Abstract
In recent years, advances in technology have provided us with tools to quantify the expression of multiple genes in individual cells. The ability to simultaneously measure multiple genes on the same cell is necessary to resolve the incredible diversity of cell subsets, as well as to define their function in the host. Fluorescence-based flow cytometry is the benchmark for this; with it, we can quantify 18 proteins per cell, at >10,000 cells per second. “Mass cytometry” is a new technology that promises to significantly extend these capabilities. Immunophenotyping by mass spectrometry provides the ability to measure more than three dozen proteins at a rate of 1,000 cells per second. We review these cytometric technologies, capable of high-content, high-throughput single-cell assays.
Keywords: Fluorescence, Inductively Coupled Plasma Mass Spectrometry, Single Cell Analysis, Immunophenotyping, Data Analysis
The Case for Deep Profiling
To understand the biological actions of cells, and their mechanisms of differentiation, we must understand how phenotype and function are structured across diverse cell types and tissues. This structure can be perturbed by innate or infectious sources, which may drive disease pathogenesis; therefore, understanding it is critical for identifying treatments and preventions. Incredible cellular diversity underlies this organization, so measurements taken at the single-cell level that encompass RNA, protein, and glycan species (“high content”) across a large number of cells (“high throughput”) will greatly aid our formulation of a more comprehensive understanding. In many respects this is walking the path previously trod by genomics and proteomics—long accustomed to thinking about many events per experiment. However, traditional single-cell analysis has focused on many cells and a few parameters per experiment. As we delve into more complex cellular systems, such as cellular signaling networks or T-cell functional responses, we must reorient this thinking to consider many parameters on many cells – in essence, “deep profiling” every single cell from a representative population of cells.
Among well-established technologies for cellular analysis, flow cytometry is unique for its ability to rapidly interrogate multiple biologic signatures (protein epitopes, nucleic acids, ion concentrations) simultaneously within a single cell. Over the last 40 years, since the introduction of the first fluorescence-based flow cytometers, the maximum number of proteins that can be simultaneously measured has progressively increased. These advances can be attributed to parallel achievements in hardware, fluorochromes, and data analysis, and has led to state-of-the-art 20-parameter flow cytometers. Concomitant with this development, our understanding of immunology and stem cell biology has matured tremendously with the discovery of scores of functionally diverse cell populations. Here we review the development and highlight applications of polychromatic flow cytometry (PFC, 6+ colors). In addition, we review recent advances in a next-generation, “post-fluorescence” single-cell technology termed mass cytometry, which is theoretically capable of measuring 70–100 parameters. Both fluorescence and mass cytometry have unique and powerful features, as well as unique challenges and limitations. Over the next decade, these complementary technologies will play central roles in dissecting the complex interactions of cells.
The Polychromatic Era
Technical Achievements that Led to Polychromatic Flow Cytometry
The development of polychromatic flow cytometry required multiple stepwise advancements in hardware and reagents. For example, the earliest fluorescence-based cytometers used arc lamps, developed originally for microscopy, emitting light at a broad spectrum of wavelengths[1]. Because this light interfered with fluorochrome-derived signals, arc-lamps were not easily used for multi-color detection. By 1974, in the Herzenberg laboratory at Stanford, argon lasers, emitting a single wavelength (488 nm) were used as excitation sources for fluorescein[2]. The high power of these lasers dramatically increased sensitivity, allowing resolution of weakly fluorescent signals[3]. Two-color fluorescence detection, using fluorescein and rhodamine dyes, was followed by adding krypton lasers in the 1970s[4]. Over time, these expensive water-cooled lasers have been replaced with HeNe lasers[5], and eventually solid-state lasers of multiple lines. Such lasers were ideal for excitation of an important new class of fluorochromes made of phycobiliproteins, including phycoerythrin (PE) and allophycocyanin (APC)[6]. The recent use of high-powered lasers specifically tuned to excited PE and APC were critical to successful PFC, for which sensitivity is a major hurdle[7].
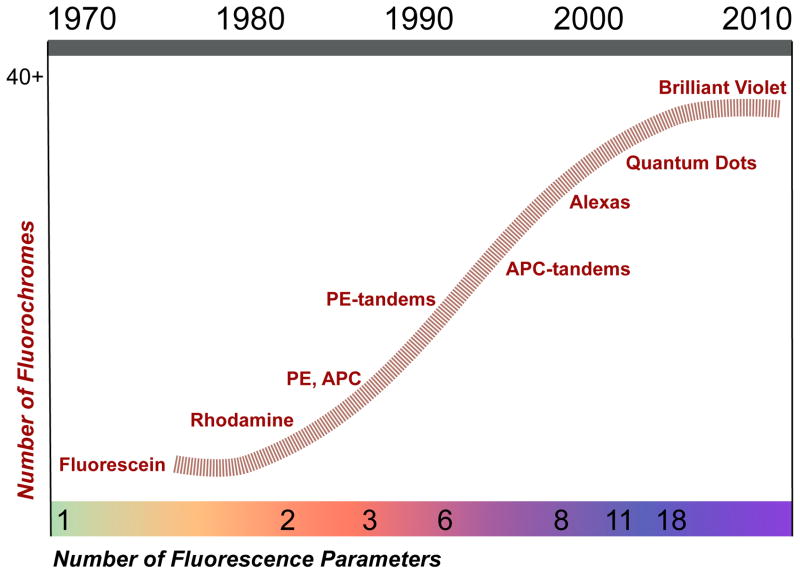
Generally, these engineering achievements slightly predated the introduction of new organic and inorganic fluorochromes. In the late 1980s (Figure 1), the remarkable ability of PE to absorb and transfer energy to other fluorescent molecules was recognized and exploited in order to produce an array of tandem dyes (e.g., PE-Texas Red, PE-Cy5, PE-Cy5.5, PE-Cy7)[8, 9]. In the 1990s, APC-based tandem dyes were synthesized[9], and a large spectrally-resolved series of small organic dyes (known as the Alexa dyes) became commercially available[10]. With this arsenal of lasers and fluorochromes, PFC graduated through 8 (1998) to 11 (2001) colors[11, 12]. During this period, violet (405 nm) lasers became available; however, there were few useful violet-excitable fluorochromes for immunophenotyping. This changed with the introduction of a series of fluorescent, inorganic semiconductor nanocrystals (called Quantum Dots) in 2004, and led to the current state-of-the-art in PFC 18-color cytometry[13]. Recently, additional violet-excitable fluorochromes were developed based on the Nobel-prize winning discovery of organic polymers that conduct electrons[14]. These dyes and their resonance-energy tandems provide additional, bright options at a variety of wavelengths – for many experiments, they are biochemically more suitable for immunophenotyping than quantum dots[15]; however, they do not increase the number of unique fluorescence parameters that can be measured, and thus 18 colors remains the current maximum.
