Abstract
Lon, ClpXP and m-AAA are the three major ATP-dependent proteases in the mitochondrial matrix. All three are involved in general quality control by degrading damaged or abnormal proteins. In addition to this role, they are predicted to serve roles in mitochondrial DNA functions including packaging and stability, replication, transcription and translation. In particular, Lon has been implicated in mtDNA metabolism in yeast, fly and humans. Here, we review the role of Lon protease in mitochondrial DNA functions, and discuss a putative physiological role for mitochondrial transcription factor A (TFAM) degradation by Lon protease. We also discuss the possible roles of m-AAA and ClpXP in mitochondrial DNA functions, and the putative candidate substrates for the three matrix proteases.
1. Overview of the AAA+ proteases in the mitochondrial matrix
Lon, ClpXP and m-AAA are the three major proteases in the animal mitochondrial matrix (Fig. 1) [1–3]. All are derived from bacterial ancestors and are highly conserved in eukaryotic cells. Lon and ClpXP localize in the mitochondrial matrix per se, whereas the m-AAA protease is membrane anchored to expose its catalytic site to the matrix space. All three proteases belong to the super family of ATPases associated with diverse cellular activities (AAA+ ATPases) and are involved in protein degradation, in which ATPase hydrolysis is utilized to unfold the substrate proteins and transfer them into the proteolytic chamber. The Lon and ClpP proteases contain serine protease domains, whereas the m-AAA protease belongs to the metalloprotease family.
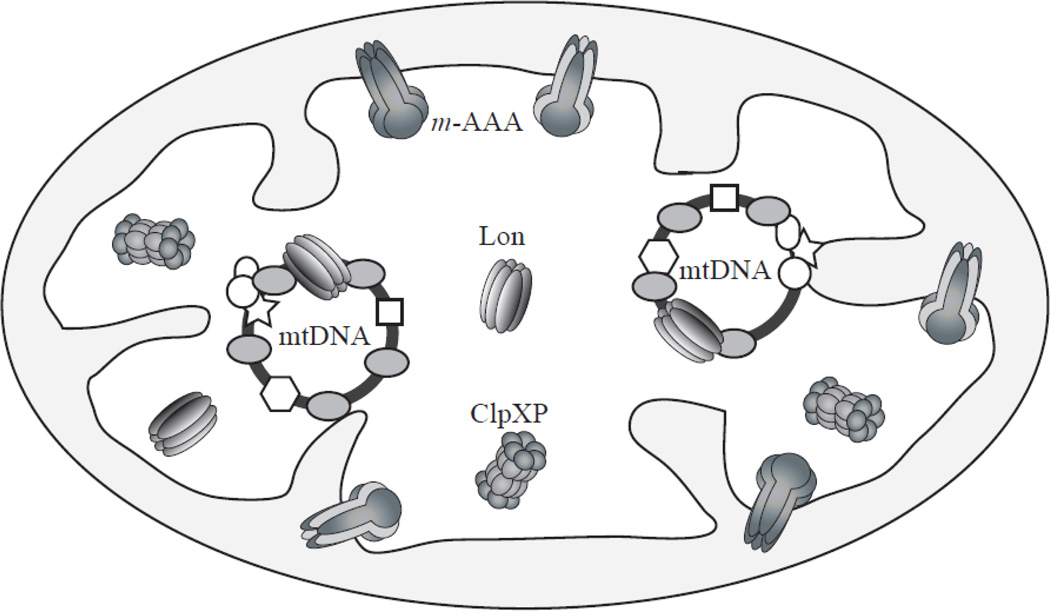
Figure 1. AAA+ proteases in the mitochondrial matrix.
Lon, ClpXP and m-AAA are the three major proteases in the animal mitochondrial matrix. Lon and ClpXP localize in the mitochondrial matrix per se, whereas the m-AAA protease is membrane anchored to expose its catalytic site to the matrix space. Lon forms a ring-shaped homo-oligomeric structure, and m-AAA forms homo- or hetero-oligomeric structures. ClpXP is composed of two subunits, ClpP and ClpX; it is a hetero-oligomeric complex in which ClpP forms a two-stack heptameric ring-shaped structure to which two hexameric ClpX rings bind on each side. Some mitochondrial proteins, such as TFAM, mtDNA replication factors, etc., bind to mtDNA to package it into a structure called the mitochondrial nucleoid. Lon is also a component of the mitochondrial nucleoid.
The matrix proteases form homo- or hetero-oligomeric, ring-shaped structures. Each subunit of Lon and m-AAA contains domains for ATPase activity and proteolysis (Fig. 2). Lon protease forms a homo-oligomeric ring-shaped structure. As with bacterial Lon, human mitochondrial Lon protease likely forms a hexamer [4], whereas Saccharomyces cerevisiae (S. cerevisiae) mitochondrial Lon forms a heptamer [5]. In contrast, m-AAA comprises two subunits, paraplegin and an ATPase family gene 3-like 2 polypeptide (AFG3L2) and in humans, can form homo-hexameric structures of AFG3L2 only, or hetero-hexamers of AFG3L2 and paraplegin [6]. Notably, mutations in paraplegin cause hereditary spastic paraplegia, whereas mutations in AFG3L2 cause hereditary spinocerebellar ataxia (SCA28) [7, 8]. Differing from Lon and m-AAA, ClpXP is composed of a proteolytic subunit, ClpP, and a chaperone-like subunit, ClpX, which carries a AAA+ domain. ClpXP is a barrel-shaped, hetero-oligomeric complex in which ClpP forms a two-stack heptameric ring-shaped structure to which two hexameric ClpX rings bind on each side [9]. Interestingly, the ClpP subunit is absent in some fungi such as S. cerevisiae and Schizosaccharomyces pombe (S. pombe) [10]. Surprisingly, the ClpX subunit is present in S. cerevisiae though not in S. pombe, and the ClpX polypeptide, known as Mcx1p (mitochondrial ClpX), lacks the interaction domain with ClpP [11]. The function of Mcx1p is not clear, but it is proposed to serve as a chaperone in the mitochondrial matrix. The matrix proteases are encoded in the nuclear genome as are most proteins in mitochondria, and function to degrade misfolded or damaged proteins (e.g., oxidized) to protect the cell from accumulating defective proteins [1–3]. There is some apparent overlap in their substrate specificities, though each of the three major proteases has a number of highly-specific substrates [12].
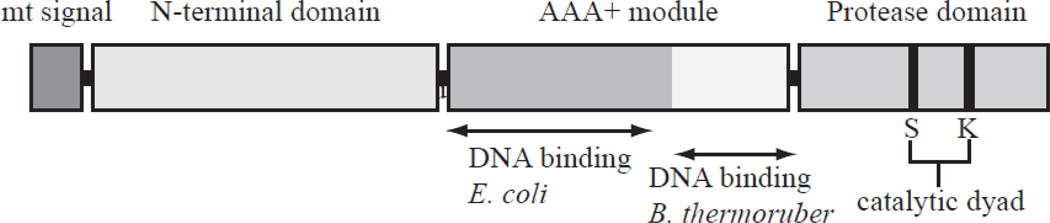
Figure 2. Domain strucure of the Lon protease.
