antibody, MC192, and brain-derived neurotrophic factor inhibit nerve growth factor-dependent neurite growth from adult sensory neurons. Neuroscience. 1999;93:253–263. doi: 10.1016/s0306-4522(99)00156-6. [DOI] [PubMed] [Google Scholar]
- Kostenko S, Dumitriu G, Laegreid KJ, Moens U. Physiological roles of mitogen-activated-protein-kinase-activated p38-regulated/activated protein kinase. World J Biol Chem. 2011;2:73–89. doi: 10.4331/wjbc.v2.i5.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Li T-K, McBride WJ. Pharmacogenetic models of alcoholism. Clin Neurosci. 1995;3:182–188. [PubMed] [Google Scholar]
- Li T-K, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genetics. 1993;23:163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- Li J, Li YH, Yuan XR. Changes of phosphorylation of cAMP response element binding protein in rat nucleus accumbens after chronic ethanol intake: naloxone reversal. Acta Pharmacol Sin. 2003;24:930–936. [PubMed] [Google Scholar]
- Li DQ, Zhang L, Pflugfelder SC, De Paiva CS, Zhang X, Zhao G, Zheng X, Su A, Qu Y. Short ragweed pollen triggers allergic inflammation through toll-like receptor 4-dependent thymic stromal lymphopoietin/OX40 ligand/OX40 signaling pathways. J Allergy Clin Immunol. 2011 doi: 10.1016/j.jaci.2011.06.041. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Habegger K, Spence JP, Foroud T, Ellison JA, Lumeng L, Li T-K, Carr LG. Glutathione S-transferase 8–8 expression is lower in alcohol-preferring than in alcohol-nonpreferring rats. Alcohol Clin Exp Res. 2004;28:1622–1628. doi: 10.1097/01.alc.0000145686.79141.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CS, Park T, Chen ZP, Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J Biol Chem. 1993;268:2781–2792. [PubMed] [Google Scholar]
- Lumeng L, Hawkins TD, Li T-K. New strains of rats with aldohol preference and non-preference. In: Thurman RG, Williamson JR, Drott H, Chance B, editors. Alcohol and Aldehyde Metabolizing Systems. III. Academic Press; New York: 1977. pp. 537–544. [Google Scholar]
- Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch. 2004;447:784–795. doi: 10.1007/s00424-003-1117-9. [DOI] [PubMed] [Google Scholar]
- Maher P. The effects of stress and aging on glutathione metabolism. Ageing Res Rev. 2005;4:288–314. doi: 10.1016/j.arr.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Mates JM, Sequra JA, Campos-Sandoval JA, Lobo C, Alonso L, Alonso FJ, Marquez J. Glutamine homeostasis and mitochondrial dynamics. Int J Biochem Cell Biol. 2009;41:2051–2061. doi: 10.1016/j.biocel.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Neuroprotective signal transduction: relevance to stroke. Neurosci Biohav Rev. 1997;21:193–206. doi: 10.1016/s0149-7634(96)00010-3. [DOI] [PubMed] [Google Scholar]
- Mayfield RA, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Murphy JM, Ikemoto S. Localization of brain reinforcement mechanism: intracranial self-administration and intracranial place-conditioning studies. Behav Brain Res. 1999;101:129–152. doi: 10.1016/s0166-4328(99)00022-4. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:1–15. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra K, Pandey S. The decreased cyclic-AMP dependent protein kinase A function in the nucleus accumbens: a role in alcohol drinking but not in anxiety-like behavior in rats. Neuropsychopharmacology. 2006;31:1406–1419. doi: 10.1038/sj.npp.1300900. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6388–73. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai KK, Pasquale EB. Eph receptors, ephrins and synaptic function. Neuroscientist. 2004;10:304–14. doi: 10.1177/1073858403262221. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li T-K. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genetics. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O. Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:347–355. doi: 10.1093/dnares/7.6.347. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology and disease. Ann Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak KL, McBride WJ, Lumeng L, Li T-K. Involvement of dopamine D2 autoreceptors in the ventral tegmental area on alcohol and saccharin intake of the alcohol-preferring P rat. Alcohol Clin Exp Res. 2000;24:476–483. [PubMed] [Google Scholar]
- Obara I, Bell RL, Goulding SP, Reyes CM, Larson LA, Ary AW, Truitt WA, Szumlinski KK. Differential effects of chronic ethanol consumption and withdrawal on Homer/glutamate receptor expression in subregions of the accumbens and amygdala of P rats. Alcohol Clin Exp Res. 2009;33:1924–1934. doi: 10.1111/j.1530-0277.2009.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Oron A, Jiang Z, Gentleman R. Gene set enrichment analysis using linear models and diagnostics. Bioinformatics. 2008;24:2586–2591. doi: 10.1093/bioinformatics/btn465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Pariante CM, Miller AH. Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol Psychiatry. 2001;49:391–404. doi: 10.1016/s0006-3223(00)01088-x. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4. Academic Press; New York: 1998. [Google Scholar]
- Pickens RW, Svidis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Arch Gen Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Potts JR, Campbell ID. Structure and function of fibronectin modules. Matrix Biol. 1996;15:313–320. doi: 10.1016/s0945-053x(96)90133-x. [DOI] [PubMed] [Google Scholar]