Figure 1. History of Fluorochrome Development.
A timeline showing when the major fluorochromes were introduced, and how this related to the maximum number parameters that could be simultaneously measured at that time.
It is noteworthy that development of 18-color flow cytometry did not arise solely from achievements in laser and fluorochrome technology. Engineering advances in the optics (optimal light collection and delivery to detectors) and signal processing (digital electronics) were important, and continue to be areas of active development. In addition, software was developed to rapidly process the raw data and correct fluorescence spillover between dyes (a process known as “compensation”). Similarly, tools to display and analyze 20-parameter data, and aggregate complex analyses across hundreds or thousands of specimens have been developed.
In summary, development of PFC required improvements in nearly every component of flow cytometry technology, including chemistry (dye development), hardware, and software. All areas are still targeted for continued improvement.
Applications of PFC
Technical advances in flow cytometry have come hand-in-hand with a deeper understanding of hematopoietic cell types and function. The earliest cytometers resolved major cell lineages, such as T- and B-cells, allowing characterization of cell-mediated and humoral immunity[16, 17]. Later, with the development of monoclonal antibodies in the 1970s[18] and the emergence of the HIV epidemic in the 1980s[19], three-color measurements of T-cell subsets (CD4+ helper and CD8+ cytotoxic) became important clinical research and diagnostic tools. Similarly, leukemia and lymphoma typing and staging have become reliant on flow cytometry – first as two-color panels, but now using 8–10 for better fidelity[20, 21]. In the 1990s, with the availability of additional fluorochromes, naïve and various memory T-cell subsets were distinguished[22], and cytokine production was quantified[23] More recently, advances in PFC instrumentation provide unprecedented resolution of immune system cells, e.g. regulatory T-cells[24], follicular helper T-cells[25], and T helper 17 cells[26], to name a few. The availability of increasingly complex flow cytometry technology drove a progressively more detailed understanding of immune cell subsets and functions.
A recent testament to the utility of 15-color experiments is the identification of memory T-cells with stem-like properties[27]. These “TSCM” are phenotypically identical to naïve cells with respect to a number of markers (CD45RA+, CD45RO−, CCR7+, CD62L+, CD27+, CD28+, and CD127+), but the cells express mildly higher levels of CD95 and IL2Rb; identification and characterization was only possible by the simultaneous measurement of a dozen cell-surface markers. TSCM have enormous proliferative capacity, can reconstitute immunodeficient hosts, and mediate anti-tumor responses in a humanized mouse model[27]. These properties suggest that TSCM are important in the maintenance of immunological memory.
Indeed, the utility of advanced PFC (15+ colors) is apparent in a variety of other biological settings; however, designing the complex staining panels required is difficult[28]. And yet, there is a demand for even more measurements to be performed on a cell-by-cell basis – e.g. the need to characterize the expression of multiple chemokine receptors on TSCM will require adding those markers to an already complex 15-color panel. Similarly, the use of barcoding schemes[29], which distinguishes cells from different samples or stimulation conditions in a mixture based labeling with unique fluorescence identifiers, is important for multiplexed analysis of cell signaling profiles in leukocytes, leukemias and lymphomas, but requires at least 2–3 additional parameters.
The need for a higher level of multi-parametric analysis of single cells cannot currently be met with fluorescence technologies because of the limitation of the number of spectrally resolvable fluorochromes. This creates an opportunity for new technologies to complement PFC for cellular analysis.
The Post-Fluorescence Era: Mass Cytometry
A new platform has been developed that couples flow cytometry with mass spectrometry. This technology, known as mass cytometry, offers single-cell analysis of at least 45 simultaneous parameters without fluorescent agents or interference from spectral overlap (Figure 2). For this, stable (non-radioactive) isotopes of non-biological, rare earth metals are employed as reporters. By exploiting the resolution, sensitivity, and dynamic range of mass spectrometry on a time-scale that allows the measurement of 1000 individual cells per second, this configuration offers a new approach to high content cytometric analysis.
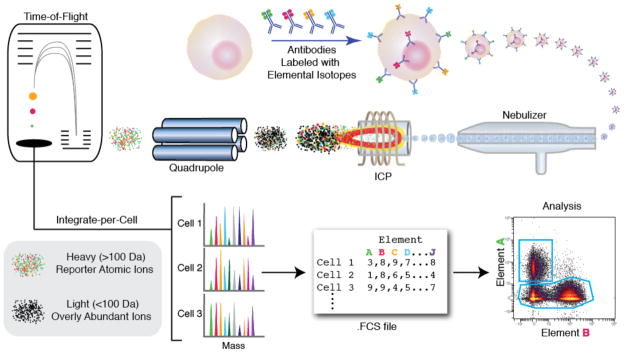
Figure 2. Mass cytometry allows single-cell atomic mass spectrometry of heavy elemental (> 100 Da) reporters.
Schematic of ICP-MS-based analysis of cellular markers. An affinity product (e.g., antibody) tagged with a specific element binds to the cellular epitope. The cell is introduced into the ICP by droplet nebulization. Each cell is atomized, ionized, overly abundant ions removed, and the elemental composition of remaining heavy elements (reporters) is determined. Signals corresponding to each elemental tag are then correlated with the presence of the respective marker and analyzed using conventional cytometry platforms.
Elemental Mass Spectrometry
Inductively coupled plasma mass spectrometry (ICP-MS) is the most advanced and sensitive means of determining the elemental composition of materials[30]. Classically, it has been used for ultra-trace (10−15 g per mL) detection of metals and other elements in both environmental (waters, soils, air) and clinical (blood, urine) samples. The central component of this system is a high-temperature plasma (~7000 K), which vaporizes the sample, breaks all molecular bonds, and strips one electron from each atom. This creates a cloud of elemental ions; after introduction into an ICP-MS, so that the relative abundance of elemental isotope can be determined. The ability to simultaneously detect and quantify trace-levels of non-biological elements from complex matrices makes ICP-MS an ideal detection tool for bioanalysis[31–38].