Lon comprises three domains, an N-terminal domain, central AAA+ ATPase domain, and a C-terminal protease domain. The N-terminal domain is involved in oligomerization and protein substrate binding. The AAA+ domain that contributes ATP binding and hydrolysis consists of two sub-domains, an α/β domain and an α domain. The DNA-binding domains in E. coli and B. thermoruber Lon reside in different regions within the AAA+ module, and that in mitochondrial Lon has not been localized. The C-terminal protease domain contains a serine and lysine dyad in the active site.
2. Functions of the mitochondrial Lon protease
2.1. Quality control of mitochondrial proteins by Lon protease
Lon is well conserved among species from bacteria to man. It comprises three domains, an N-terminal domain, central ATPase domain, and a C-terminal protease domain (Fig. 2) [13, 14]. The N-terminal domain is involved in oligomerization and protein substrate binding. The AAA+ domain that contributes ATP binding and hydrolysis localizes in the central domain and consists of two sub-domains, an α/β domain and an α domain. The α/β domain is involved in ATP binding and the α domain contributes to ATP hydrolysis. The C-terminal domain carries the protease domain, which contains a serine and lysine dyad in the active site. Protease activity is dependent on ATP hydrolysis in the AAA+ domain [15]. In Escherichia coli (E. coli) and S. cerevisiae, mutations in the ATP binding site of Lon result in loss of protease activity, and those in the protease catalytic site abolish ATPase activity [15–17].
Lon contributes to protein quality control surveillance in mitochondria by degrading preferentially oxidatively-modified or misfolded proteins before they aggregate [18]. Knockdown of Lon in human and Drosophila cultured cells results in the increase of oxidized mitochondrial proteins [19, 20]. In S. cerevisiae, a number of oxidized mitochondrial proteins, including stress-related proteins, metabolic enzymes and respiratory chain subunits, have been identified as substrates for Lon using proteomics approaches [21, 22]. In animal mitochondria, aconitase, the steroidogenic acute regulatory protein, and mitochondrial transcription factor A (TFAM, also called mtTFA) have been identified as specific substrates [19, 23, 24].
Depletion of Lon shows a variety of cellular phenotypes. In human fibroblast cells, depletion of Lon over 4 days resulted in apoptotic cell death with highly abnormal mitochondrial function and morphology [25, 26]. In contrast, Lon knockdown in human colon carcinoma cells allows survival for at least 15 days [25, 26], and knockdown in Drosophila cultured cell lines expressing <10% of endogenous levels allows growth for at least six months [19]. In S. cerevisiae, loss of Lon leads to defects in mtDNA and results in a respiratory phenotype [27, 28]. Moreover, Lon is required for the expression of intron-containing mtDNA-encoded genes in S. cerevisiae [29]. The variable effects observed upon Lon depletion may reflect species- or cell type- specificity. In that regard, accompanying increases in the levels of oxidized proteins might suggest that variability in cell viability as a result of Lon depletion results from variable cellular tolerances for oxidative damage to mitochondrial proteins. In any case, the molecular mechanisms responsible for these functions remain unclear.
Lon is also known as a stress protein. In yeast and animals, its gene expression profile changes under various stress conditions, such as heat shock, oxidative stress, and serum starvation [28, 30, 31]. Interestingly, the expression of Lon shows age-related decline. In mouse skeletal muscle, Lon expression declines in an age-related manner, and in rat liver, Lon protease activity decreases and oxidized proteins accumulate with age [32, 33]. By contrast, constitutive overexpression of Lon results in an extended lifespan in the fungus, Podospora anserina [34]. These data argue that Lon and other AAA+ proteases in mitochondria may be involved in the longevity of various organisms.
2.2. Lon is a conserved DNA- binding protein
Lon protease exhibits a DNA-binding ability that is also conserved from bacteria to man. Several groups have shown that E. coli Lon binds preferentially to double-stranded DNA (dsDNA) in a nonspecific manner [35–37]. Interestingly, mammalian Lon protease binds preferentially to single-stranded DNA (ssDNA) [38–40]. Mammalian Lon recognizes ssDNA in both mitochondrial promoter regions, and binds specifically to the G- or GT-rich element in the light-strand promoter (LSP), and also to the RNA produced from LSP; its binding activity is enhanced or inhibited in the presence of protein substrates or nucleotides, respectively [38–41]. Moreover, it was reported that Lon interacts with mtDNA in cultured cells, binding not only to the control region including the mitochondrial promoters, but also to many regions containing G-rich elements [26].
Although Lon has been shown to bind dsDNA, ssDNA and RNA, there are no reported studies of the DNA-binding domain in mitochondrial Lon. However, analysis of recombinant deletion variants of E. coli Lon localized the DNA-binding domain within the α/β domain in the AAA+ module (Fig. 2) [42]. Another study identified the α domain within the AAA+ module as binding DNA using recombinant forms of Brevibacillus thermoruber Lon (Fig. 2) [43, 44]. From these studies, it appears that the DNA-binding domain of Lon is located in AAA+ module, but the sub-domain involved may vary among organisms. Interestingly, a human Lon variant lacking both ATPase and protease activity still retains DNA-binding activity [39]. Therefore, ATP hydrolysis and protease activity per se are not critical for DNA binding. Even though the DNA-binding ability of Lon has been known for 30 years, its physiological role remains elusive.
3. Potential roles for Lon in mtDNA transactions
3.1. Regulation of TFAM by Lon protease
TFAM was originally identified as a mitochondrial transcription factor, and is an abundant high mobility group (HMG) DNA-binding protein [45]. In transcriptional initiation, TFAM was proposed to bind mitochondrial promoters to stimulate mitochondrial transcription by mitochondrial RNA polymerase and mitochondrial transcription factor B2 (mtTFB2, also called TFB2M) [46]. However, recent studies argue that TFAM might not be a core transcription factor, because a mtTFB2/mitochondrial RNA polymerase complex can interact directly with mitochondrial promoters, and mtTFB2 is required for promoter melting in vitro [47]. Two additional in vitro studies extend this finding, showing that only mitochondrial RNA polymerase and mtTFB2 form the core transcription initiation complex, although TFAM stimulates transcription [48, 49].
In addition to its promoter binding activity, TFAM also binds DNA nonspecifically, a property that is essential for mtDNA packaging [50]. mtDNA is packaged with specific core proteins in a complex called the mitochondrial nucleoid [51, 52]. The major protein components of the animal mitochondrial nucleoid are DNA replication and transcription factors such as TFAM, the mtDNA helicase Twinkle, mitochondrial DNA polymerase (pol γ), mitochondrial single-stranded DNA-binding protein (mtSSB), mitochondrial transcription factor B1 (mtTFB1, also called TFB1M), and mtTFB2 [53–55]. Additionally, some mitochondrial metabolic enzymes, e.g., subunits of NADH dehydrogenase and ATP synthase, have been identified as components of the mitochondrial nuceloid [55]. Notably, mitochondrial Lon protease has also been demonstrated to be a component of mitochondrial nucleoids [55].
