Abstract
Binding of urokinase-type plasminogen activator (uPA)1 to its receptor, uPAR, in estrogen receptor-α (ERα) expressing breast cancer cells, transiently activates ERK downstream of FAK, Src family kinases, and H-Ras. Herein, we show that when uPAR is over-expressed, in two separate ERα-positive breast cancer cell lines, ERK activation occurs autonomously of uPA and is sustained. Autonomous ERK activation by uPAR requires H-Ras and Rac1. A mutated form of uPAR, which does not bind vitronectin (uPAR-W32A), failed to induce autonomous ERK activation. Expression of human uPAR or mouse uPAR but not uPARW32A in MCF-7 cells provided a selection advantage when these cells were deprived of estrogen in cell culture for two weeks. Similarly, MCF-7 cells that express mouse uPAR formed xenografts in SCID mice that survived and increased in volume in the absence of estrogen supplementation, probably reflecting the pro-survival activity of phospho-ERK. Autonomous uPAR signaling to ERK was sensitive to the EGFR tyrosine kinase inhibitors, Erlotinib and Gefitinib. The transition in uPAR signaling from uPA-dependent and transient to autonomous and sustained is reminiscent of the transformation in ErbB2/ HER2 signaling observed when this gene is amplified in breast cancer. uPAR over-expression may provide a pathway for escape of breast cancer cells from ERα-targeting therapeutics.
Keywords: urokinase-type plasminogen activator, uPAR, ERK, EGF Receptor, estrogen receptor, breast cancer
1. Introduction
Estrogen receptor-α (ERα) is expressed in up to 75% of all cases of adenocarcinoma of the breast [1-2]. These tumors are frequently treated with agents that inhibit ERα activation, down-regulate ERα, or reduce estrogen synthesis [3-4]. Unfortunately, in many cases, tumor cells acquire molecular changes that allow resistance to anti-estrogens. The resulting cancers are frequently aggressive and rapidly progressing. Previously described receptors that may become activated and allow escape from ERα-targeting drugs include the EGF receptor (EGFR), ErbB2/HER2, and Insulin-like Growth Factor Receptor-1 (IGF1R) [2, 5-7]. In addition to its function as a transcription factor, ERα activates Src family kinases (SFKs), ERK, and phosphatidylinositol 3-kinase (PI3K) [8]. Thus, it is reasonable to hypothesize that other receptors, which activate the same cell-signaling pathways, may offset the requirement for estrogen in breast cancer cells.
The urokinase receptor (uPAR) gene may be amplified in cancer of the breast and pancreas [9-10]. In diverse solid tumors, uPAR expression is correlated with disease progression [11-15]. Although uPAR plays a pivotal role in activation of protease cascades at the cell surface, important processes in cancer have been linked to uPAR-initiated cell-signaling, including cell migration, survival, release from states of dormancy, epithelial-mesenchymal transition (EMT), and cancer stem cell-like behavior [16-20]. In mice, uPAR promotes cancer metastasis independently of urokinase-type plasminogen activator (uPA), implicating pathways other than or in addition to protease activation [21].
uPAR is GPI-anchored and thus, utilizes a system of co-receptors and transactivation pathways to trigger cell-signaling [18]. uPAR-initiated cell-signaling is regulated by two ligands, uPA and vitronectin, which interact with distinct binding sites [22-26]. Some signaling pathways activated downstream of uPAR, such as that leading to Rac1, do not require uPA [27-29]. Whether uPA is necessary for uPAR-dependent ERK activation is less clear. There is evidence that ERK may be activated in the absence of uPA; however, uPA produced endogenously by the cells may be involved [17, 26, 30]. It is extremely important to understand the mechanism by which uPAR activates ERK because in cancer cells, phospho-ERK is a potent cell-survival factor [31]. In ERα-expressing MCF-7 breast cancer cells, uPAR-dependent ERK activation is strictly dependent on uPA [32-34]. This pathway also requires focal adhesion kinase (FAK) and SFKs [16-17, 33-34]. EGFR transactivation may be involved [30, 35].
When expressed at high levels, receptor tyrosine kinases (RTKs) may signal independently of ligands. One example involves ErbB2/HER2, which dimerizes and signals in the absence of ligand in breast cancer cells [36, 37]. In this study, we show that over-expression of uPAR in two distinct ERα-expressing breast cancer cell types transforms the uPAR signaling mechanism so that ERK is activated autonomously of uPA. Autonomous uPAR signaling to ERK occurs downstream of H-Ras and Rac1, unlike uPA-induced signaling, which occurs downstream of HRas alone [32-34]. Activation of autonomous uPAR signaling provides a selection advantage for ERα-expressing breast cancer cells when estrogen is unavailable in vitro and in vivo. Because uPAR-initiated cell-signaling has been reported to involve receptor dimerization or oligomerization [38, 39], we hypothesize that the transformation in uPAR signaling mechanism reported here is similar to that previously reported for ErbB2/HER2 [36].
2. Materials and Methods
2.1. Reagents
17β-estradiol (E2) was from Sigma-Aldrich (St. Louis, MO). The PDGF receptor kinase-selective inhibitor, Tyrphostin AG1296, and the SFK inhibitor, PP2, were from EMD Biosciences (Gibbstown, NJ). Erlotinib and Gefitinib were from LC Laboratories (Woburn, MA). Expression constructs encoding dominant-negative Rac1 (DN-Rac1/Rac1-S17N), dominant-negative HRas (DN-H-Ras/Ras-S17N), dominant-negative focal adhesion kinase (FAK) (DN-FAK/FAKY397F), wild-type FAK, and hemagglutinin (HA)-tagged ERK1 are previously described [29, 33, 34, 40]. Expression constructs encoding human uPAR and mouse uPAR also are previously described [21, 41]. The full-length human uPAR cDNA in pCDNA3.1 was mutated at a single base-pair to generate uPAR-W32A, using the Quick-Change Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Human uPAR-specific antibody was from Molecular Diagnostics (Stamford, CT). Allophycocyanin (APC)-conjugated human uPAR-specific antibody and isotype-matched control antibody for flow cytometry were from eBioscience (San Diego, CA). Mouse uPAR-specific antiserum was generously provided by Dr. Andrew Mazar (Northwestern University, Evanston, IL). Phospho-ERK-specific antibody was from Cell Signaling Technologies (Danvers, MA). Rac/Cdc42 assay reagent (PAK1-PBD), which includes residues 67-150 of p21-activated kinase fused to glutathione-S-transferase and coupled to glutathione-Sepharose, was from Millipore (Billerica, MA), as was the antibody that detects total ERK. Rac1-specific antibody was from BD Biosciences (Franklin Lakes, NJ). Horseradish peroxidase-conjugated antibodies specific for mouse or rabbit IgG were from GE Healthcare (Piscataway, NJ). qPCR reagents, including primers and probes, were from Applied Biosystems (Foster City, CA).
2.2. Cell Culture
Low passage MCF-7 and MDA-MB 231 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Hyclone) supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 μg/ml). MDA-MB 361 cells (ATCC) were maintained in RPMI-1640 medium supplemented with 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml). MCF-7 cells were transfected to express mouse uPAR using lipofectamine 2000 (Invitrogen), selected in hygromycin (0.4 mg/ml), and single-cell cloned. MCF-7 cells that over-express human uPAR are previously described [20].
uPAR-specific siRNA smart pool and siCONTROL non-targeting control (NTC) siRNA pool were obtained from Dharmacon. siRNA transfection was performed with Lipofectamine 2000 in serum-free medium (SFM). siRNAs were introduced twice, at 24 and 72 h. Mouse uPAR mRNA expression was determined by qPCR and immunoblot analysis.