Mass Cytometry
Mass cytometry is the adaptation of ICP-MS to single-cell analysis[39], based on the concept that a purified single isotope could be used to tag antibodies and that these conjugates could be quantified in an ICP-MS detection system. Mass cytometry has essentially the same workflow as conventional flow cytometry (Figure 2). Cells are stained with target-specific antibodies labeled with metal isotopes (typically lanthanide metals)[37, 40]; these are the same antibody clones used in conventional cytometry. Cells are also stained with Rh- or Ir- conjugated DNA intercalators, providing a baseline for detection and information about DNA content[33]. The use of differential intercalator staining[36] as well as chemical labeling with chelated metals[41, 42] provides a viability measure as well. In the instrument, stained cells are nebulized into single-cell droplets and introduced into the plasma. The resulting charged atomic ion clouds are immediately transferred into the high vacuum of the mass spectrometer.
Because all cellular material is ionized, atomic ions are produced from elements common in cells (such as carbon, nitrogen, oxygen) along with ions from the argon plasma itself. To resolve the probe ions (e.g. lanthanides) from these overly abundant ions, the mass cytometer is configured as a quadrupole-time-of-flight (qTOF) instrument[30]. The quadrupole acts as a filter allowing only the heavier elemental ions, which consist primarily of the reporter ‘masses’, to be quantitated by TOF mass analysis.
For a typical cell, the ion cloud has a lifetime of ~300 μs over which it is measured (scanned) 20–30 times by TOF-MS. This lifetime precludes analysis of more than 1000 cells per second, which would result in the inability to resolve single cells. Since the elemental reporters used are uncommon in a biological context or laboratory environment, there is little measurable signal (background) observed between each cell. The amount of each isotopic reporter is quantified for each cell’s ion cloud by integrating across all scans for that cell; resulting summary data is recorded in an FCS file format, so that it can be visualized with conventional flow cytometry software. Originally developed at the University of Toronto[39], the first commercial version of the mass cytometer, called the CyTOF, as well as associated reagents, are produced and distributed by DVS Sciences (www.dvssciences.com).
Comparisons of Mass Cytometry vs. PFC
Although mass cytometry offers a number of unique features compared to PFC, the technology is relatively new and encompasses unique hurdles. PFC has the unique capability to work on live cells, and to be able to viably recover analyzed cells. Beyond this obvious difference, the two technologies are complementary – while there is overlap, each is well-suited to addressing a particular set of questions. Below, the features of each technology are compared and summarized in Table 1.
Table 1.
Comparison of utility and performance of state of the art commercial fluorescence flow cytometry and mass cytometry single-cell analysis platforms.
| Technology | Fluorescence Flow Cytometry | Mass Cytometry | |
|---|---|---|---|
|
| |||
| Measurement basis | Fluorescent probes | Stable mass isotope probes | |
|
| |||
|
Experimental design
| |||
| Max # of measurements | 20 (18 fluorescence) | 37 (including DNA) | |
|
| |||
| Theoretical # of subsets* | 2.6 × 105 | 1.4 × 1011 | |
|
| |||
| Panel design complexity (# of probes) | Easy: | <8 | 37 |
| Moderate: | 8–12 | ||
| Hard: | 12–18 | ||
|
| |||
| Sensitivity range for different probes** | 0.1 – 10 | 1 – 2 | |
|
| |||
|
Sample Throughput
| |||
| Sampling Efficiency | > 95% | < 30% | |
|
| |||
| Measured Cells/second | 25,000 | 500 – 1,000 | |
|
| |||
| Cells/hour | 25 – 60 million | 2 million | |
|
| |||
|
Commercial reagent cost
| |||
| Per probe per test*** | $2 – 8 | $1.50 – 3 | |
Theoretical number of subsets is the number of distinct cell types determinable, assuming only “on” or “off” for each marker; i.e., 2colors.
Sensitivity range is in arbitrary units, and compares the rough sensitivity for different probes (fluorescence or ICP-MS) to detect a given epitope on a cell by immunophenotyipng.
Estimated based on the price of commercially conjugated reagents or unconjugated antibodies and commercial conjugation kits.
Dimensionality
Years of hardware and reagent advances preceded the complex, state-of-the-art PFC experiments reported in the last decade. In contrast, the learning curve for the first large-scale mass cytometry experiment was much less steep[42]. This study—predicated on the fact that mass cytometry could exploit and adapt many established PFC principles - examined regulatory cell signaling behavior across hematopoietic cells using two 34-parameter panels, each of which included 31 antibody targets, a DNA intercalator, and measures of viability and cell size. One panel employed 31 channels for cellular phenotyping, while the other simultaneously analyzed 18 intracellular phosphorylation responses and 13 cell surface phenotypic markers in response to a variety of immunological perturbations. Less than a year later, a 37-parameter study of virus-specific T-cell function and phenotype was reported[41]. These experiments used the maximum number of parameters currently accessible by mass cytometry.
To date, conventional PFC has shouldered a major burden in immunology. For instance, mapping the complex immune system requires measurement of a carefully selected set of 12+ markers and fluorophores matched with laser lines to call out the many attendant cell subpopulations of interest. But as the number of markers reaches 15, designing PFC panels becomes laborious—with a current limit of 18 markers measured simultaneously. In cases in which multiple intracellular events (pathways) must be tracked, PFC cannot simultaneously detail multiple pathways across multiple cell subsets (i.e., on a cell-by-cell basis).
Therefore, the ability of mass cytometry to measure so many parameters, without the loss of sensitivity accompanying compensation, is an important advantage. However, there are a number of cellular qualities mass cytometry cannot yet measure. For example, forward and side scatter (FSC and SSC) are light-based measures of cell size and granularity commonly employed in flow cytometry to discriminate large granular leukocytes, lymphocytes, doublets of cells, and cellular debris. These measures, which are used to filter out experimental artifacts or provide broad definitions of cell subsets, are not currently available for first generation mass cytometers. Additionally, small molecule fluorescent reporters for Ca2+ flux[43], mitochondrial permeation[44], and cell division (CFSE)[45] do not have metal-reporter equivalents. On the other hand, there are also opportunities to measure novel metal parameters at the single-cell level including: platinum (cisplatin – a cancer drug), barium (an MRI imaging contrast reagent), iodine (radioactive iodine therapy – for thyroid ablation), and gold (for experimental autoimmune therapy).