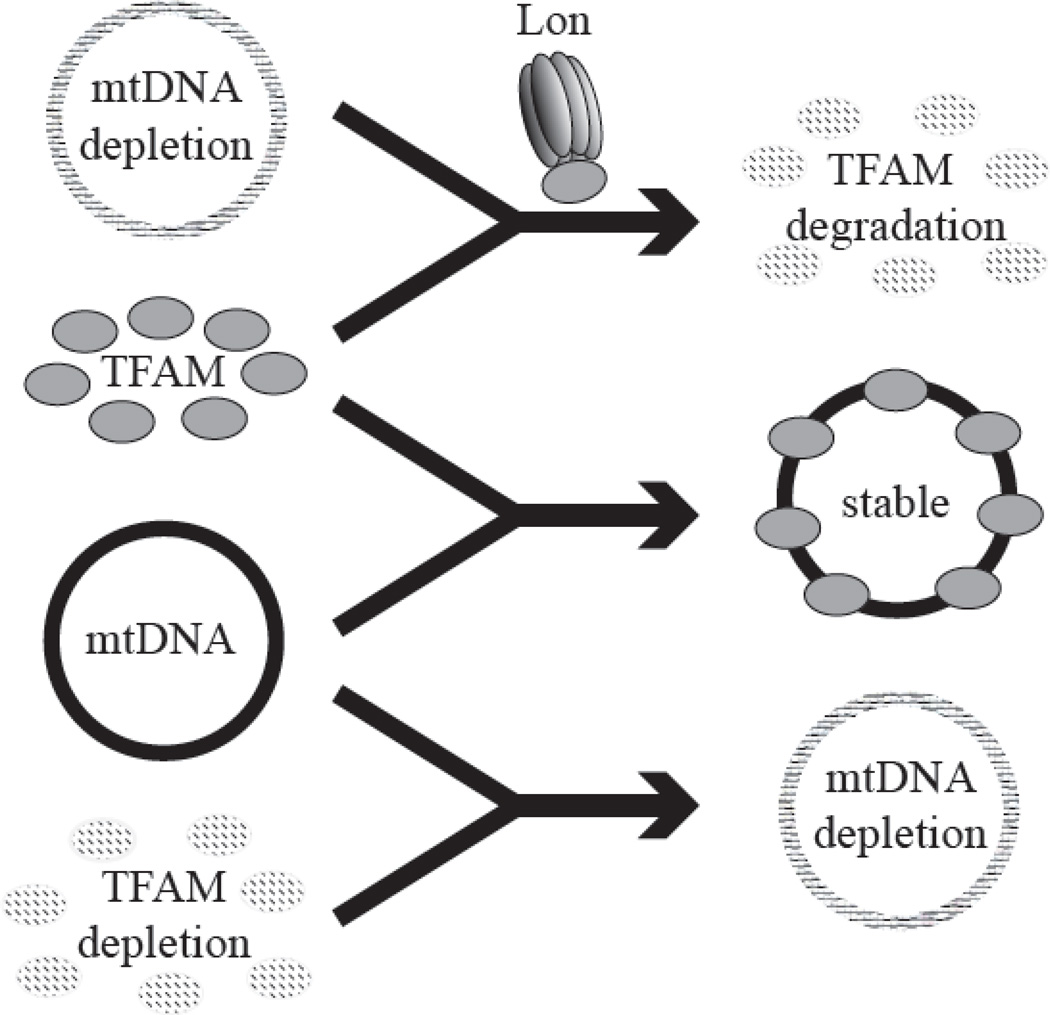
Although more than 30 proteins have been identified as components of mitochondrial nucleoids, TFAM is the key component and serves a crucial role in mtDNA maintenance. mtDNA copy number in cells changes in parallel with the relative levels of TFAM protein [56–59]. Moreover, mtDNA and TFAM levels are interdependent, such that knockdown of TFAM results in mtDNA depletion, and reduction of mtDNA copy number results in degradation of TFAM (Fig. 3) [19, 60]. Degradation upon mtDNA depletion is specific for TFAM, whereas the protein levels of other nucleoid factors including mtDNA helicase, the catalytic subunit of pol γ, and mtTFB2 are unchanged [19].
Figure 3. mtDNA and TFAM function in an interdependent manner.
Upper panel, Upon mtDNA depletion, TFAM is degraded by the Lon protease; middle panel, mtDNA and TFAM stabilize each other; lower panel, reduction of TFAM results in depletion of mtDNA.
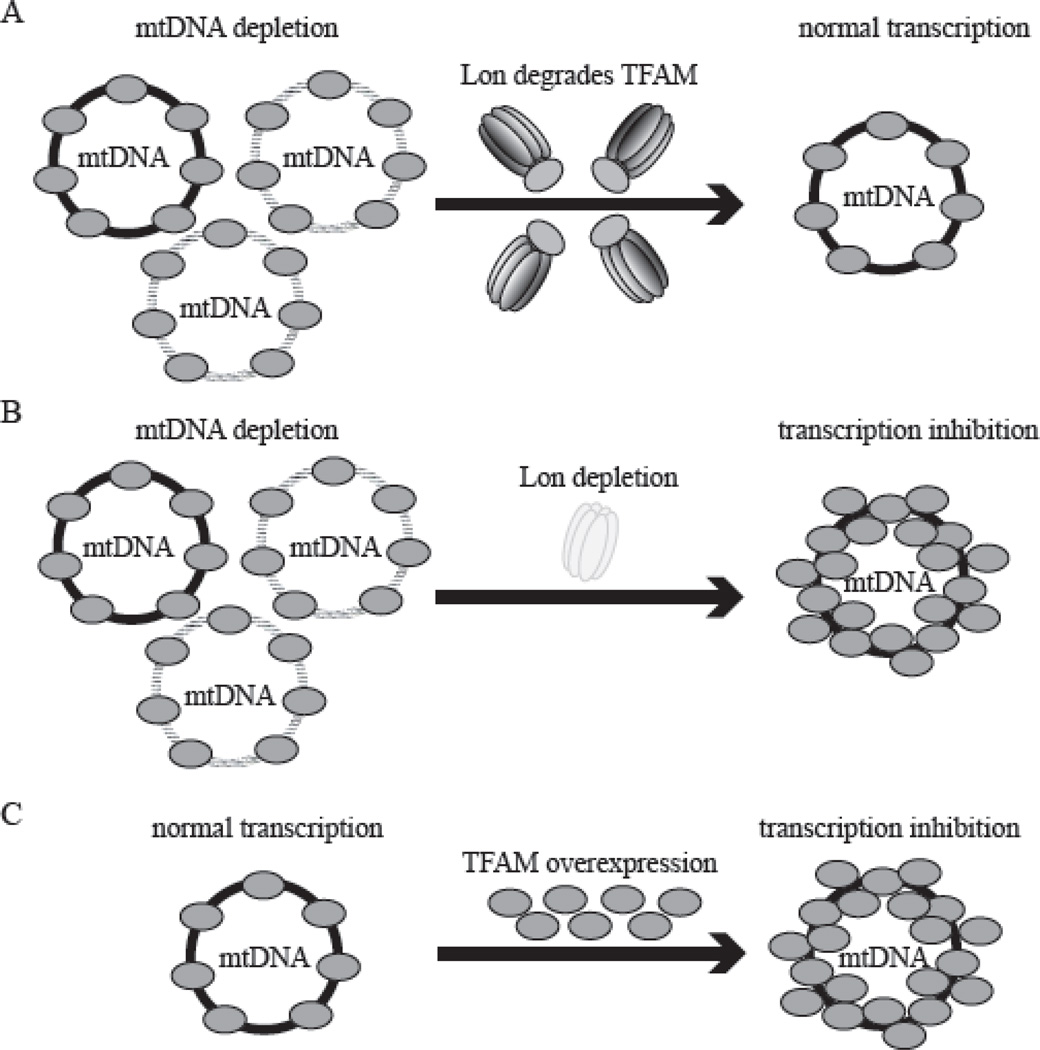
We recently demonstrated that the Lon protease is responsible for degradation of TFAM upon mtDNA depletion in Drosophila cells [19]. In Lon knockdown cells, the relative levels of TFAM protein are not reduced upon by mtDNA depletion, in contrast to the depletion of TFAM protein upon mtDNA depletion in control cells (Fig. 3, Fig. 4A and 4B), implying that Lon degrades unbound TFAM. Moreover, Lon may be also responsible for TFAM turnover under normal conditions, because the levels of TFAM and mtDNA inversely relate to the amount of cellular Lon. Furthermore, the elevation of TFAM protein results in increased mtDNA copy number, and TFAM down-regulation leads to mtDNA depletion. Hence, the TFAM: mtDNA ratio is nearly constant regardless of the specific mtDNA copy number in a cell. In contrast, the induction of mtDNA depletion in Lon depleted cells does not result in degradation of TFAM, and as a result there is a dramatic increase in the TFAM: mtDNA ratio (Fig. 4B) [19]. Thus, Lon stabilizes the TFAM: mtDNA ratio in mitochondria by selective degradation of TFAM.
Figure 4. Lon protease degrades TFAM to stabilize the TFAM: mtDNA ratio.
A, Upon reduction of mtDNA copy number in normal cells, Lon degrades TFAM to normalize the TFAM: mtDNA ratio. As a result of this process, mtDNA transcription occurs normally. B, Upon mtDNA reduction in Lon-depleted cells, TFAM is not degraded, resulting in a dramatic increase in the TFAM: mtDNA ratio. This results in a severe inhibition in mtDNA transcription, which is likely caused by mtDNA overpackaging by TFAM. C, Excess TFAM overexpression leads to an increase in the TFAM: mtDNA ratio, resulting in a severe inhibition in mtDNA transcription.
3.2. The TFAM: mtDNA ratio is critical for mtDNA transcription