2.3. Real-time qPCR
Total RNA was isolated from cells in culture using the NucleoSpin RNA II kit (Machery-Nagel). cDNA was synthesized using the iScript cDNA synthesis kit (BioRad). qPCR was performed using a StepOnePlus instrument (Applied Biosystems) and a one-step program: 95° C, 20s; 95° C, 1s; and 60° C, 20s for 40 cycles. HPRT-1 gene expression was measured as a normalizer. Results were analyzed by the relative quantity (ΔΔCt) method. Experiments were performed in duplicate with internal triplicate determinations.
2.4. Analysis of cell-signaling
Cell extracts were prepared in radioimmune precipitation assay (RIPA) buffer (20 mM sodium phosphate, 150 mM NaCl, pH 7.4, 1% Triton, 0.1% SDS, 0.5% sodium deoxycholate) containing complete protease inhibitor mixture (Roche Applied Science) and 1 mM sodium orthovanadate. Protein concentrations were determined by bicinchoninic acid assay (Sigma-Aldrich). Equal amounts of cell extract were subjected to SDS-PAGE, electrotransferred to PVDF membranes, and probed with primary antibodies to detect phospho-ERK and total ERK.
In experiments in which cells were transfected with constructs encoding wild-type FAK, DNFAK, DN-Rac1, or DN-H-Ras, the construct encoding HA-ERK1 was introduced simultaneously using Lipofectamine 2000 or FugeneHD (Roche), so that ERK activation could be monitored exclusively in the transfected cells [33, 34]. HA-ERK1 also was introduced in transient transfection studies with the construct encoding mouse uPAR, human uPAR, or uPAR-W32A. Cell extracts were prepared and HA-ERK1 was immunoprecipitated from equal amounts of cell extracts (500 μg) using HA antibody-conjugated agarose beads (Sigma-Aldrich). Immunoprecipitates were subjected to immunoblot analysis and probed with antibodies specific for phospho-ERK and total ERK.
GTP-loaded Rac1 was determined by affinity-precipitation using PAK1-PBD, which recognizes only the GTP-bound forms of Rac1 and Cdc42, as previously described [21, 29]. Mouse uPAR-expressing and control MCF-7 cells were cultured in 10 cm plates for 16 h. Cultures were washed with ice-cold PBS and extracted in the supplied buffer supplemented with protease inhibitor cocktail and 1 mM sodium orthovanadate. The extracts were incubated with 15 μg of PAK1-PBD reagent for 45 min at 4°C. The glutathione-Sepharose was washed four times and then treated with SDS sample buffer to dissociate the PAK1-PBD and associated proteins. Immunoblot analysis was performed to detect Rac1. Samples of each cell extract also were subjected to immunoblot analysis before incubation with PAK1-PBD to determine total Rac1. Immunoblots were digitized and quantified using Quantity One 1-D Analysis Software (BioRad).
2.5. Flow cytometry
Cells (3 × 105) were plated in 6-well plates and co-transfected to express wild-type uPAR or uPAR-W32A. Cells were co-transfected with pEGFP to express green fluorescent protein (GFP). After 24 h, cells were suspended 2% bovine serum albumin (BSA) in PBS and then incubated with APC-conjugated uPAR-specific antibody (0.5 μg/1 × 105 cells) or isotype-matched IgG for 60 min at 4° C. Flow cytometry was performed using a FACSCanto II flow cytometer (BD Biosciences). Results were analyzed with FlowJo software.
2.6. Orthotopic xenografts
Animal experiments were performed in accordance with protocols approved by the University of California San Diego Animal Care Program. Anesthetized 8-week-old C.B-17/lcrCrl-scid-BR mice (Charles River Laboratories) were inoculated bilaterally in the fourth mammary fat pad with M3, M4 or control MCF-7 cells (1 × 106), which were transfected with empty vector (EV),suspended in 50 μl of Matrigel (Sigma). Primary tumor growth was monitored weekly. Ten weeks after tumor cell injection, the mice were euthanized and the mammary fat pads were visually inspected for tumor. Tumor formation was confirmed by histological analysis. Xeno-graft tumor volumes (vol) were calculated using the formula: Vol = (4/3)π × ((largest radius + smaller radius)/2)3. Data processing and statistical analysis were performed using GraphPad Prism (GraphPad Software, Inc.) and Microsoft Excel (Microsoft Corporation).
2.7. Histology and immunohistochemistry analysis of mouse tissues
Formalin-fixed tissue was paraffin-embedded. Serial 4 μm sections were stained with hematoxylin and eosin. Immunohistochemistry (IHC) was performed using the Vantana Discovery® XT System (Vantana). Sections were pretreated with citric acid buffer and then incubated with polyclonal antibody specific for mouse uPAR (1:200) or phospho-ERK followed by peroxidaseconjugated secondary antibody. Peroxidase activity was imaged using 3,3′-diaminobenzidine. Slides were examined using a Leica DM2500 light microscope. Images were acquired using Leica DFC420 digital camera and Leica Application Suite software.
3. Results
3.1.uPAR over-expression induces uPA-independent ERK activation
MCF-7 breast cancer cells express low levels of uPAR and undetectable levels of uPA [32]. ERK activation downstream of uPAR is entirely dependent on exogenously-added uPA [32-33]. To test whether the level of uPAR expression affects the mechanism by which uPAR activates ERK, we over-expressed human uPAR in MCF-7 cells and derived two cloned cell lines (H1 and H5). Fig. 1A shows that uPAR expression was substantially increased in the H1 and H5 cells. The level of phospho-ERK, observed in the absence of added uPA, also was increased in H1 and H5 cells, compared with the level observed in control (EV) cells that were transfected with empty vector.
Figure 1.
High levels of uPAR expression induce autonomous signaling to ERK. A, Cell extracts from H1, H5 and control/EV MCF-7 cells were subjected to immunoblot analysis to detect human uPAR, phospho-ERK (p-ERK), total ERK, and tubulin as a loading control. B, MCF-7 cells were transiently co-transfected to express human uPAR or empty vector (EV) and HA-ERK1. Cell extracts were immuneprecipitated with HA-specific antibody and subjected to immunoblot analysis to detect phospho-ERK and total ERK. C, Cell extracts from M3, M4, and control/EV MCF-7 cells were subjected to immunoblot analysis to detect mouse uPAR, phospho-ERK, total ERK, and tubulin. D, MCF-7 cells were transiently co-transfected to express HA-ERK1 and mouse uPAR or control vector (EV). Cell extracts were immuno-precipitated with HA-specific antibody and subjected to immunoblot analysis to detect phospho-ERK and total ERK. E, Mouse uPAR was silenced in M3, M4, and control/EV MCF-7 cells. Cell extracts were subjected to immunoblot analysis to detect mouse uPAR, phospho-ERK (pERK) and total ERK. F, Cell extracts from MDA-MB-231 cells and H5 cells were subjected to immunoblot analysis to detect human uPAR and total ERK.
To confirm that the increase in phospho-ERK was not an artifact resulting from single-cell cloning, we examined MCF-7 cells that were transiently transfected to over-express human uPAR. The cells were co-transfected to express HA-tagged ERK1, to permit analysis of ERK phosphorylation selectively in the transfected cells. Fig. 1B shows that HA-ERK1 activation was increased by uPAR over-expression, in the absence of exogenously added uPA.