Sensitivity
Currently, the sensitivity of lanthanide-tagged antibodies is lower than that of the most popular fluorescent reporters[36, 42] (Figure 3). The primary reason for this limitation is the chelating polymer[37, 40] common to the commercially available probes. This polymer allows a maximum of around 100 metal reporter ions (M3+) to be attached to an antibody molecule, creating a ceiling on signal levels until alternative probes can be developed.
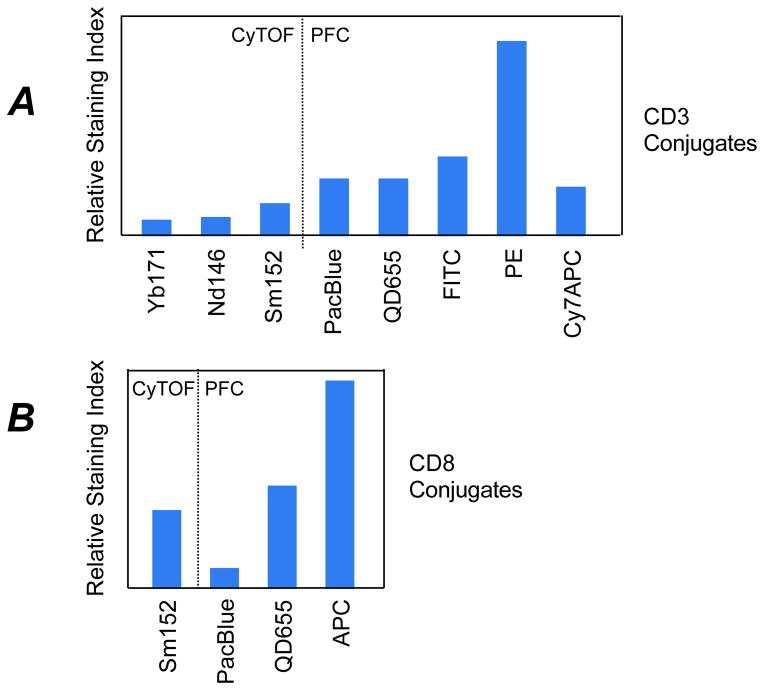
Figure 3. Performance of various metal isotopes and fluorochromes.
(a) Relative staining indices, calculated as described in [15], for a CD3 antibody conjugated to Yb171, Neodynium 146 (Nd146), or Samarium 152 (Sm152) and analyzed by CyTOF, or Pacific Blue (PacBlue), Quantum Dot 655 (QD655), FITC, PE, or Cy7APC and analyzed by PFC. Staining indices by CyTOF are low. (b) Relative staining indices for mouse anti-human CD8 (clone RPA-T8) conjugated to Sm152 and analyzed by CyTOF, and conjugated to PacBlue, QD655, or APC and analyzed by PFC. Staining indices for CD8 conjugates were considerably higher than anti-CD3 antibodies, and CyTOF conjugates had better performance than PacBlue (PFC) conjugate. Note, low staining indices may be acceptable for many applications, because background staining can be low by CyTOF.
While this has not prevented the measurement of many popular cytometric targets[36, 41, 42], it could preclude analysis of those with low signal-to-noise ratios. On the other hand, the low background signals in mass cytometry (where elemental isotopes are not naturally found in cells) compared to flow cytometry (with inherent auto-fluorescence of cells), can balance this deficiency and thereby have a significant advantage in multiplexed measurement of subtle regulatory changes (e.g.. protein phosphorylation[42]). In any event, an important advantage of mass cytometry is the similarity in the sensitivity across all lanthanide-based reporters—varying only 2-fold in sensitivity across isotopes from the lower to upper mass range. In contrast, the sensitivity of various fluorochromes can differ widely (10–50 fold) in flow cytometry and is a primary hurdle to development of multicolor panels[28].
Usability
The CyTOF mass cytometer provides three orders of magnitude of resolution between adjacent detection mass channels; pragmatically, two adjacent metal isotopes can differ in abundance by ~103 before spectral overlap arising from imperfect resolution of masses needs to be corrected by compensation (i.e., 0.1% spillover). As noted, the signal response for the majority of the lanthanide metal isotope reporters falls within approximately 2-fold of one another. Compared to fluorescence, where changing a single parameter might require redesign of an entire analysis panel to avoid spectral overlap issues, these qualities of mass cytometry significantly simplify experimental panel design. Although mass cytometry all but eliminates spectral overlap issues, there can still be signal interference/overlap from isotopic impurities in the metal reporters (usually + and/or – 1 Da) and oxidation (+16 Da) of the reporter ions during analysis[30, 34]. Notably, a wider variety of tagged antibodies are available for flow cytometry than for CyTOF; however, kits are available for in-house conjugation of even small quantities of purified antibody.
Quantitation
Both fluorescence[46] and mass-based[30, 36, 47] measurements are quantitative when the proper controls are employed. The CyTOF mass cytometer is linear across almost four orders of magnitude regardless of the number of parameters measured. Fluorescence flow cytometers typically have a range of at least five orders of magnitude where linearity is often a function of the photon amplifiers. Practically speaking, this working dynamic range for fluorescence detection can be compromised both by autofluorescence as well as spectral spillover. Nevertheless, both mass and fluorescence cytometers use detection technologies that are highly mature, and thus are highly accurate. In terms of precision, for any given reagent, fluorescence measurements will be somewhat better than mass cytometry in theory; however, in practice this precision is already better than biological variation and so is likely irrelevant.
Sampling and Throughput
Mass cytometry throughput is limited to about 1000 cells per second[39]; commercial fluorescence cytometers can operate at rates 25–50 times faster. In addition, cell injection and cleaning routines are time-consuming, increasing run times per sample. Moreover, unlike flow cytometry, where nearly all the cells introduced into the instrument are analyzed, the nebulization of single cell droplets into the ICP is currently inefficient, allowing the measurement of about 30% of the sample’s cells. However, this loss can be overcome when a high number of analytical parameters are desired per sample – perhaps requiring one sample by mass cytometry and multiple runs on PFC. Moreover, this loss is stochastic so it does not appear to introduce sampling bias. Overall, the current generation mass cytometer can process about 8 samples per hour, with about 250,000 processed events per sample (i.e., 2 million events per hour). This analysis rate defines the lower limit for rare event detection that can be achieved, though as shown in Bendall et al. [42] this does not compromise detection of important, rare cell populations if one is willing to collect enough cells over a sufficient time frame.