Why is it important that Lon stabilizes the TFAM: mtDNA ratio? The molecular ratio of TFAM relative to mtDNA has been reported to be ~900:1 in human placental mitochondria [53, 61]. Because a dimer of TFAM binds at 35–40 bp intervals, ~900 molecules of TFAM is just sufficient to coat the 16.6 Kbp circular human mtDNA [62]. Thus, any TFAM in excess would not bind mtDNA directly under normal conditions. Indeed, overexpression of TFAM results in a dramatic increase in the TFAM: mtDNA ratio and as a result, mitochondrial transcription is suppressed severely (Fig. 4C) [19]. In cultured human and Drosophila cells, a >2-fold increase in the TFAM: mtDNA ratio causes a reduction in both mitochondrial transcription and mtDNA replication [19, 63]. Similarly, in vitro experiments also show that excess TFAM inhibits mitochondrial transcription [49, 64]. The inhibitory effect on mitochondrial transcription upon TFAM overexpression in cells likely results from overpackaging of mtDNA (Figure 4).
3.3. Mitochondrial Lon (Lon1/ PIM1) in S. cerevisiae and S. pombe
In S. cerevisiae, PIM1 is the ortholog of animal Lon protease and is also localized in the mitochondrial matrix [27, 28]. As in animal cells, induction of PIM1/ Lon is observed under stress conditions [28]. In contrast, S. cerevisiae does not have the ClpP subunit of the ClpXP protease [10]. Loss of PIM1/ Lon causes mtDNA deletion, impairs mitochondrial gene expression and results in respiratory deficiency. Interestingly, PIM1/Lon is essential for the splicing of introns contained in the mtDNA-encoded mRNAs for cytochrome c oxidase subunit I and cytochrome b [29]. In addition, a role for PIM1 has been postulated in mtDNA replication, gene expression, and/ or maintenance, but this has not yet been demonstrated.
The yeast homologue of TFAM, Abf2, is an abundant protein whose primary role is to stabilize and compact mtDNA. Yeast mitochondria are estimated to contain one molecule of Abf2 protein for every 15 bp of mtDNA, similar to the molecular ratio of TFAM to mtDNA in human mitochondria [53, 61, 65]. Excess overexpression of Abf2 also causes mtDNA depletion in yeast [66]. However, it is not clear that Abf2p is a substrate for PIM1. Surprisingly, unlike TFAM, Abf2 is not essential for either maintenance or transcription of yeast mtDNA, although it does serve a role in mtDNA maintenance [65]. Indeed, abf2Δ cells can maintain mtDNA when grown on a non-fermentable carbon source, but mtDNA is rapidly lost from abf2Δ cells grown on a fermentable carbon source. This establishes a role for Abf2 in mtDNA metabolism, though different from that of TFAM in animal cells. Aconitase (Aco1) and acetohydrogenase reductoisomerase (Ilv5) can substitute for Abf2 function in mtDNA maintenance in S. cerevisiae [67–69]. Aco1p and Ilv5p are metabolic enzymes in mitochondria and also exhibit DNA-binding activity [68, 70]. Interestingly, these proteins are known substrates for PIM1/ Lon [21, 22, 71, 72]. As with animal DNA-binding proteins in the mitochondrial nucleoid, excess overexpression of Aco1p and Ilv5p might cause impairment of mtDNA transcription and/ or replication and as a result, PIM1 may degrade these proteins to sustain appropriate mtDNA function. Additional novel protein substrates for PIM1/ Lon have also recently been identified among the mitochondrial nucleoid proteins. These include subunits of pyruvate dehydrogenase, dihydrolipoyl transsuccinylase, mitochondrial aldehyde dehydrogenase, etc. [21, 22]. Thus, regulation of mitochondrial nucleoid proteins by PIM1/ Lon may represent an important level of control in S. cerevisiae.