In control qPCR and immunoblotting experiments, we confirmed that H1 and H5 cells do not express uPA, like the parental MCF-7 cells (Supplementary Fig. 1). Thus, our results suggested that uPAR over-expression in MCF-7 cells induces ERK activation autonomously of uPA. To further test this hypothesis, we transfected MCF-7 cells to express mouse uPAR. uPA-binding to uPAR is highly species-specific [21, 42, 43], precluding ligation of mouse uPAR by trace levels of human uPA, which may have been produced by the MCF-7 cells. As shown in Fig. 1C, ERK was activated, in the absence of exogenously added uPA, in two cloned cell lines that express mouse uPAR (M3 and M4). MCF-7 cells that were transiently transfected to express mouse uPAR and HA-ERK1 also demonstrated increased HA-ERK1 activation, in the absence of exogenously added uPA (Fig. 1D).
To confirm that the increase in ERK activation, observed when uPAR was over-expressed, was due to uPAR, we silenced uPAR gene expression in M3 and M4 cells. The extent of silencing was nearly complete, as determined by qPCR (Supplementary Fig. 2) and by immunoblot analysis (Fig. 1E). Phospho-ERK was decreased to the level observed in control MCF-7 cells when mouse uPAR expression was silenced with siRNA.
To estimate the extent of uPAR over-expression in our transfected cell lines, we compared the abundance of uPAR in H5 cells and wild-type MDA-MB 231 breast cancer cells. MDA-MB 231 cells are highly aggressive cancer cells that metastasize readily in animal model systems [44, 45]. uPAR signaling in MDA-MB 231 cells occurs independently of exogenously-added uPA [17]. By immunoblot analysis and densitometry, the level of uPAR in H5 cells was only 25% higher than that detected in MDA-MB 231 cells (Fig. 1F). Thus, the transformation in uPAR signaling mechanism, observed in transfected MCF-7 cells, reflects a level of uPAR that may be found naturally in breast cancer cells, especially when uPAR gene amplification occurs [9, 10].
3.2.UPAR regulates ERK activation only in the absence of E2
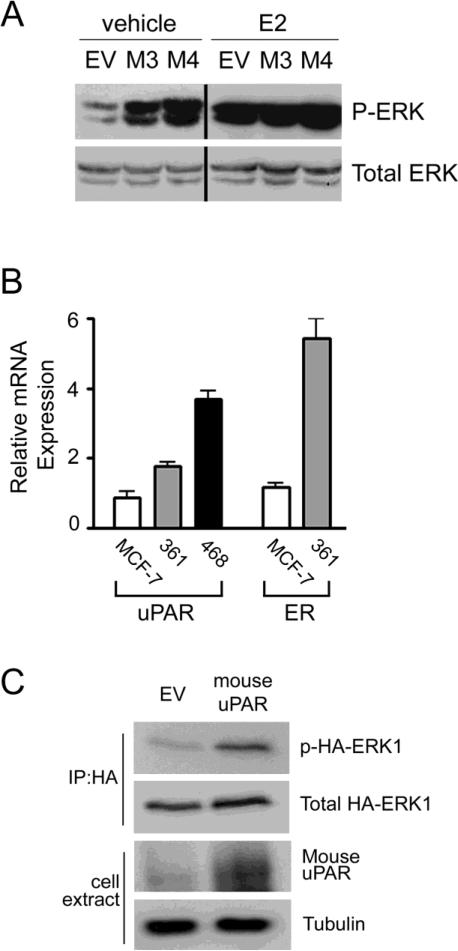
In the studies presented thus far, cells were cultured in SFM for 18 h before analyzing ERK activation. Limited ERα activation was possible due to phenol red in the medium [46]. In Fig. 2A, mouse uPAR-expressing and control MCF-7 cells were cultured for 18 h in SFM, in the presence or absence of E2 (20 nM). Although ERK activation was substantially increased in M3 and M4 cells in the absence of E2, the difference was neutralized by E2 supplementation. These results suggest that uPAR may control ERK activation in ERα-positive breast cancer cells, principally when E2 is absent or when drugs that inhibit the E2-ERα signaling system are introduced.
Figure 2.
Autonomous uPAR signaling in the presence and absence of E2. A, M3, M4, and EV/control MCF-7 cells were cultured in SFM for 18 h and treated with 20 nM E2 or vehicle in SFM for an additional 18 h. Cell extracts were subjected to immunoblot analysis to detect phospho-ERK (p-ERK) and total ERK. B, Relative mRNA expression was determined for uPAR and ERα in MCF-7 cells, MDA-MB-361 cells, and MDA-MB-468 cells (mean ± SEM, n=3). C, MDA-MB-361 cells were transiently co-transfected to express HA-ERK1 and mouse uPAR or control vector (EV). Cell extracts were immunoprecipitated with HA-specific antibody and subjected to immunoblot analysis to detect phospho-ERK (p-ERK) and total ERK. Total cell extracts were subjected to immunoblot analysis to detect mouse uPAR and tubulin.
To test whether uPAR over-expression activates ERK autonomously of uPA in a second ERα-positive breast cancer cell line, we studied MDA-MB 361 cells. These cells express more uPAR mRNA than MCF-7 cells but less than MDA-MB 468 cells (Fig. 2B). When MDA-MB 361 cells were transiently transfected to express mouse uPAR, the basal level of phospho-HA-ERK1 was increased (Fig. 2C). MDA-MB 361 cells express low levels of uPA (results not shown); however, because this is a human cell line, uPA that is produced endogenously should not bind significantly to mouse uPAR [21, 42, 43].
3.3. Common factors in the pathway by which uPAR activates ERK, autonomously and in response to uPA
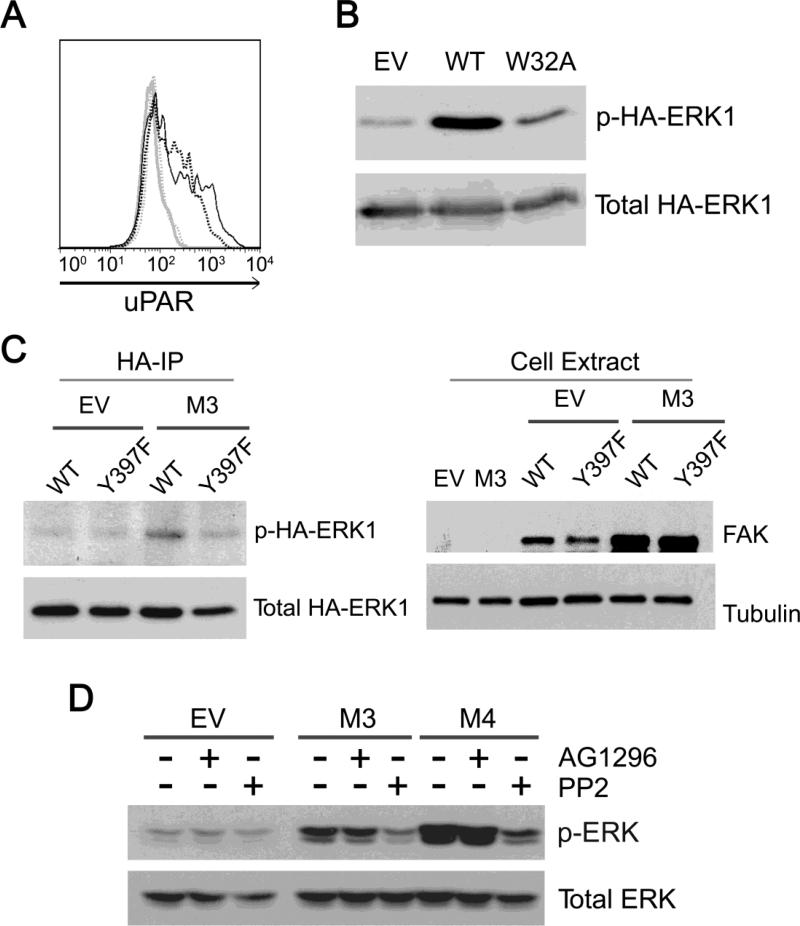
When cells are cultured in serum-containing medium, vitronectin is the major protein that coats tissue-culture plastic [47]. A single mutation in the structure of uPAR (W32A) blocks the interaction of uPAR with vitronectin [26]. To assess whether sustained ERK activation, resulting from uPAR over-expression, requires uPAR-binding to vitronectin, MCF-7 cells were transfected to express uPAR-W32A or wild-type uPAR and GFP. Flow cytometry studies were performed to detect cell-surface uPAR in the GFP-gated population. Fig. 3A shows that the level of cell-surface uPAR was similar in cells that expressed wild-type uPAR or uPAR-W32A.