Experimental
Overall, the current reagents available for mass cytometry may be utilized best for investigating intracellular regulatory molecules (where autofluorescence can be highly confounding), or in situations where a very high number of simultaneous measurements are needed. Rare event analysis, detailed phenotyping requiring the measurement of low abundant cellular targets or light-scatter properties and/or rapid analysis of individual samples for the time being may be better suited for fluorescence cytometry. For instance, improvements such as in situ single cell amplification techniques using DNA branch chain[48, 49], polymers with more chelator sites allowed per Ab or lanthanide nanocrystals might overcome certain sensitivity issues, while more efficient sampling interfaces with integrated optics for measuring light-based reporters are already on the horizon (see below).
Future Developments in the Post-Fluorescence Era
As has occurred throughout the history of flow cytometry, improvements in mass cytometry are likely to dramatically increase its utility. Current efforts are focused on a number of areas: throughput, mass and dynamic range, and development of additional and more sensitive metal isotope probes. While the low cellular acquisition rate is a physical limitation of mass cytometry, a prototype auto-sampler has been introduced. This will help wash-out time between samples and allow automated acquisition. In addition, sample multiplexing techniques currently used in flow cytometry, such as fluorescent cell barcoding (FCB)[29, 50], are also being adapted to mass cytometry. Use of barcoding, even in a binary approach, allows up to 128 conditions to be multiplexed using seven parameters (27), potentially leaving 30 or more parameters for single-cell measurement.
Conventional commercial ICP-MS instruments have a linear range of almost 109; however, for the CyTOF mass cytometer the range is 104 across a measurement window large enough to include all Lanthanide isotopes. Interestingly, this is not an instrument limitation, but a limitation in the speed of computer hardware[40]. Without limitations in the speed of data digitization, the current CyTOF mass cytometer could actually measure all known non-biological transition metals with a linear dynamic range of ~106 per cell. Future generations of instrumentation should be much improved in this regard.
Similarly, there are enough unique non-biological elemental isotopes to move mass cytometry measurements well beyond 50 simultaneous parameters per cell (Figure 4) in the future. Two important hurdles accompany such development efforts. First is the effective attachment of the reporter isotopes to antibodies. Currently, reagents employ polymeric chelators, like DTPA and DOTA, which show a preference for metals with a positive charge of three in solution (e.g., the lanthanide series). The utilization of chelators targeting +2 or +4 metal ions (i.e., the non-biological transition metals Cd, Te, Pd, Ag, Sn, Os, Hf) could provide more than 20 additional measurement parameters. Secondly is the availability of pure forms of additional isotopes. For many elemental isotopes, purifying out contaminating isotopes becomes prohibitive, and many are available only with >5% contamination. While this contamination does not impact the ability to quantify the probe, it does re-introduce the need for compensation and quite likely associated difficulties in panel development.
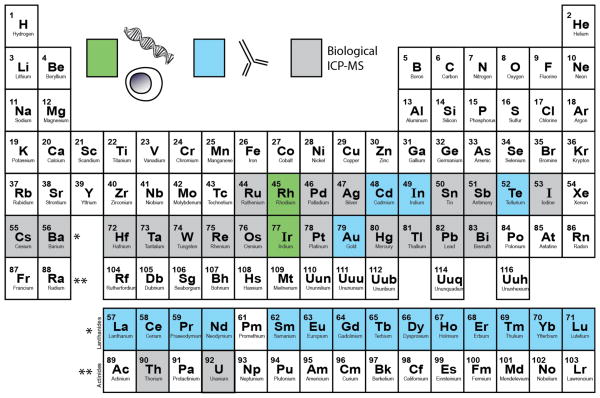
Figure 4. Periodic table summarizing the feasible elemental reporters for single-cell mass cytometry measurement in a biological matrix.
Colored elements (green, blue, grey) are those with at least one (relatively) stable isotope having an atomic mass greater than 100 daltons. Green elements have been demonstrated in estimating DNA content and cell size[33, 42], blue elements have been conjugated to antibodies for cell-based mass cytometry measurements using either a chelating polymer[37, 40] or semiconductor nanocrystals – Qdots[42], grey elements have not been published in mass cytometry studies yet but are readily analyzed by ICP-MS. These are future development targets.
Increased sensitivity of mass cytometry instrumentation, as with all mass spectrometers, is expected to improve incrementally with subsequent generations. Improvement of sensitivity through reagent technology represents the most immediately promising area of mass cytometry analysis. With commercial chelating polymers it is possible to attach on the order of 100 metal atoms per antibody. Given the low ion transmission efficiency (1 in 104 ions reach the detector) at least 100 molecules must be present (theoretically) on a cell before a signal is observed. Though, in practice, this limit of detection (LOD) is considerably higher. However, probes can be constructed with substantially higher metal content, reducing the LOD drastically. For example, solid metal nanoparticles, like quantum dots[51], can serve as reporters in mass cytometry as well, and can contain as many as 105 atoms of a given metal. With the utility of quantum dots[42] and lanthanide nanoparticles[52, 53] already demonstrated in mass cytometry, their optimized application could increase the number of atoms bound per antibody, thus lowering the LOD, increasing signal intensity and providing better resolution of cell populations with low levels of target protein expression. Ultimately, tools like these may allow detection of single molecules on single cells.
Analysis of Multiplexed, Multiparametric Data
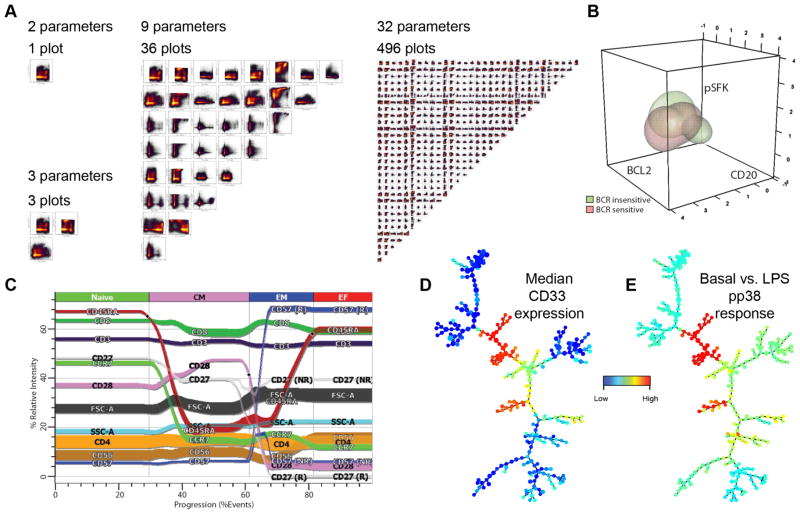
Rapid increases in the numbers of measurable single-cell parameters, both in flow and mass cytometry, have brought a daunting increase in the complexity of the data. Analysis of flow cytometry data is typically manual, performed in one or two dimensions at a time by selecting subsets of interest from parent populations. This approach is not scalable, though, and suffers from individual user bias (Figure 5A). Moreover, it requires a prior knowledge of the cell-type of interest, so unexpected cell types may be overlooked.