In S. pombe, Lon1 is the ortholog of animal Lon protease and is localized in the mitochondrial matrix space. As in S. cerevisiae, S. pombe lacks the ClpXP protease in mitochondria [10]. Very recently, it was reported that S. pombe lacking the Lon1 gene shows respiratory deficiency due to mitochondrial dysfunction as in S. cerevisiae [73]. In contrast to the Δpim1 mutant in S. cerevisiae, the Δlon1 mutant retains mtDNA and normal mitochondrial transcription in S. pombe. In addition, because there are no introns within the mtDNA-encoded genes, Lon1 is not essential for the splicing process in S. pombe mitochondria. Thus, in this organism it appears that Lon1 is essential for some mitochondrial functions, but not for mtDNA metabolism. Surprisingly, S. pombe lacks a gene for Abf2, so other nucleoid proteins such as Aco1p and Ilv5p may sustain mtDNA functions [74]. Altogether, the multi-faceted roles of the Lon protease may differ among fungi and also from those in animal mitochondria.
4. Do ClpXP and m-AAA proteases affect mtDNA functions?
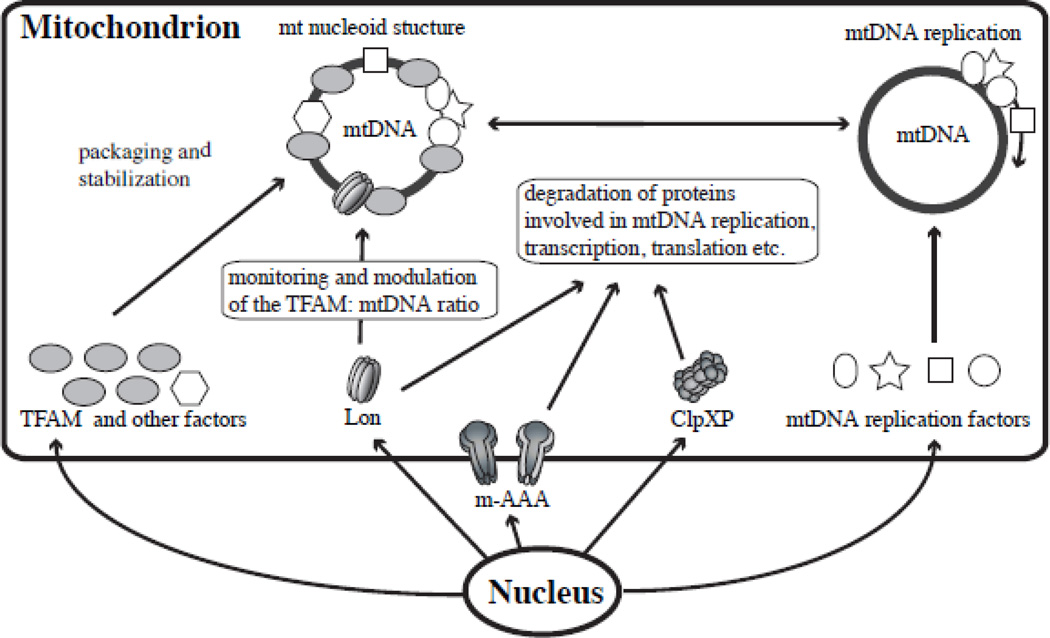
ClpXP and m-AAA might affect mtDNA function (Fig. 5). However, there is no evidence to date that m-AAA and ClpXP regulate mtDNA replication, maintenance or gene expression by their proteolytic activities. Although ClpXP contributes to the degradation of misfolded proteins, few of its substrates have been identified [75]. Some substrates have been identified for m-AAA, and most of them are membrane proteins [6, 76]. These include the prohibitin proteins (PHB1 and PHB2) in S. cerevisiae that are ubiquitous, highly-conserved proteins localized in the mitochondrial inner membrane [77]. Prohibitins contribute to multiple processes including cell cycle progression, apoptosis, and protein stability [78]. Interestingly, prohibitins are also among those proteins associated with mitochondrial nucleoids [55]. A recent study shows that prohibitin 1 contributes to maintenance of mtDNA and TFAM levels: depletion of PHB1 results in the reduction of TFAM and mtDNA [79]. Taken together it seems plausible that PHB1 turnover by m-AAA protease plays a role in mtDNA metabolism. Furthermore, m-AAA protease is essential for the maturation of mitochondrial ribosomal protein MRPL32 [6, 80]. The N-terminal region of MRPL32 is removed by m-AAA protease, and this maturation step is well conserved from yeast to man. Maturation of MRPL32 is essential for its assembly into mitochondrial ribosomes, which are essential for translation of the mtDNA-encoded respiratory subunits. Thus in this regard, m-AAA regulates mitochondrial gene expression by its proteolytic activity.
Figure 5. Potential physiological roles of the AAA+ proteases in mtDNA function.
The model shows that the three AAA+ proteases in the mitochondrial matrix regulate mitochondrial protein levels to sustain proper mtDNA functions, such as mtDNA packaging and stability, replication, transcription and translation.
5. Potential physiological roles of matrix AAA+ proteases in mtDNA functions
5.1. What is the physiological role of TFAM degradation by Lon protease in mtDNA function?
Lon is expressed ubiquitously, and is induced transiently up to ~8 fold under stress conditions [31]. However, a 5-fold increase of Lon results only in a modest reduction of mtDNA copy number (70% of that in control cells), and this does not influence the levels of mtDNA transcripts or of the translated proteins [19, 56]. Thus, Lon protein levels per se may not be a crucial factor in mtDNA function.
Knockdown of DNA replication factors or TFAM results in a dramatic reduction of mtDNA copy number, and increases in their levels can also result in modest increases in mtDNA levels. For example, the exogenous expression of TFAM or mtDNA helicase results in increased mtDNA copy number [58, 59, 81–84]. Notably, co-overexpression of TFAM and mtDNA helicase show additive effects in increased mtDNA copy number in mice [84]. This is reasonable because the increases result from their influence at different levels in mtDNA maintenance: mtDNA helicase increases the mtDNA copy number through increased replication of mtDNA, whereas TFAM increases mtDNA stability by DNA binding and packaging. In general, the rate of mtDNA synthesis is controlled by the level and activity of DNA replication factors, and its overall stability is determined by TFAM levels and perhaps those of other nucleoid factors. Interestingly, an inverse relationship between mtDNA copy number and mtDNA size was shown in an earlier study evaluating normal and aberrant mtDNA molecules containing deletions and duplications, such that a constant mtDNA mass was maintained [85]. A ‘quantity control’ process such as Lon regulation of TFAM levels provides a reasonable explanation for this phenomenon.
The availability of DNA replication factors and TFAM is regulated by the same or similar transcription factors (such as nuclear respiratory factors, NRF-1 and 2), and co-activators of the PCG-family (such as PGC-1α) in mammals, and by the DNA replication-related element-binding factor (DREF) in Drosophila [46, 86–89]. Despite this coordinate control, calibration of cellular TFAM levels may require additional mechanisms of regulation. Indeed in some cases, TFAM expression may be in excess over newly-synthesized mtDNA, and in other cases, the newly-synthesized mtDNA may be in excess over TFAM. This is the case in particular when external factors change the balance between the amount of mtDNA, and the protein levels of TFAM. For example, some anti-viral drugs such as AZT and ddC inhibit mtDNA replication, such that the ratio of TFAM: mtDNA is increased transiently [90]. Here, we would propose that Lon monitors and adjusts the TFAM: mtDNA ratio to sustain appropriate mtDNA transcription levels by selective degradation of TFAM (Fig. 5).