- Rebecchi MJ, Scarlata S. Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Melendez RI, Kuc KA, Lumeng L, Li T-K, Murphy JM, McBride WJ. Comparison of intracranial self-administration of ethanol within the posterior ventral tegmental area between alcohol-preferring and Wistar rats. Alcohol Clin Exp Res. 2004;28:1212–1219. doi: 10.1097/01.alc.0000134401.30394.7f. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, McKinzie DL, Crile RS, Murphy JM, McBride WJ. Regional heterogeneity for the intracranial self-administration of ethanol within the ventral tegmental area of female Wistar rats. Psychopharmacology. 2000;149:217–224. doi: 10.1007/s002139900347. [DOI] [PubMed] [Google Scholar]
- Shinozaki Y, Sato Y, Koizumi S, Ohno Y, Nagao T, Inoue K. Retinoic acids acting through reinoid receptors protect hippocampal neurons from oxygen-glucose deprivation-mediated cell death by inhibition of c-jun-N-terminal kinase and p38 mitogen-activated protein kinase. Neuroscience. 2007;147:153–63. doi: 10.1016/j.neuroscience.2007.04.032. [DOI] [PubMed] [Google Scholar]
- Sigvardsson S, Bohman M, Cloninger CR. Replication of the Stockholm adoption study of alcoholism. Arch Gen Psychiatry. 1996;53:681–687. doi: 10.1001/archpsyc.1996.01830080033007. [DOI] [PubMed] [Google Scholar]
- Sinclair JD, Le AD, Kiianmaa K. The AA and ANA rat lines, selected for differences in voluntary alcohol consumption. Experientia. 1989;45:798–805. doi: 10.1007/BF01954055. [DOI] [PubMed] [Google Scholar]
- Sommer W, Hyytia P, Kiianmaa K. The alcohol-preferring AA and alcohol-avoiding ANA rats: meurobiology of the regulation of alcohol drinking. Addict Biol. 2006;11:289–309. doi: 10.1111/j.1369-1600.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Bartsch D, Brors B, Dahmen N, Deussing J, Eils R, Ende G, Gallinot J, Gebicke-Haerter P, Heinz A, Kiefer F, Jager W, Mann K, Matthaus F, Nothen M, Rietschel M, Sartorius A, Schutz G, Sommer WH, Sprengel R, Walter H, Wichmann E, Wienker T, Wurst W, Zimmer A. An integrated genome research network for studying the genetics of alcohol addiction. Addict Biol. 2010;15:369–379. doi: 10.1111/j.1369-1600.2010.00276.x. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc Series B Stat Methodol. 2004;66:187–205. [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Hnasko TS, Britt JP, Edwards RH, Bonci A. Dopamineric terminals in the nucleus accumbens but not the dorsal striatum co-release glutamate. J Neurosci. 2010;30:8229–8233. doi: 10.1523/JNEUROSCI.1754-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez I, Bodega G, Fernandez B. Glutamine synthetase in brain: effect of ammonia. Neurochem Int. 2002;41:123–42. doi: 10.1016/s0197-0186(02)00033-5. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlinsk K, Kalivas PW, Woorley P. Homer proteins: implications for neuropsychiatric disorders. Curr Opin Neurobiol. 2006;16:251–257. doi: 10.1016/j.conb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mamm Genome. 2008;19:352–385. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70–78. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault C, Lai C, Wilke N, Duong B, Olive MF, Rahman S, Dong H, Hodge CW, Lockhart DJ, Miles MF. Expression profiling of neural cells reveals specific patterns of ethanol-responsive gene expression. Mol Pharmacol. 2000;58:1593–1600. doi: 10.1124/mol.58.6.1593. [DOI] [PubMed] [Google Scholar]
- Toyota T, Yamada K, Saito K, Detera-Wadleigh SD, Yoshikawa T. Association analysis of adenylate cyclase type 9 gene using pedigree disequilibrium test in bipolar disorder. Mol Psychiatry. 2002;7:450–452. doi: 10.1038/sj.mp.4000992. [DOI] [PubMed] [Google Scholar]
- Tsatsanis C, Androulidaki A, Venihaki M, Margioris AN. Signaling networks regulating cyclooxygenase-2. Int J Biochem Cell Biol. 2006;38:1654–61. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Tunbridge EM, Harrison PJ, Weinberger DR. Catechol-O-methyltransferase, cognition and psychosis: val158Met and beyond. Biol Psychiatry. 2005;60:141–151. doi: 10.1016/j.biopsych.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Turner PR, O’Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Van Neerven S, Kampmann E, Mey J. RAR/RXR and PPAR/RXR signaling in neurological and psychiatric diseases. Prog Neurobiol. 2008;85:433–451. doi: 10.1016/j.pneurobio.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Yang X, Horn K, Baraban JM, Wand GS. Chronic ethanol administration decreases phosphorylation of cyclic AMP response element-binding protein in granule cells of rat cerebellum. J Neurochem. 1998;70:224–232. doi: 10.1046/j.1471-4159.1998.70010224.x. [DOI] [PubMed] [Google Scholar]
- Zhai H, Li Y, Lu L. Drug-induced alterations in the extracellular signal-regulated kinase (ERK) signaling pathway: implications for reinforcement and reinstatement. Cell Mol Neurobiol. 2008;28:157–172. doi: 10.1007/s10571-007-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Crews F. Induction of innate immune gene expression cascades in brain slice cultures by ethanol: key role of NF-kB and proinflammatory cytokines. Alcohol Clin Exp Res. 2010;34:777–789. doi: 10.1111/j.1530-0277.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- Zou JY, Crews FT. TNF alpha potentiates glutamate neurotoxicity by inhibiting glutamate uptake in organotypic brain slice cultures: neuroprotection by NF kappa B inhibition. Brain Res. 2005;1034:11–24. doi: 10.1016/j.brainres.2004.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.