Figure 5. Complexity of multidimensional single-cell analysis.
(a) “Human interpretable” two-(D)imensional scatter plots are not a scalable solution in single-cell analysis. As the number of single cell parameters increases, the number of unique 2D plots increases exponentially. (b) FLAME analysis of B lymphocytes. Clustered cell events graphically displayed for visual comparison of receptor signaling cell populations. (c) Gemstone analysis of CD8 T cells showing progression of phenotypic markers including branching expression of markers like CD57 D–E) SPADE plots representing clusters of cells in normal human bone marrow. D) is the expression of CD33 (monocytes) across all cell clusters. (e) is the level of phosphor p38 (pp38) measured in response to LPS stimulation and normalized to an unstimulated (basal) control.
For such analyses, Flowjo (www.treestar.com) is a popular stand-alone software package for flow cytometry. It contains features that simplify and standardize multi-sample analyses where samples can be grouped and analyzed together, with tools like density-based (magnetic) gating to reduce the effects of staining variability. It also includes a wide variety of graphical outputs for visually summarizing multi-dimensional experiments (e.g., polychromatic plots). Cytobank (www.cytobank.org) is a collaboration-centric, web-based analysis platform that has features considered common to flow analysis platforms, has a web-based data sharing and repository function for community-based analysis[54], and ‘omics-styled heat map features for integrated analysis of phospho-flow studies—essential for intracellular signaling systems.
While these latter platforms provide the means to manually analyze and summarize large sets of samples, in terms of population frequencies and expression levels, they do not provide a mechanism to identify overall trends. To address this, SPICE[55] goes a step further to help identify larger trends across user-defined cell populations in large study groups. Still, this approach relies on prior knowledge of existing cell populations. This is problematic for identifying unexpected changes in unanticipated cell populations within complex systems.
To this end, a number of analysis platforms have recently been developed to assist the search for small changes in n-dimensional datasets. The problem is illustrated in Figure 5A where, as the number of measured parameters increases, “seeing” the shape of the information requires huge sets of conventional 2-dimensional plots. It is akin to the problem of the blind men and the elephant—each seeing with their hands only a small part (a tail, the trunk) of the whole elephant and thereby not encompassing a view of the entire elephant. Now, imagine the problem with a 40 dimensional elephant wherein we are equally blind since we cannot humanly “see” in more than 3 dimensions at best. Approaches, therefore, are required to enable ‘human interpretable’ two-dimensional visualizations that require minimal or readily managed user supervision.
Samspectral[56] and density reduction clustering[57] approach this problem by offering different approaches to cluster cells based on the expression levels of various parameters. However, unlike SPICE, they do not provide mechanisms to directly compare these clusters across large sample sets. FLAME - Flow Analysis with Automated Multivariate Estimation[58] - (Figure 5B) also, performs unsupervised clustering, providing tabular summaries tools to visually compare clusters between samples. Most recently, spanning-tree progression analysis of density-normalized events (SPADE) (www.cytospade.org) provides a platform to analyze large cohorts of samples where cells are clustered in multidimensional space and then reduced to a two-dimensional representation using a minimum spanning tree algorithm[59] (Figure 5D–E). This 2D model represents the relative relationship between each cell cluster in all samples, can be used to interrogate the expression of various parameters between clusters, and provides the ability to then compare clusters across samples. SPADE is now an integrated application in Cytobank.
Instead of clustering, there are other approaches that can leverage the complexity of n-dimensional single-cell datasets in order to lower dimensionality to biologically meaningful observations – for example, identifying the simplest combination of markers with biological or clinical relevance. Principle components analysis (PCA) has been used classically to calculate linear vectors through all measured parameters, thus identifying those combinations that describe the most variance in the data and relationships between samples. However, this method is not generally useful to immunophenotyping data, because of the general lack of correlations of expression in most markers. To address this, FlowType[60], a new R-package, takes n-dimensional data, automatically defines populations, and exhaustively stratifies all possible combinations of markers, comparing them across different samples. The idea is to identify the simplest population from the n-dimensional dataset that differs between samples. This, SPADE, and similar packages are available through Bioconductor project (www.bioconductor.org), and are being evaluated using common datasets against pre-defined performance metrics through the FlowCAP (Flow Cyotmetry: Critical Assessment of Population Identification Methods -http://flowcap.flowsite.org/) initiative. Similarly the parameters expressed by a single cell can be linked to each other, on a cell-by-cell basis, to construct relationship networks or classifiers. This approach, which employs Bayesian inference, has been particularly useful to examine T cell receptor signaling, revealing interactions between regulatory phospho-proteins without biochemical interrogation [61] as well as investigating cell signaling feedback mechanisms[62, 63].
Lastly, Gemstone (www.vsh.com) works differently to create a 2D summary of n-dimensional single cell datasets – exploiting the continuous expression patterns of various parameters (Figure 5). It employs probability state modeling to organize and visualize cell populations relative to one another[64, 65]. While this analysis usually requires a priori knowledge of the relationship between at least some of the markers measured, it still visually summarizes all cells in a given sample and can reveal cell subsets and relationships that other tools may not.
Many of these approaches take into account an important problem in multi-parametric analysis: the fact that simultaneous measurement of multiple markers can provide more information than is ultimately necessary for understanding the biology of a disease. Cells could be parsed too finely, into functionally redundant subsets. Given this possibility, it is important to consider how many parameters must be measured simultaneously to effectively address a particular hypothesis. To arrive at such conclusions, however, high dimensional, hypothesis generating experiments, and the tools to analyze and distill them, will be necessary.