Figure 3.
Molecular mechanisms of autonomous uPAR signaling to ERK. A, MCF-7 cells were transiently co-transfected to express GFP and wild-type uPAR (dark solid tracing), uPAR-W32A (dark broken tracing), or control vector (light solid tracing). Cell-surface uPAR expression was determined by flow cytometry. The isotype control is shown with the light broken tracing. B, MCF-7 cells were transiently co-transfected to express HA-ERK1 and wild-type uPAR, uPARW32A or control vector (EV). Cell extracts were immunoprecipitated with HA-specific antibody and subjected to immunoblot analysis for phospho-ERK (p-ERK) and total ERK. C, M3 and control EV cells were transiently co-transfected to express HA-ERK1 and DN-FAK or wild-type (WT) FAK. Cell extracts were immunoprecipitated with HA-specific antibody and subjected to immunoblot analysis. Total cellular extracts of M3 and EV cells, which were transfected with wild-type FAK or DN-FAK, and control (non-transfected) cells were subjected to immunoblot analysis to detect FAK and tubulin, as a loading control. D, M3, M4, and EV/ control MCF-7 cells were treated with the SFK inhibitor (PP2, 1 μM), the PDGF receptor inhibitor (AG1296, 10 μM), or vehicle for 18 h in SFM. Extracts were analyzed for phospho-ERK and total ERK.
Next, MCF-7 cells were transfected to express wild-type uPAR or uPAR-W32A and HA-ERK1. Fig. 3B shows that HA-ERK1 was phosphorylated in cells that express wild-type uPAR but not in cells that express uPAR-W32A. Thus, association of uPAR with vitronectin appears to be necessary for autonomous uPAR signaling to ERK.
SFKs and FAK are required for uPA-induced ERK activation in MCF-7 cells [33, 34]. SFKs also have been implicated in the uPA-independent pathway by which vitronectin-binding to uPAR leads to Rac1 activation [27, 28]. To test the role of FAK in autonomous uPAR signaling to ERK, we transfected M3 cells and control EV cells to express DN-FAK or wild-type FAK. Cells were co-transfected to express HA-ERK1. The level of phospho-HA-ERK1 was approximately equivalent in control EV cells that were transfected with wild-type FAK or DN-FAK (Fig. 3C). By contrast, in M3 cells, DN-FAK decreased the level of phospho-HA-ERK1, suggesting that FAK is necessary for autonomous ERK activation in these cells. In control experiments, we confirmed that wild-type FAK and DN-FAK were expressed at similar levels in both EV and M3 cells. With both constructs, the transfection efficiency was higher in M3 cells; however, the HA-ERK1 co-transfection procedure corrects for differences in transfection efficiency [33, 34].
The SFK-selective pharmacological inhibitor, PP2, also substantially decreased phospho-ERK in M3 and M4 cells, suggesting an important role for SFKs in autonomous uPAR signaling to ERK (Fig. 3D). As a control, we examined the PDGF receptor-selective tyrosine kinase inhibitor (TKI), AG1296. ERK phosphorylation in M3 or M4 cells was unchanged by AG1296.
3.4.Rac1 and H-Ras cooperate in autonomous uPAR signaling to ERK
uPAR over-expression activates Rac1 in multiple cell types [21, 27-29]. Fig. 4A shows that the level of GTP-loaded Rac1 was increased in both M3 and M4 cells, as anticipated. To test whether the increase in phospho-ERK occurs downstream of activated Rac1 in mouse uPAR-expressing MCF-7 cells, M3, M4, and EV cells were transfected to express DN-Rac1 [29] and HA-ERK1. DN-Rac1 did not significantly change the level of phospho-HA-ERK1 in EV cells (Fig. 4B). By contrast, in both M3 and M4 cells, DN-Rac1 decreased phospho-HA-ERK1, albeit incompletely. To confirm that DN-Rac1 was expressed in M3, M4 and EV cells, we performed immunoblotting experiments to detect total Rac1 and DN-Rac1, which is GFP-tagged. DN-Rac1 was detected in all three cell lines.
Figure 4.
Rac1 and H-Ras cooperate to induce autonomous uPAR signaling to ERK. A, M3, M4, and EV/control MCF-7 cells extracts were affinity precipitated with PAK-1 PBD and subjected to immunoblot analysis to detect GTP-bound Rac1. The original cell extracts were also subjected to immunoblot analysis to determine total Rac1 as a loading control. B, M3, M4, and EV MCF-7 cells were transiently co-transfected to express HA-ERK1 and DN-Rac1 or control vector. Extracts were immunoprecipitated with HA-specific antibody and subjected to immunoblot analysis to detect phospho-ERK (p-ERK) and total ERK. Total cellular extracts were subjected to immunoblot analysis to detect GFP-tagged DN-Rac1 and total (untagged) Rac1. C, M3, M4, and EV MCF-7 cells were transiently co-transfected to express HA-ERK1 and DN-H-Ras or control vector. Extracts were immunoprecipitated with HA-specific antibody and subjected to immunoblot analysis to detect phospho-ERK and total ERK. Ratios of phospho-HA-ERK1 to total-HA-ERK1 were determined by densitometry and are reported under the immunoblots.
To study the role of H-Ras in autonomous uPAR signaling to ERK, cells were co-transfected to express DN-H-Ras and HA-ERK1. DN-H-Ras is previously described [40]. In control EV cells, DN-H-Ras did not significantly affect phospho-HA-ERK1. By contrast, DN-H-Ras decreased phospho-HA-ERK1 in both M3 and M4 cells (Fig. 4C). Like DN-Rac1, the effects of DN-H-Ras were incomplete. These results support a model in which autonomous uPAR signaling to ERK reflects the activity of two separate pathways, involving H-Ras and Rac1, which converge at the level of ERK. Unfortunately, we were unable to simultaneously express DN-Rac1 and DN-HRas in MCF-7 cells, due to the decreased viability of dually transfected cells.