How does Lon recognize and degrade excess TFAM? While the mechanism by which mitochondrial Lon recognizes its target proteins remains unclear, bacterial Lon was shown to recognize specific aromatic residue-rich sequences that are hidden in the hydrophobic cores of native structures, but are accessible in unfolded structures [91]. Notably, the HMG boxes in TFAM contain four conserved aromatic residues within a hydrophobic core, and these residues may be masked when TFAM binds mtDNA [50, 57, 58]. When TFAM is not bound to mtDNA, Lon may recognize the exposed hydrophobic core to degrade it. Our current hypothesis is that excess, free TFAM is degraded by Lon before it can bind DNA. Interestingly, both TFAM and Lon bind preferentially to the control region of mtDNA; it seems plausible that Lon may interact with TFAM to form a complex poised for degradation when TFAM is released from mtDNA. Another possible explanation is that TFAM not bound to mtDNA becomes exposed to oxidative stress, whereas that bound to mtDNA comprising part of the core of the mitochondrial nucleoid is stabilized [55]. Because Lon shows a specificity for oxidatively-damaged proteins, oxidized TFAM would be degraded.
In Drosophila cells, it takes about 6 days to normalize the TFAM: mtDNA ratio after EtBr treatment [19]. This, and the finding that excess TFAM present after overexpression is not degraded efficiently to normalize the TFAM: mtDNA ratio [19], suggests that the cellular capacity for TFAM degradation by Lon is limited. At the same time, this level of regulation of TFAM is critical for the maintenance of mtDNA transcription.
5.2. Is there a quantity control degradation pathway for other mitochondrial nucleoid proteins?
Does overexpression of other mitochondrial nucleoid proteins cause impairment of the replication, maintenance and transcription of mtDNA? In Drosophila cultured cells, a 50-fold increase in mtTFB1 shows no negative effects on mtDNA copy number or its transcription, whereas a 40-fold increase in mtTFB2 results in a 2-fold increase of mtDNA and its transcription levels [82, 92]. Similar results were obtained from HeLa cells overexpressing human mtTFB1 or mtTFB2 [93]. Moreover, upon 15-fold overexpression of the wild-type d-mtDNA helicase, mtDNA copy number is increased in Drosophila cultured cells [83]. Similarly, overexpression of either the catalytic core of human mitochondrial DNA polymerase (pol γ–α) or mtDNA helicase shows no negative effects on mtDNA replication in human cultured cells [94, 95]. Thus, although Lon interacts with mtDNA helicase and pol γ–α in human cultured cells [39], it may function in their degradation but perhaps not to regulate their relative abundance.
Overexpression of some mitochondrial nucleoid proteins results in defects in mtDNA replication. For example, overexpression of human mitochondrial transcription termination factor MTERF (also called mTERF1), which is a component of mitochondrial nucleoid, increases pausing in mtDNA replication [96]. Mammalian MTERF binds a specific site located downstream of the 3’-end of the mitochondrial 16S rRNA gene to terminate mitochondrial transcription [97]. Overexpressed MTERF also binds at other sites in human mtDNA [96]. Likewise, overexpression of two members of the MTERF family, MTERFD1 (also called mTERF3) and MTERFD3 (also called mTERF2), show similar results in human cultured cells [98]. Because MTERFD1 and MTERFD3 do not show strong DNA-binding specificity, it seems likely that overexpressed forms are able to bind multiple regions of mtDNA [99, 100]. Similarly, excess overexpression of TFAM causes impairment in mtDNA replication in human cells [63]. In composite, these data argue that increased levels of non-specific DNA-binding proteins inhibit mtDNA replication, suggesting that the levels of mitochondrial nucleoid proteins that could inhibit mtDNA replication may need to be regulated in a ‘quantity control’ manner by Lon and/ or other mitochondrial matrix AAA+ proteases (Fig. 5).
6. Concluding remarks
Lon, ClpXP and m-AAA are the major proteases in the mitochondrial matrix and involved in general protein quality control. Clearly, most proteins involved in mtDNA functions, including DNA packaging and stability, replication, transcription and translation, are likely degraded by these AAA+ proteases in their individual quality control roles yet importantly, their levels may also be regulated by them in specific quantity control roles (Fig. 5). We have described here the regulation of TFAM levels by Lon in such a ‘quantity control’ manner, and the potential physiological roles of the three proteases in mtDNA functions. Although information on the specific substrates for the matrix proteases remains very limited, and their roles in mtDNA functions are only beginning to be elucidated, their likely impact on mitochondrial biogenesis is already apparent, and further investigation is warranted to unravel their overall contributions to mitochondrial metabolism.
Highlights.
We review the structure and function of mitochondrial matrix proteases.
We explore the roles of the matrix proteases in regulation of mtDNA functions.
We propose a physiological role for TFAM regulation by Lon protease.