Conclusions
Over the last 40 years, a continual improvement in single-cell analysis technologies has driven our investigation and understanding of immunology and stem cell biology. Pushing the multi-parameter limits of fluorescence-based analysis has led to unprecedented studies of regulatory signaling in both the healthy and diseased hematopoietic system. It has also identified an astounding number of distinct immune cell subsets—most of which have no assigned function. Now, next-generation, mass cytometry instrumentation will likely drive the next stage of “deep profiling” in mapping biological mechanisms of normal development, the role of multiple cell subsets in carrying out appropriate immune responses, and how any malfunction in these causes disease. Both fluorescence and mass cytometry will continue to be critical tools in cell biology for the foreseeable future; their complementarity with other single-cell applications and future improvements has the greatest promise for future discovery.
Acknowledgments
M.R. and P.K.C. are supported by the Intramural Research Program of the NIAID, NIH, and by the Collaboration for AIDS Vaccine Discovery (CAVD), Grant #OPP1032325, from the Bill and Melinda Gates Foundation. S.C.B. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017-09). G.P.N. is supported by the Rachford and Carlota A. Harris Endowed Professorship and grants from U19 AI057229, P01 CA034233, HHSN272200700038C, 1R01CA130826, CIRM DR1-01477 and RB2-01592, NCI RFA CA 09-011, NHLBI-HV-10-05(2), European Commission HEALTH.2010.1.2-1, and the Bill and Melinda Gates Foundation (GF12141-137101).
Footnotes
Conflict of Interest Disclosure. M.R. receives royalties on the sale of FlowJo software and Cy7APC fluorescent reagents. G.P.N. owns stock and is a paid consultant with DVS Sciences (CyTOF manufacturer) and is a paid consultant with Becton Dickenson, a purveyor of reagents central to both cytometry platforms.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Dilla M, et al. Los Alamos Scientifi Laboratory Annual Report of the Biological and Medical Research Group. 1967. The fluorescent cell photometer: A new method for the rapid measurement of biological cells stained with fluorescent dyes. [Google Scholar]
- 2.Hulett HR, et al. Development and application of a rapid cell sorter. Clin Chem. 1973;19:813–816. [PubMed] [Google Scholar]
- 3.Kamentsky LA. Future directions for flow cytometry. J Histochem Cytochem. 1979;27:1649–1654. doi: 10.1177/27.12.391998. [DOI] [PubMed] [Google Scholar]
- 4.Steinkamp JA, et al. Dual-laser flow cytometry of single mammalian cells. J Histochem Cytochem. 1979;27:273–276. doi: 10.1177/27.1.374585. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro HM, et al. Immunofluorescence measurement in a flow cytometer using low-power helium-neon laser excitation. Cytometry. 1983;4:276–279. doi: 10.1002/cyto.990040314. [DOI] [PubMed] [Google Scholar]
- 6.Oi VT, et al. Fluorescent phycobiliprotein conjugates for analyses of cells and molecules. J Cell Biol. 1982;93:981–986. doi: 10.1083/jcb.93.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telford WG. Lasers in flow cytometry. Methods Cell Biol. 2011;102:375–409. doi: 10.1016/B978-0-12-374912-3.00015-8. [DOI] [PubMed] [Google Scholar]
- 8.Lansdorp PM, et al. Single laser three color immunofluorescence staining procedures based on energy transfer between phycoerythrin and cyanine 5. Cytometry. 1991;12:723–730. doi: 10.1002/cyto.990120806. [DOI] [PubMed] [Google Scholar]
- 9.Roederer M, et al. Cy7PE and Cy7APC: bright new probes for immunofluorescence. Cytometry. 1996;24:191–197. doi: 10.1002/(SICI)1097-0320(19960701)24:3<191::AID-CYTO1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 10.Berlier JE, et al. Quantitative comparison of long-wavelength Alexa Fluor dyes to Cy dyes: fluorescence of the dyes and their bioconjugates. J Histochem Cytochem. 2003;51:1699–1712. doi: 10.1177/002215540305101214. [DOI] [PubMed] [Google Scholar]
- 11.Roederer M, et al. 8 color, 10-parameter flow cytometry to elucidate complex leukocyte heterogeneity. Cytometry. 1997;29:328–339. doi: 10.1002/(sici)1097-0320(19971201)29:4<328::aid-cyto10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 12.De Rosa SC, et al. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 13.Chattopadhyay PK, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 14.The Nobel Prize in Chemistry 2000 - Scientific Background. 2011 http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2000/advanced.html.