3.5. uPAR over-expression provides a selection advantage for MCF-7 cells in the absence of E2 in vitro and in vivo
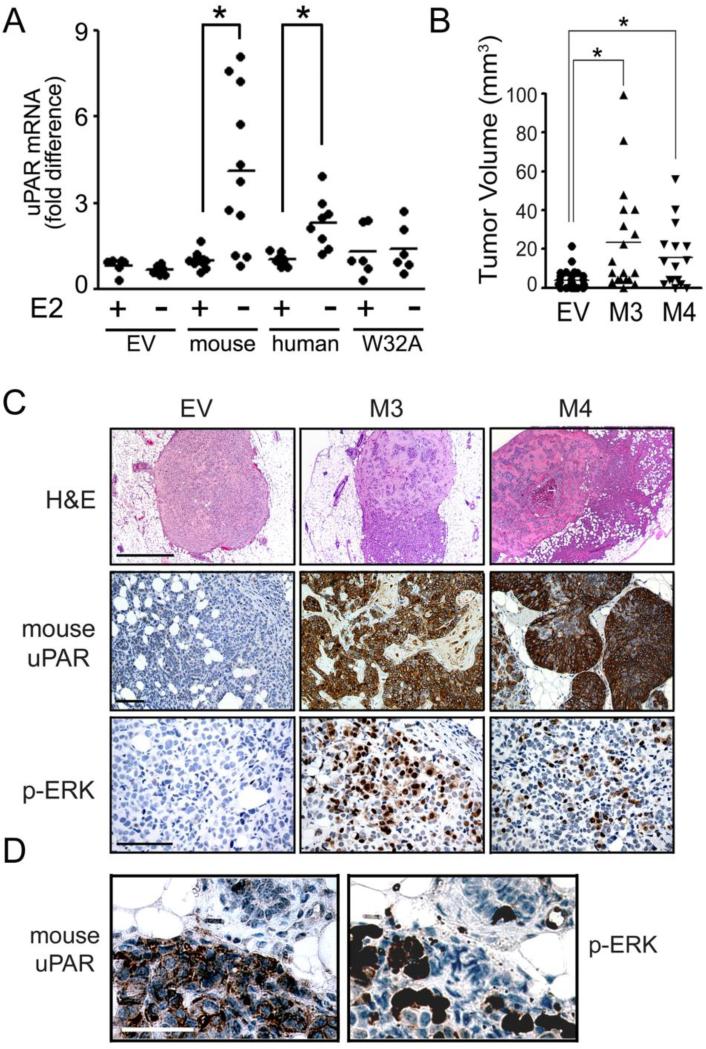
To test whether uPAR expression provides a survival advantage for ERα-positive cells when E2 is not available, we transfected MCF-7 cells to express human uPAR or mouse uPAR [20]. Instead of selecting the cells with antibiotics, cultures were maintained in medium that was supplemented with charcoal-treated serum (CTS) and E2 (20 nM) or vehicle for 2 weeks. As shown in Fig. 5A, cells that were E2-deprived demonstrated mouse uPAR mRNA levels that were increased about 4-fold, on average, compared with E2-treated cells (p<0.05, Student's t-test). In cells that were transfected to over-express human uPAR, E2 deprivation increased uPAR mRNA levels 2.5-fold (p<0.05). We interpret these results to indicate that, in the absence of E2, uPAR over-expression provides a growth/survival advantage and cells which express higher levels of uPAR are selectively recovered.
Figure 5.
E2 deficiency selects for uPAR-expressing MCF-7 cells in vitro and in orthotopic xenografts in vivo. A, MCF-7 cells were transfected to express mouse uPAR, human uPAR, uPARW32A, or empty vector (EV) and cultured in the presence or absence of 20 nM E2 for two weeks in phenol red-free DMEM supplemented with CTS. Relative uPAR mRNA expression was determined by qPCR (*, p<0.05, Student's t-test). B, SCID mice were injected with M3, M4 or control/EV MCF-7 cells. Tumor volume was determined after surgical resection (mean, *, p< 0.05, Mann-Whitney rank sum test). C, Images of tumors formed by M3, M4, and EV cells include representative H&E-stained sections of the tumors (first row; 5x, bar, 100 μm), IHC to detect mouse uPAR (second row; 20x, bar 10 μm) and phospho-ERK (third row; 40x, bar 10 μm). D, Adjacent sections of a representative tumor were stained to detect mouse uPAR and phospho-ERK (40x, bar, 10 μm).
We performed the equivalent experiment with cells that were transfected to express uPARW32A because this form of uPAR does not support cell-signaling. uPAR-W32A failed to provide a selection advantage when cells were deprived of E2 for 2 weeks (Fig. 5A). In additional control experiments, E2 deprivation did not significantly affect uPAR mRNA expression when MCF-7 cells were transfected with empty vector.
MCF-7 cells typically form tumors in SCID mice only when estrogen supplementation is provided [6, 46, 48]. We previously demonstrated that human uPAR over-expression in MCF-7 cells significantly increases the frequency of tumor formation and growth in the absence of estrogen supplementation [20]. The mechanism was not determined. In new studies, we inoculated mouse uPAR-expressing and control MCF-7 cells into mammary fat pads in SCID mice (106 cells/injection). Tumor formation and volume were assessed at 10 weeks.
M3 and M4 cells formed tumors that were significantly increased in volume (p<0.05), compared with the tumors formed by control cells (Fig. 5B). Representative H&E-stained sections of recovered tumors are shown in Fig. 5C. Mouse uPAR-expressing cells tended to invade outside the Matrigel capsule, as anticipated. In IHC studies, mouse uPAR was clearly detected in both M3 and M4 cells in vivo. The cancer cells in tumors formed by EV cells were mouse uPAR-immunonegative, as anticipated. We also performed IHC studies to detect phospho-ERK in vivo in tumors formed by EV, M3, and M4 cells. Foci of robustly phospho-ERK-positive cancer cells were abundant in tumors formed by M3 and M4 cells. Tumors formed by control EV cells were phospho-ERK negative at the level of sensitivity of the antibody. These results confirm that the increase in ERK phosphorylation, observed in M3 and M4 cells in vitro, is retained in vivo and may be responsible for the increase in tumor volume observed in the absence of E2 supplementation.
By staining adjacent sections from individual tumors, we showed that mouse uPAR- immuno-positive cells were frequently but not uniformly immunopositive for phospho-ERK (Fig. 5D). Factors that may have influenced whether cells were phospho-ERK-immunopositive include cell cycle phase and the cellular microenvironment.
3.6. EGFR TKIs block autonomous uPAR signaling to ERK
EGFR-specific TKIs are efficacious in the treatment of a number of cancers in which EGFR gene amplification and/or mutations are prevalent [49-52]. Recent studies suggest that EGFR TKIs may be useful in the treatment of breast cancer [53-55]. Because EGFR co-receptor activity has been implicated in the pathway by which uPA-binding to uPAR activates ERK [30, 35, 41], we studied the effects of two EGFR TKIs on ERK activation in uPAR over-expressing MCF-7 cells.
M3, M4, and EV cells were treated with Erlotinib or Gefitinib for 24 h. Fig. 6A shows that both TKIs almost entirely neutralized the increase in ERK activation in M3 cells. In M4 cells, the decrease in ERK activation was less complete, but still substantial. To further test the activity of EGFR TKIs in uPAR over-expressing MCF-7 cells, we applied a transient transfection strategy. MCF-7 cells were transfected to express mouse uPAR and HA-ERK1 and treated with Erlotinib or Gefitinib for 24 h. Fig 6B shows that the EGFR TKIs blocked the increase in HA-ERK1 associated with transient mouse uPAR expression.
Figure 6.