Acknowledgements
This work was supported by National Institutes of Health Grant GM45295 (to L.S.K.), and a Grant-in-Aid for Scientific Research (C) from Japan Society for the Promotion of Science (to Y.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ugarte N, Petropoulos I, Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid Redox Signal. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 2.Truscott KN, Lowth BR, Strack PR, Dougan DA. Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem Cell Biol. 2010;88:97–108. doi: 10.1139/o09-167. [DOI] [PubMed] [Google Scholar]
- 3.Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010;19:987–999. doi: 10.1002/pro.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahlberg H, Kutejova E, Suda K, Wolpensinger B, Lustig A, Schatz G, Engel A, Suzuki CK. Mitochondrial Lon of Saccharomyces cerevisiae is a ring-shaped protease with seven flexible subunits. Proc Natl Acad Sci U S A. 1999;96:6787–6790. doi: 10.1073/pnas.96.12.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsuta T, Langer T. AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res Microbiol. 2009;160:711–717. doi: 10.1016/j.resmic.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 8.Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, Veneziano L, Sacco T, Boda E, Brussino A, Bonn F, Castellotti B, Baratta S, Mariotti C, Gellera C, Fracasso V, Magri S, Langer T, Plevani P, Di Donato S, Muzi-Falconi M, Taroni F. Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat Genet. 2010;42:313–321. doi: 10.1038/ng.544. [DOI] [PubMed] [Google Scholar]
- 9.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojtyra UA, Thibault G, Tuite A, Houry WA. The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J Biol Chem. 2003;278:48981–48990. doi: 10.1074/jbc.M307825200. [DOI] [PubMed] [Google Scholar]
- 11.van Dyck L, Dembowski M, Neupert W, Langer T. Mcx1p, a ClpX homologue in mitochondria of Saccharomyces cerevisiae. FEBS Lett. 1998;438:250–254. doi: 10.1016/s0014-5793(98)01310-6. [DOI] [PubMed] [Google Scholar]
- 12.Savel'ev AS, Novikova LA, Kovaleva IE, Luzikov VN, Neupert W, Langer T. ATP-dependent proteolysis in mitochondria. m-AAA protease and PIM1 protease exert overlapping substrate specificities and cooperate with the mtHsp70 system. J Biol Chem. 1998;273:20596–20602. doi: 10.1074/jbc.273.32.20596. [DOI] [PubMed] [Google Scholar]
- 13.Lee I, Suzuki CK. Functional mechanics of the ATP-dependent Lon protease-lessons from endogenous protein and synthetic peptide substrates. Biochim Biophys Acta. 2008;1784:727–735. doi: 10.1016/j.bbapap.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ugarte N, Petropoulos I, Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid Redox Signal. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 15.van Dijl JM, Kutejova E, Suda K, Perecko D, Schatz G, Suzuki CK. The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc Natl Acad Sci U S A. 1998;95:10584–10589. doi: 10.1073/pnas.95.18.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer H, Glockshuber R. A point mutation within the ATP-binding site inactivates both catalytic functions of the ATP-dependent protease La (Lon) from Escherichia coli. FEBS Lett. 1994;356:101–103. doi: 10.1016/0014-5793(94)01244-x. [DOI] [PubMed] [Google Scholar]
- 17.Starkova NN, Koroleva EP, Rumsh LD, Ginodman LM, Rotanova TV. Mutations in the proteolytic domain of Escherichia coli protease Lon impair the ATPase activity of the enzyme. FEBS Lett. 1998;422:218–220. doi: 10.1016/s0014-5793(98)00012-x. [DOI] [PubMed] [Google Scholar]
- 18.Bender T, Lewrenz I, Franken S, Baitzel C, Voos W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell. 2011;22:541–554. doi: 10.1091/mbc.E10-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc Natl Acad Sci U S A. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngo JK, Davies KJ. Importance of the lon protease in mitochondrial maintenance and the significance of declining lon in aging. Ann N Y Acad Sci. 2007;1119:78–87. doi: 10.1196/annals.1404.015. [DOI] [PubMed] [Google Scholar]
- 21.Bayot A, Gareil M, Rogowska-Wrzesinska A, Roepstorff P, Friguet B, Bulteau AL. Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1. J Biol Chem. 2010;285:11445–11457. doi: 10.1074/jbc.M109.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender T, Leidhold C, Ruppert T, Franken S, Voos W. The role of protein quality control in mitochondrial protein homeostasis under oxidative stress. Proteomics. 2010;10:1426–1443. doi: 10.1002/pmic.200800619. [DOI] [PubMed] [Google Scholar]
- 23.Granot Z, Kobiler O, Melamed-Book N, Eimerl S, Bahat A, Lu B, Braun S, Maurizi MR, Suzuki CK, Oppenheim AB, Orly J. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by Lon protease: the unexpected effect of proteasome inhibitors. Mol Endocrinol. 2007;21:2164–2177. doi: 10.1210/me.2005-0458. [DOI] [PubMed] [Google Scholar]
- 24.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 25.Bota DA, Ngo JK, Davies KJ. Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic Biol Med. 2005;38:665–677. doi: 10.1016/j.freeradbiomed.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 26.Lu B, Yadav S, Shah PG, Liu T, Tian B, Pukszta S, Villaluna N, Kutejova E, Newlon CS, Santos JH, Suzuki CK. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J Biol Chem. 2007;282:17363–17374. doi: 10.1074/jbc.M611540200. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki CK, Suda K, Wang N, Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264:891. doi: 10.1126/science.8178144. [DOI] [PubMed] [Google Scholar]
- 28.Van Dyck L, Pearce DA, Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- 29.van Dyck L, Neupert W, Langer T. The ATP-dependent PIM1 protease is required for the expression of intron-containing genes in mitochondria. Genes Dev. 1998;12:1515–1524. doi: 10.1101/gad.12.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hori O, Ichinoda F, Tamatani T, Yamaguchi A, Sato N, Ozawa K, Kitao Y, Miyazaki M, Harding HP, Ron D, Tohyama M, D MS, Ogawa S. Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J Cell Biol. 2002;157:1151–1160. doi: 10.1083/jcb.200108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngo JK, Davies KJ. Mitochondrial Lon protease is a human stress protein. Free Radic Biol Med. 2009;46:1042–1048. doi: 10.1016/j.freeradbiomed.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakala H, Delaval E, Hamelin M, Bismuth J, Borot-Laloi C, Corman B, Friguet B. Changes in rat liver mitochondria with aging. Lon protease-like reactivity and N(epsilon)-carboxymethyllysine accumulation in the matrix. Eur J Biochem. 2003;270:2295–2302. doi: 10.1046/j.1432-1033.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 34.Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol. 2009;11:852–858. doi: 10.1038/ncb1893. [DOI] [PubMed] [Google Scholar]
- 35.Charette MF, Henderson GW, Doane LL, Markovitz A. DNA-stimulated ATPase activity on the lon (CapR) protein. J Bacteriol. 1984;158:195–201. doi: 10.1128/jb.158.1.195-201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung CH, Goldberg AL. DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc Natl Acad Sci U S A. 1982;79:795–799. doi: 10.1073/pnas.79.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonezaki S, Okita K, Oba T, Ishii Y, Kondo A, Kato Y. Protein substrates and heat shock reduce the DNA-binding ability of Escherichia coli Lon protease. Appl Microbiol Biotechnol. 1995;44:484–488. doi: 10.1007/BF00169948. [DOI] [PubMed] [Google Scholar]
- 38.Fu GK, Markovitz DM. The human LON protease binds to mitochondrial promoters in a single-stranded, site-specific, strand-specific manner. Biochemistry. 1998;37:1905–1909. doi: 10.1021/bi970928c. [DOI] [PubMed] [Google Scholar]
- 39.Liu T, Lu B, Lee I, Ondrovicova G, Kutejova E, Suzuki CK. DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J Biol Chem. 2004;279:13902–13910. doi: 10.1074/jbc.M309642200. [DOI] [PubMed] [Google Scholar]
- 40.Lu B, Liu T, Crosby JA, Thomas-Wohlever J, Lee I, Suzuki CK. The ATP-dependent Lon protease of Mus musculus is a DNA-binding protein that is functionally conserved between yeast and mammals. Gene. 2003;306:45–55. doi: 10.1016/s0378-1119(03)00403-7. [DOI] [PubMed] [Google Scholar]