- 15.Chattopadhyay PK, Gaylord B, Palmer A, Jiang N, Raven MA, Lewis G, Reuter MA, Nurur R, Price DA, Betts MR, Roederer M. Brilliant Violet Fluorophores: A New Class of Ultrabright Fluorescent Compounds for Immunofluorescence Experiments. 2011. Submitted. [DOI] [PubMed] [Google Scholar]
- 16.Hulett HR, et al. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–749. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- 17.Wilder ME, Cram LS. Differential fluorochromasia of human lymphocytes as measured by flow cytometry. J Histochem Cytochem. 1977;25:888–891. doi: 10.1177/25.7.70458. [DOI] [PubMed] [Google Scholar]
- 18.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 19.Update on acquired immune deficiency syndrome (AIDS)--United States. MMWR Morb Mortal Wkly Rep. 1982;31:507–508. 513–504. [PubMed] [Google Scholar]
- 20.Peters JM, Ansari MQ. Multiparameter flow cytometry in the diagnosis and management of acute leukemia. Arch Pathol Lab Med. 2011;135:44–54. doi: 10.5858/2010-0387-RAR.1. [DOI] [PubMed] [Google Scholar]
- 21.Craig FE, Foon KA. Flow cytometric immunophenotyping for hematologic neoplasms. Blood. 2008;111:3941–3967. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 22.Festin R, et al. Single laser flow cytometric detection of lymphocytes binding three antibodies labelled with fluorescein, phycoerythrin and a novel tandem fluorochrome conjugate. J Immunol Methods. 1990;126:69–78. doi: 10.1016/0022-1759(90)90013-l. [DOI] [PubMed] [Google Scholar]
- 23.Picker LJ, et al. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood. 1995;86:1408–1419. [PubMed] [Google Scholar]
- 24.Borsellino G, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. 2007;110:1225–1232. doi: 10.1182/blood-2006-12-064527. [DOI] [PubMed] [Google Scholar]
- 25.Deenick EK, Ma CS. The regulation and role of T follicular helper cells in immunity. Immunology. 2011;134:361–367. doi: 10.1111/j.1365-2567.2011.03487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAleer JP, Kolls JK. Mechanisms controlling Th17 cytokine expression and host defense. J Leukoc Biol. 2011;90:263–270. doi: 10.1189/jlb.0211099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahnke Y, et al. Publication of optimized multicolor immunofluorescence panels. Cytometry A. 77:814–818. doi: 10.1002/cyto.a.20916. [DOI] [PubMed] [Google Scholar]
- 29.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 30.Thomas R. Practical Guide to ICP-MS: A Tutorial for Beginners. 2. CRC Press; 2008. [Google Scholar]
- 31.Tanner SD, et al. Multiplex bio-assay with inductively coupled plasma mass spectrometry: Towards a massively multivariate single-cell technology. Spectrochimica Acta Part B: Atomic Spectroscopy. 2007;62:188–195. [Google Scholar]
- 32.Razumienko E, et al. Element-tagged immunoassay with ICP-MS detection: evaluation and comparison to conventional immunoassays. J Immunol Methods. 2008;336:56–63. doi: 10.1016/j.jim.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ornatsky OI, et al. Study of cell antigens and intracellular DNA by identification of element-containing labels and metallointercalators using inductively coupled plasma mass spectrometry. Anal Chem. 2008;80:2539–2547. doi: 10.1021/ac702128m. [DOI] [PubMed] [Google Scholar]
- 34.Ornatsky OI, et al. Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2008;23:463–469. doi: 10.1039/b710510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ornatsky OI, et al. Messenger RNA Detection in Leukemia Cell lines by Novel Metal-Tagged in situ Hybridization using Inductively Coupled Plasma Mass Spectrometry. Translational Oncogenomics. 2006;1:1. [PMC free article] [PubMed] [Google Scholar]
- 36.Ornatsky O, et al. Multiple cellular antigen detection by ICP-MS. J Immunol Methods. 2006;308:68–76. doi: 10.1016/j.jim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Lou X, et al. Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Engl. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baranov VI, et al. A sensitive and quantitative element-tagged immunoassay with ICPMS detection. Anal Chem. 2002;74:1629–1636. doi: 10.1021/ac0110350. [DOI] [PubMed] [Google Scholar]
- 39.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 40.Majonis D, et al. Synthesis of a Functional Metal-Chelating Polymer and Steps toward Quantitative Mass Cytometry Bioassays. Anal Chem. doi: 10.1021/ac101901x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newell EW, NS, Bendall SC, Nolan GP, Davis MM. Cytometry by Time-Of-Flight shows Combinatorial Cytokine Expression and Virus-Specific Cell Niches within a Continuum of CD8+ T Cell Phenotypes. Immunity. 2012 doi: 10.1016/j.immuni.2012.01.002. In Press, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.June CH, Moore JS. Measurement of intracellular ions by flow cytometry. Curr Protoc Immunol. 2004;Chapter 5(Unit 5):5. doi: 10.1002/0471142735.im0505s64. [DOI] [PubMed] [Google Scholar]
- 44.Cottet-Rousselle C, et al. Cytometric assessment of mitochondria using fluorescent probes. Cytometry A. 79:405–425. doi: 10.1002/cyto.a.21061. [DOI] [PubMed] [Google Scholar]
- 45.Ingulli E. Tracing tolerance and immunity in vivo by CFSE-labeling of administered cells. Methods Mol Biol. 2007;380:365–376. doi: 10.1007/978-1-59745-395-0_23. [DOI] [PubMed] [Google Scholar]
- 46.Iyer SB, et al. Quantitation of CD38 expression using QuantiBRITE beads. Cytometry. 1998;33:206–212. doi: 10.1002/(sici)1097-0320(19981001)33:2<206::aid-cyto15>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 47.Abdelrahman AI, et al. Metal-Containing Polystyrene Beads as Standards for Mass Cytometry. J Anal At Spectrom. 25:260–268. doi: 10.1039/b921770c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsson C, et al. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat Methods. 2004;1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 49.Larsson C, et al. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 50.Krutzik PO, et al. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 51.Chattopadhyay PK. Quantum dot technology in flow cytometry. Methods Cell Biol. 2011;102:463–477. doi: 10.1016/B978-0-12-374912-3.00018-3. [DOI] [PubMed] [Google Scholar]
- 52.Pich A, et al. Biocompatible hybrid nanogels. Small. 2008;4:2171–2175. doi: 10.1002/smll.200801159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vancaeyzeele C, et al. Lanthanide-containing polymer nanoparticles for biological tagging applications: nonspecific endocytosis and cell adhesion. J Am Chem Soc. 2007;129:13653–13660. doi: 10.1021/ja073970w. [DOI] [PubMed] [Google Scholar]
- 54.Kotecha N, et al. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010;(Unit 10.17) doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roederer M, et al. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011 doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zare H, et al. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinformatics. 11:403. doi: 10.1186/1471-2105-11-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walther G, et al. Automatic Clustering of Flow Cytometry Data with Density-Based Merging. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pyne S, et al. Automated high-dimensional flow cytometric data analysis. Proc Natl Acad Sci U S A. 2009;106:8519–8524. doi: 10.1073/pnas.0903028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aghaeepour NCP, Ganesan A, O’Neill K, Zare H, Jalali A, Hoos HH, Roederer M, Brinkman RR. Early Immunologic Correlates of HIV Protection Can Be Identified from Computational Analysis of Complex Multivariate T-cell Flow Cytometry Assays. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts082. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sachs K, et al. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 62.Sachs K, et al. Characterization of patient specific signaling via augmentation of Bayesian networks with disease and patient state nodes. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:6624–6627. doi: 10.1109/IEMBS.2009.5332563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sachs K, et al. Learning cyclic signaling pathway structures while minimizing data requirements. Pac Symp Biocomput. 2009:63–74. doi: 10.1142/9789812836939_0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bagwell CB. Breaking the dimensionality barrier. Methods Mol Biol. 699:31–51. doi: 10.1007/978-1-61737-950-5_2. [DOI] [PubMed] [Google Scholar]
- 65.Bagwell CB. Probability state models. p. 7653509. [Google Scholar]