EGFR-specific TKIs block autonomous uPAR signaling in MCF-7 cells. A, M3, M4, and EV cells were treated with the EGFR inhibitors, Gefitinib (1 μM) and Erlotinib (1 μM), or with vehicle for 24 h in SFM. Extracts were analyzed for phospho-ERK and total ERK. B, MCF-7 cells were transiently co-transfected to express mouse uPAR or empty vector (EV) and HA-ERK1 and treated with Gefitinib (1 μM), Erlotinib (1 μM), or with vehicle for 24 h in SFM. Cell extracts were immunoprecipitated with HA-specific antibody and subjected to immunoblot analysis to detect phospho-ERK and total ERK. Ratios of phospho-ERK to total ERK or phospho-HA-ERK1 to total-HA-ERK1 were determined by densitometry and are reported under the immunoblots.
4. Discussion
The mechanism by which uPAR triggers cell-signaling remains incompletely understood. Coreceptors, including integrins and FPRL1, and transactivation pathways involving RTKs have been implicated [18]. The role of ligand-binding to uPAR in cell-signaling also is incompletely understood. The pathway that leads to activation of Rac1, downstream of p130Cas and DOCK180, is strictly dependent on uPAR-binding to vitronectin [27, 28]. Although the literature regarding activation of the H-Ras-ERK pathway is less clear, in MCF-7 cells, ERK activation requires uPA-binding to uPAR [33, 34].
In this study, we have shown that increased uPAR expression transforms the mechanism of ERK activation downstream of uPAR so that it occurs autonomously of uPA and is sustained. Given the central role of ERK in important cellular processes, including proliferation, survival, and cell migration [31], autonomous uPAR signaling to ERK has substantial potential to impact breast cancer cell physiology. The transition in uPAR-dependent cell-signaling from ligand (uPA)-dependent to -independent is analogous to the paradigm observed with ErbB2/HER2 in breast cancer cells [36]. The genes for ErbB2/HER2 and uPAR may be amplified in the same human breast cancer cells [9].
Unlike uPA-initiated ERK activation, which is entirely dependent on H-Ras [33], autonomous uPAR signaling to ERK apparently occurs downstream of Rac1 and H-Ras. Pathways by which Rac1 promote ERK activation are previously reported. For example, p21-activated kinase promotes ERK activation by increasing association of MEK1 with ERK [56].
One mechanism accounting for the transient nature of ERK activation, in uPA-treated cells, is MEK-dependent SOS phosphorylation, which promotes SOS dissociation from Grb2 and Shc [33, 57]. It is also possible that uPA-binding to uPAR triggers pulsatile cell-signaling events and that uPAR recycling is necessary for sustained cell-signaling [58]. Autonomous uPAR signaling to ERK was observed in cells that were cultured in SFM for 18 h. Thus, the level of activated ERK detected represents a steady-state. Compared with transient ERK activation, sustained ERK activation, observed with uPAR over-expression, is more likely to impact processes such as gene transcription and cell growth [58-62].
It has been proposed that uPAR-initiated cell-signaling requires uPAR dimerization or oligomerization [38-39]. If this model is correct, an explanation may be presented for the transformation in uPAR signaling mechanism observed here. When uPAR is present at low abundance, uPA is needed to promote uPAR dimerization or oligomerization [63]. However, with increased uPAR expression, oligomerization of uPAR in the absence of uPA should be favored, triggering autonomous signaling.
uPA and its inhibitor, PAI-1, have been implicated in development of resistance to the anti-estrogen drug, Tamoxifen, in breast cancer patients [64]. This is important because uPA and PAI-1 form a complex, which still binds to uPAR and induces sustained ERK activation unlike free uPA, which induces transient ERK activation [58]. As a result, uPA-PAI-1 complex may selectively promote cancer cell survival [58]. Autonomous uPAR signaling provides a selection advantage for MCF-7 cells, in the absence of uPA and PAI-1, in vitro and in vivo. When MCF-7 cells were transfected to express mouse or human uPAR and cultured in E2-deficient or -replete medium (no prior antibiotic selection), E2 deficiency selected for cells with higher levels of uPAR. These results may be explained by the ability of uPAR to promote ERK activation when E2 is absent.
The xenografting experiments performed in this study utilized MCF-7 cells that express mouse uPAR. The improvement in survival and growth of the MCF-7 cell orthotopic xenografts was similar to that previously observed when MCF-7 cells were transfected to over-express human uPAR [20]. Unlike mouse uPAR, human uPAR cannot bind uPA that is produced by non-malignant cells in the tumor microenvironment. Thus, the previously reported growth advantage of human uPAR over-expressing MCF-7 cells in vivo was unexplained. From the studies reported here, we now understand that uPA is not required for activation of the H-Ras-ERK pathway in human uPAR-over-expressing MCF-7 cells. Furthermore, for the first time in this study, we have demonstrated that the ability of uPAR to activate ERK in breast cancer cells is retained when the cells are implanted in mammary fat pads in vivo in mice. Autonomous uPAR signaling to ERK occurs in the microenvironment of a tumor.
To determine whether the level of uPAR expression in our transfected MCF-7 cells was substantially higher than what may be encountered in wild-type cells, we compared H5 cells and MDA-MB 231 cells. The uPAR protein level was only 25% higher in the H5 cells. We therefore conclude that the transformation in uPAR signaling mechanism described here may occur in breast cancer cells without genetic modification. Autonomous uPAR signaling may provide a pathway for breast cancer cell survival when estrogen is absent or in patients that are treated with drugs that antagonize ERα [2, 5-7]. uPAR is expressed at increased levels in hypoxia, which gradually develops as tumors enlarge [19]. Thus, intrinsic to the process of tumor growth may be a pathway for increased uPAR expression. uPAR gene amplification also may increase uPAR expression to a level that is sufficient for autonomous signaling to ERK.
Autonomous uPAR signaling was apparently dependent on EGFR co-receptor activity because the TKIs, Erlotinib and Gefitinib, inhibited ERK activation. Although EGFR co-receptor function has been observed in cells that are treated with uPA, the ability of uPA to induce ERK activation is not strictly dependent on the EGFR because responses are detected in EGFR-deficient cells [30, 35]. EGFR and uPAR also collaborate to promote activation of the mitogenic transcription factor, STAT5b [35, 41, 65]. Although EGFR inhibitors are not routinely used in breast cancer therapy, new studies suggest that these TKIs may be effective in cancers that relapse after treatment of ERα antagonists, such as Tamoxifen [53, 55]. By inhibiting autonomous uPAR signaling, EGFR TKIs may counteract the pro-survival advantage imparted by uPAR in ERα-positive cells, under estrogen deprivation conditions. Furthermore, the ability of Erlotinib and Gefitinib to inhibit autonomous uPAR signaling may explain why these drugs show efficacy in some patients with Tamoxifen-resistant breast cancer.
From these studies, we propose a model for uPAR signaling to ERK in which the uPAR concentration in the plasma membrane is critical. As the uPAR expression level increases, for example with increasing tumor hypoxia, a transformation in the mechanism of uPAR signaling may occur, triggering autonomous and sustained cell-signaling to ERK in the absence of uPA. Rac1, H-Ras, and the EGFR cooperate to induce these changes. Further work will be required to determine the effects of uPAR over-expression on other signaling pathways known to be activated downstream of uPAR.