- 41.Chen SH, Suzuki CK, Wu SH. Thermodynamic characterization of specific interactions between the human Lon protease and G-quartet DNA. Nucleic Acids Res. 2008;36:1273–1287. doi: 10.1093/nar/gkm1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomura K, Kato J, Takiguchi N, Ohtake H, Kuroda A. Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J Biol Chem. 2004;279:34406–34410. doi: 10.1074/jbc.M404725200. [DOI] [PubMed] [Google Scholar]
- 43.Lin YC, Lee HC, Wang I, Hsu CH, Liao JH, Lee AY, Chen C, Wu SH. DNA-binding specificity of the Lon protease alpha-domain from Brevibacillus thermoruber WR-249. Biochem Biophys Res Commun. 2009;388:62–66. doi: 10.1016/j.bbrc.2009.07.118. [DOI] [PubMed] [Google Scholar]
- 44.Lee AY, Hsu CH, Wu SH. Functional domains of Brevibacillus thermoruber lon protease for oligomerization and DNA binding: role of N-terminal and sensor and substrate discrimination domains. J Biol Chem. 2004;279:34903–34912. doi: 10.1074/jbc.M403562200. [DOI] [PubMed] [Google Scholar]
- 45.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 46.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 47.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shutt TE, Bestwick M, Shadel GS. The core human mitochondrial transcription initiation complex: It only takes two to tango. Transcription. 2011;2:55–59. doi: 10.4161/trns.2.2.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci U S A. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill ME. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilkerson RW. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int J Biochem Cell Biol. 2009;41:1899–1906. doi: 10.1016/j.biocel.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 52.Rebelo AP, Dillon LM, Moraes CT. Mitochondrial DNA transcription regulation and nucleoid organization. J Inherit Metab Dis. 2011;34:941–951. doi: 10.1007/s10545-011-9330-8. [DOI] [PubMed] [Google Scholar]
- 53.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 56.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 57.Goto A, Matsushima Y, Kadowaki T, Kitagawa Y. Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem J. 2001;354:243–248. doi: 10.1042/0264-6021:3540243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsushima Y, Matsumura K, Ishii S, Inagaki H, Suzuki T, Matsuda Y, Beck K, Kitagawa Y. Functional domains of chicken mitochondrial transcription factor A for the maintenance of mitochondrial DNA copy number in lymphoma cell line DT40. J Biol Chem. 2003;278:31149–31158. doi: 10.1074/jbc.M303842200. [DOI] [PubMed] [Google Scholar]
- 59.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pohjoismaki JL, Wanrooij S, Hyvarinen AK, Goffart S, Holt IJ, Spelbrink JN, Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 65.Diffley JF, Stillman B. A close relative of the nuclear, chromosomal highmobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci U S A. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 68.Macierzanka M, Plotka M, Pryputniewicz-Drobinska D, Lewandowska A, Lightowlers R, Marszalek J. Maintenance and stabilization of mtDNA can be facilitated by the DNA-binding activity of Ilv5p. Biochim Biophys Acta. 2008;1783:107–117. doi: 10.1016/j.bbamcr.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 69.Zelenaya-Troitskaya O, Perlman PS, Butow RA. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 1995;14:3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen XJ, Wang X, Butow RA. Yeast aconitase binds and provides metabolically coupled protection to mitochondrial DNA. Proc Natl Acad Sci U S A. 2007;104:13738–13743. doi: 10.1073/pnas.0703078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bateman JM, Iacovino M, Perlman PS, Butow RA. Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and the Pim1p protease. J Biol Chem. 2002;277:47946–47953. doi: 10.1074/jbc.M209071200. [DOI] [PubMed] [Google Scholar]
- 72.Major T, von Janowsky B, Ruppert T, Mogk A, Voos W. Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease pim1. Mol Cell Biol. 2006;26:762–776. doi: 10.1128/MCB.26.3.762-776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guha S, Lopez-Maury L, Shaw M, Bahler J, Norbury CJ, Agashe VR. Transcriptional and cellular responses to defective mitochondrial proteolysis in fission yeast. J Mol Biol. 2011;408:222–237. doi: 10.1016/j.jmb.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 74.Nosek J, Tomaska L, Bolotin-Fukuhara M, Miyakawa I. Mitochondrial chromosome structure: an insight from analysis of complete yeast genomes. FEMS Yeast Res. 2006;6:356–370. doi: 10.1111/j.1567-1364.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- 75.Hansen J, Gregersen N, Bross P. Differential degradation of variant medium-chain acyl-CoA dehydrogenase by the protein quality control proteases Lon and ClpXP. Biochem Biophys Res Commun. 2005;333:1160–1170. doi: 10.1016/j.bbrc.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 76.Stiburek L, Zeman J. Assembly factors and ATP-dependent proteases in cytochrome c oxidase biogenesis. Biochim Biophys Acta. 2010;1797:1149–1158. doi: 10.1016/j.bbabio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 77.Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol. 1999;19:3435–3442. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol Metab. 2009;20:394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 79.Kasashima K, Sumitani M, Satoh M, Endo H. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids. Exp Cell Res. 2008;314:988–996. doi: 10.1016/j.yexcr.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 80.Martinelli P, Rugarli EI. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim Biophys Acta. 2010;1797:1–10. doi: 10.1016/j.bbabio.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 81.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 82.Matsushima Y, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells. J Biol Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 83.Matsushima Y, Kaguni LS. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J Biol Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- 85.Tang Y, Schon EA, Wilichowski E, Vazquez-Memije ME, Davidson E, King MP. Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol Biol Cell. 2000;11:1471–1485. doi: 10.1091/mbc.11.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bruni F, Polosa PL, Gadaleta MN, Cantatore P, Roberti M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J Biol Chem. 2010;285:3939–3948. doi: 10.1074/jbc.M109.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fernandez-Moreno MA, Bruni F, Adan C, Sierra RH, Polosa PL, Cantatore P, Garesse R, Roberti M. The Drosophila nuclear factor DREF positively regulates the expression of the mitochondrial transcription termination factor DmTTF. Biochem J. 2009;418:453–462. doi: 10.1042/BJ20081174. [DOI] [PubMed] [Google Scholar]
- 88.Garesse R, Kaguni LS. A Drosophila model of mitochondrial DNA replication: proteins, genes and regulation. IUBMB Life. 2005;57:555–561. doi: 10.1080/15216540500215572. [DOI] [PubMed] [Google Scholar]
- 89.Kim YS, Shin MJ, Yang DJ, Yamaguchi M, Park SY, Yoo MA. Transcriptional regulation of the Drosophila ANT gene by the DRE/DREF system. Genes Cells. 2007;12:569–579. doi: 10.1111/j.1365-2443.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 90.Bailey CM, Anderson KS. A mechanistic view of human mitochondrial DNA polymerase gamma: providing insight into drug toxicity and mitochondrial disease. Biochim Biophys Acta. 2010;1804:1213–1222. doi: 10.1016/j.bbapap.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc Natl Acad Sci U S A. 2009;106:18503–18508. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsushima Y, Adan C, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J Biol Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 93.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, Jacobs HT. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 95.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Byrnes J, Garcia-Diaz M. Mitochondrial transcription: How does it end? Transcr. 2011;2:32–36. doi: 10.4161/trns.2.1.14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hyvarinen AK, Pohjoismaki JL, Holt IJ, Jacobs HT. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Mol Biol Rep. 2011;38:1321–1328. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 99.Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 100.Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta. 2009;1787:296–302. doi: 10.1016/j.bbabio.2009.01.018. [DOI] [PubMed] [Google Scholar]