Conclusions
We demonstrate that the requirement for uPA to initiate cell-signaling downstream of uPAR, in ERα-expressing breast cancer cells, depends on the uPAR expression level. At high expression levels, uPAR signals autonomously to ERK and this pathway provides a selection advantage for breast cancer cells in the absence of estrogen. Autonomous uPAR signaling to ERK occurs downstream of H-Ras and Rac1, unlike uPA-initiated cell-signaling, which occurs downstream of H-Ras alone. The EGFR-selective TKIs, Erlotinib and Gefitinib, inhibit autonomous uPAR signaling.
Supplementary Material
Supplementary Figure 1. uPAR over-expression does not increase uPA expression in MCF-7 cells. A, Relative uPA mRNA expression was determined in human uPAR over-expressing H1 and H5 cells and in control EV cells (mean ± SEM, n=3). B, Conditioned SFM was collected from cultures of EV, H5, and MDA-MB-231 cells at equivalent confluency and concentrated 10-fold. The conditioned medium samples were subjected to immunoblot analysis to detect uPA. Cell extracts were subjected to immunoblot analysis to detect tubulin.
Supplementary Figure 2. Efficiency of mouse uPAR gene-silencing. Mouse uPAR was silenced in M3, M4, and control/EV MCF-7 cells. Relative mouse uPAR mRNA expression was determined by qPCR (mean ± SEM, n=3).
Highlights.
Increased uPAR expression transforms signaling to ERK, so that it is autonomous of uPA
Autonomous uPAR Signaling is H-Ras and Rac1-dependent
EGFR is an important co-receptor for autonomous uPAR signaling
High uPAR expression provides a selection advantage for ERA+ breast cancer cells
Acknowledgements
This work is supported by NIH grant R01 CA94900
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The abbreviations used are: DN-FAK, dominant-negative FAK; DN-Rac1, dominant-negative Rac1; DN-Ras, dominant-negative Ras; E2, 17β-estradiol; EGFR, epidermal growth factor receptor; ERα, estrogen receptor-α; IHC, immunohistochemistry; SFK, Src family kinase; SFM, serum-free medium; uPA, urokinase-type plasminogen activator; uPAR, urokinase-type plasminogen activator receptor.
Authorship
BME, MJ, and SLG designed research; BME, MJ, ST, and DLW performed research; BME, MJ, and SLG analyzed data; BME, MJ, and SLG wrote the paper. All authors have read and approved the final manuscript.
Disclosure statement
The authors have no conflict of interest
References
- 1.Allred DC, Brown P, Medina D. Breast Cancer Res. 2004;6:240–245. doi: 10.1186/bcr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massarweh S, Schiff R. Endocr Relat Cancer. 2006;(Suppl 1):S15–24. doi: 10.1677/erc.1.01273. [DOI] [PubMed] [Google Scholar]
- 3.Johnston SR, Dowsett M. Nat Rev Cancer. 2003;3:821–831. doi: 10.1038/nrc1211. [DOI] [PubMed] [Google Scholar]
- 4.Riggs BL, Hartmann LC. N Engl J Med. 2003;348:618–629. doi: 10.1056/NEJMra022219. [DOI] [PubMed] [Google Scholar]
- 5.Fagan DH, Yee D. J Mammary Gland Biol Neoplasia. 2008;13:423–429. doi: 10.1007/s10911-008-9098-0. [DOI] [PubMed] [Google Scholar]
- 6.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, Weiss H, Rimawi M, Schiff R. Cancer Res. 2008;68:826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 7.Johnston SR. Clin Cancer Res. 2010;16:1979–1987. doi: 10.1158/1078-0432.CCR-09-1823. [DOI] [PubMed] [Google Scholar]
- 8.Fox EM, Andrade J, Shupnik MA. Steroids. 2009;74:622–627. doi: 10.1016/j.steroids.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng S, Tripathy D, Shete S, Ashfaq R, Saboorian H, Haley B, Frenkel E, Euhus D, Leitch M, Osborne C, Clifford E, Perkins S, Beitsch P, Khan A, Morrison L, Herlyn D, Terstappen LW, Lane N, Wang J, Uhr J. Proc Natl Acad Sci U S A. 2006;103:17361–17365. doi: 10.1073/pnas.0608113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildenbrand R, Niedergethmann M, Marx A, Belharazem D, Allgayer H, Schleger C, Strobel P. Am J Pathol. 2009;174:2246–2253. doi: 10.2353/ajpath.2009.080785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JL, Seetoo D, Wang Y, Ranson M, Berney CR, Ham JM, Russell PJ, Crowe PJ. Int J Cancer. 2000;89:431–439. doi: 10.1002/1097-0215(20000920)89:5<431::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 12.Memarzadeh S, Kozak KR, Chang L, Natarajan S, Shintaku P, Reddy ST, Farias-Eisner R. Proc Natl Acad Sci U S A. 2002;99:10647–10652. doi: 10.1073/pnas.152127499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, Gao Y, Ji Z, Zhang X, Xu Q, Li G, Guo Z, Zheng B, Guo X. J Pediatr Surg. 2004;39:1512–1519. doi: 10.1016/j.jpedsurg.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 14.de Bock CE, Wang Y. Med Res Rev. 2004;24:13–39. doi: 10.1002/med.10054. [DOI] [PubMed] [Google Scholar]
- 15.Mazar AP. Clin Cancer Res. 2008;14:5649–5655. doi: 10.1158/1078-0432.CCR-07-4863. [DOI] [PubMed] [Google Scholar]
- 16.Aguirre-Ghiso JA, Kovalski K, Ossowski L. J Cell Biol. 1999;147:89–104. doi: 10.1083/jcb.147.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Z, Webb DJ, Jo M, Gonias SL. J Cell Sci. 2001;114:3387–3396. doi: 10.1242/jcs.114.18.3387. [DOI] [PubMed] [Google Scholar]
- 18.Blasi F, Carmeliet P. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 19.Lester RD, Jo M, Montel V, Takimoto S, Gonias SL. J Cell Biol. 2007;178:425–436. doi: 10.1083/jcb.200701092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo M, Eastman BM, Webb DL, Stoletov K, Klemke R, Gonias SL. Cancer Res. 2010;70:8948–8958. doi: 10.1158/0008-5472.CAN-10-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo M, Takimoto S, Montel V, Gonias SL. Am J Pathol. 2009;175:190–200. doi: 10.2353/ajpath.2009.081053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Y, Waltz DA, Rao N, Drummond RJ, Rosenberg S, Chapman HA. J Biol Chem. 1994;269:32380–32388. [PubMed] [Google Scholar]
- 23.Sidenius N, Blasi F. FEBS Lett. 2000;470:40–46. doi: 10.1016/s0014-5793(00)01282-5. [DOI] [PubMed] [Google Scholar]
- 24.Liang OD, Chavakis T, Kanse SM, Preissner KT. J Biol Chem. 2001;276:28946–28953. doi: 10.1074/jbc.m011347200. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Lawrence DA, Zhang L. J Biol Chem. 2003;278:29925–29932. doi: 10.1074/jbc.M300751200. [DOI] [PubMed] [Google Scholar]
- 26.Madsen CD, Ferraris GM, Andolfo A, Cunningham O, Sidenius N. J Cell Biol. 2007;177:927–939. doi: 10.1083/jcb.200612058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjoller L, Hall A. J Cell Biol. 2001;152:1145–1157. doi: 10.1083/jcb.152.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith HW, Marra P, Marshall CJ. J Cell Biol. 2008;182:777–790. doi: 10.1083/jcb.200712050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Z, Thomas KS, Webb DJ, Moravec R, Salicioni AM, Mars WM, Gonias SL. J Cell Biol. 2002;159:1061–1070. doi: 10.1083/jcb.200207070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo M, Thomas KS, O'Donnell DM, Gonias SL. J Biol Chem. 2003;278:1642–1646. doi: 10.1074/jbc.M210877200. [DOI] [PubMed] [Google Scholar]
- 31.Roberts PJ, Der CJ. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen DH, Hussaini IM, Gonias SL. J Biol Chem. 1998;273:8502–8507. doi: 10.1074/jbc.273.14.8502. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen DH, Catling AD, Webb DJ, Sankovic M, Walker LA, Somlyo AV, Weber MJ, Gonias SL. J Cell Biol. 1999;146:149–164. doi: 10.1083/jcb.146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen DH, Webb DJ, Catling AD, Song Q, Dhakephalkar A, Weber MJ, Ravichandran KS, Gonias SL. J Biol Chem. 2000;275:19382–19388. doi: 10.1074/jbc.M909575199. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, Aguirre Ghiso J, Estrada Y, Ossowski L. Cancer Cell. 2002;1:445–457. doi: 10.1016/s1535-6108(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 36.Brennan PJ, Kumagai T, Berezov A, Murali R, Greene MI. Oncogene. 2000;19:6093–6101. doi: 10.1038/sj.onc.1203967. [DOI] [PubMed] [Google Scholar]
- 37.Lemmon MA, Schlessinger J. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunningham O, Andolfo A, Santovito ML, Iuzzolino L, Blasi F, Sidenius N. EMBO J. 2003;22:5994–6003. doi: 10.1093/emboj/cdg588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caiolfa VR, Zamai M, Malengo G, Andolfo A, Madsen CD, Sutin J, Digman MA, Gratton E, Blasi F, Sidenius N. J Cell Biol. 2007;179:1067–1082. doi: 10.1083/jcb.200702151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai H, Szeberényi J, Cooper GM. Mol Cell Biol. 1990;10:5314–5323. doi: 10.1128/mcb.10.10.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jo M, Thomas KS, Takimoto S, Gaultier A, Hsieh EH, Lester RD, Gonias SL. Oncogene. 2007;26:2585–2594. doi: 10.1038/sj.onc.1210066. [DOI] [PubMed] [Google Scholar]
- 42.Estreicher A, Wohlwend A, Belin D, Schleuning WD, Vassalli JD. J. Biol. Chem. 1989;264:1180–1189. [PubMed] [Google Scholar]
- 43.Lin L, Gårdsvoll H, Huai Q, Huang M, Ploug M. J Biol Chem. 2010;285:10982–10992. doi: 10.1074/jbc.M109.093492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 45.Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verástegui E, Zlotnik A. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 46.Welshons WV, Wolf MF, Murphy CS, Jordan VC. Mol Cell Endocrinol. 1988;57:169–178. doi: 10.1016/0303-7207(88)90072-x. [DOI] [PubMed] [Google Scholar]
- 47.Hayman EG, Pierschbacher MD, Ohgren Y, Ruoslahti E. Proc Natl Acad Sci. 1983;80:4003–4007. doi: 10.1073/pnas.80.13.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noël AC, Callé A, Emonard HP, Nusgens BV, Simar L, Foidart J, Lapiere CM, Foidart JM. Cancer Res. 1991;51:405–414. [PubMed] [Google Scholar]
- 49.Pao W, Chmielecki J. Nature Reviews Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van den Eynde M, Baurain JF, Mazzeo F, Machiels JP. Acta Clin Belg. 2011;66:10–17. doi: 10.2143/ACB.66.1.2062508. [DOI] [PubMed] [Google Scholar]
- 51.Voelzke WR, Petty WJ, Lesser GJ. Curr Treat Options Oncol. 2008;9:23–31. doi: 10.1007/s11864-008-0053-5. [DOI] [PubMed] [Google Scholar]
- 52.Ciardiello F, Tortora G. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 53.Osborne CK, Neven P, Dirix LY, Mackey JR, Robert J, Underhill C, Schiff R, Gutierrez C, Migliaccio I, Anagnostou VK, Rimm DL, Magill P, Sellers M. Clin Cancer Res. 2011;17:1147–1159. doi: 10.1158/1078-0432.CCR-10-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Massarweh S, Tham YL, Huang J, Sexton K, Weiss H, Tsimelzon A, Beyer A, Rimawi M, Cai WY, Hilsenbeck S, Fuqua S, Elledge R. Breast Cancer Res Treat. 2011;129:819–827. doi: 10.1007/s10549-011-1679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutteridge E, Agrawal A, Nicholson R, Leung Cheung K, Robertson J, Gee J. Int J Cancer. 2010;126:1806–1816. doi: 10.1002/ijc.24884. [DOI] [PubMed] [Google Scholar]
- 56.Eblen ST, Slack JK, Weber MJ, Catling AD. Mol Cell Biol. 2002;22:6023–6033. doi: 10.1128/MCB.22.17.6023-6033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langlois WJ, Sasaoka T, Saltiel AR, Olefsky JM. J Biol Chem. 1995;270:25320–25323. doi: 10.1074/jbc.270.43.25320. [DOI] [PubMed] [Google Scholar]
- 58.Webb DJ, Thomas KS, Gonias SL. J Cell Biol. 2001;152:741–752. doi: 10.1083/jcb.152.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meloche S, Seuwen K, Pagès G, Pouysségur J. Mol Endocrinol. 1992;6:845–854. doi: 10.1210/mend.6.5.1603090. [DOI] [PubMed] [Google Scholar]
- 60.Lenormand P, Sardet C, Pages G, L'Allemain G, Brunet A, Pouyssegur J. J. Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sergeant N, Lyon M, Rudland PS, Fernig DG, Delehedde M. J Biol Chem. 2000;275:17094–17099. doi: 10.1074/jbc.M000237200. [DOI] [PubMed] [Google Scholar]
- 62.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Nat Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 63.Sidenius N, Andolfo A, Fesce R, Blasi F. J Biol Chem. 2002;277:27982–27990. doi: 10.1074/jbc.M111736200. [DOI] [PubMed] [Google Scholar]
- 64.Meijer-van Gelder ME, Look MP, Peters HA, Schmitt M, Brünner N, Harbeck N, Klijn JGM, Foekens JA. Cancer Res. 2004;64:4563–4568. doi: 10.1158/0008-5472.CAN-03-3848. [DOI] [PubMed] [Google Scholar]
- 65.Hu J, Jo M, Cavenee WK, Furnari F, Vandenberg SR, Gonias SL. Proc Natl Acad Sci U S A. 2011;108:15984–15989. doi: 10.1073/pnas.1113416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. uPAR over-expression does not increase uPA expression in MCF-7 cells. A, Relative uPA mRNA expression was determined in human uPAR over-expressing H1 and H5 cells and in control EV cells (mean ± SEM, n=3). B, Conditioned SFM was collected from cultures of EV, H5, and MDA-MB-231 cells at equivalent confluency and concentrated 10-fold. The conditioned medium samples were subjected to immunoblot analysis to detect uPA. Cell extracts were subjected to immunoblot analysis to detect tubulin.
Supplementary Figure 2. Efficiency of mouse uPAR gene-silencing. Mouse uPAR was silenced in M3, M4, and control/EV MCF-7 cells. Relative mouse uPAR mRNA expression was determined by qPCR (mean ± SEM, n=3).