SUMMARY
Methylation of the N6 position of adenosine (m6A) is a post-transcriptional modification of RNA whose prevalence and physiological relevance is poorly understood. The recent discovery that FTO, an obesity risk gene, encodes an m6A demethylase implicates m6A as an important regulator of physiological processes. Here we present a method for transcriptome-wide m6A localization, which combines m6A-specific methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-Seq). We use this method to identify mRNAs of 7,676 mammalian genes that contain m6A, indicating that m6A is a common base modification of mRNA. The m6A modification exhibits tissue-specific regulation and is markedly increased throughout brain development. We find that m6A sites are enriched near stop codons and in 3' UTRs, and we uncover an association between m6A residues and microRNA binding sites within 3' UTRs. These findings provide a resource for identifying transcripts that are substrates for adenosine methylation and reveal insights into the epigenetic regulation of the mammalian transcriptome.
INTRODUCTION
The fat mass and obesity-associated (FTO) gene is a major regulator of metabolism and energy utilization (Church et al., 2009; Church et al., 2010; Fischer et al., 2009). In humans, FTO polymorphisms that increase FTO expression are associated with elevated body mass index and increased risk for obesity (reviewed in Fawcett and Barroso, 2010). FTO is a member of the Fe(II)- and oxoglutarate-dependent AlkB oxygenase family, and was originally shown to catalyze the oxidative demethylation of methylated thymidine and uracil (Gerken et al., 2007; Jia et al., 2008). However, FTO exhibits low activity toward these base modifications, and they are relatively infrequent with unclear physiological relevance (Klagsbrun, 1973). Thus, the physiologically relevant targets of FTO was unclear until recent studies that showed that FTO can demethylate N6-methyladenosine (m6A), a naturally occurring adenosine modification (Jia et al., 2011). These studies link adenosine methylation to physiological roles in human biological processes.
The distribution of m6A in RNA is poorly understood. Previous studies have found that m6A exists in RNA from a variety of unique organisms, including viruses, yeast, and mammals (Beemon and Keith, 1977; Bodi et al., 2010). m6A is found in tRNA (Saneyoshi et al., 1969), rRNA (Iwanami and Brown, 1968), and viral RNA (Beemon and Keith, 1977; Dimock and Stoltzfus, 1977). Although m6A is detectable in mRNA-enriched RNA fractions (Desrosiers et al., 1974), it has been confirmed in vivo in only one mammalian mRNA (Horowitz et al., 1984). The abundance of m6A has been shown to be 0.1–0.4% of total adenosine residues in cellular RNA (Dubin and Taylor, 1975; Perry et al., 1975; Wei et al., 1975), suggesting that this modification may be widespread throughout the transcriptome. Although the existence of m6A has been known for many years, progress in establishing the prevalence of m6A in mRNA has lagged behind that of other modified bases. This is due in large part to the lack of available methods for detecting the presence of m6A. Because methylation of adenosine does not alter its ability to base pair with thymidine or uracil, m6A is not amenable to detection with standard hybridization or sequencing-based methods.
Here, we examine the prevalence, regulation, and functional roles of m6A in the transcriptome. We show that m6A is a developmentally regulated RNA modification which is dynamically modified. Using antibodies that recognize m6A, we have developed an affinity enrichment strategy which, when coupled with next-generation sequencing, allows for the high-throughput identification of m6A sites. Using this approach, we present the first transcriptome-wide profile of m6A localization in RNA. We find that m6A is a widespread modification that is present in the mRNAs of over 7,600 genes and in over 300 noncoding RNAs. Additionally, m6A is highly enriched near the stop codon and in the 3’ UTR. Furthermore, bioinformatic analysis of m6A localization reveals consensus sites for m6A and identifies a potential interaction between m6A and microRNA pathways.
RESULTS
Detection of m6A in Mammalian mRNA
Because m6A exhibits the same base pairing as unmodified adenosine, it is not readily detectable by standard sequencing or hybridization-based approaches. Additionally, m6A is not susceptible to chemical modifications which might otherwise facilitate its detection, such as bisulfite treatment which is used to detect 5mC in DNA. The methods used thus far to detect m6A have involved treating cells with radiolabeled methionine, the precursor of the endogenous methylating agent S-adenosylmethionine, to impart radiolabeled methyl groups to adenosine (Csepany et al., 1990; Dubin and Taylor, 1975; Narayan and Rottman, 1988). Radiolabeled m6A residues are subsequently mapped with thin-layer chromatography or HPLC.
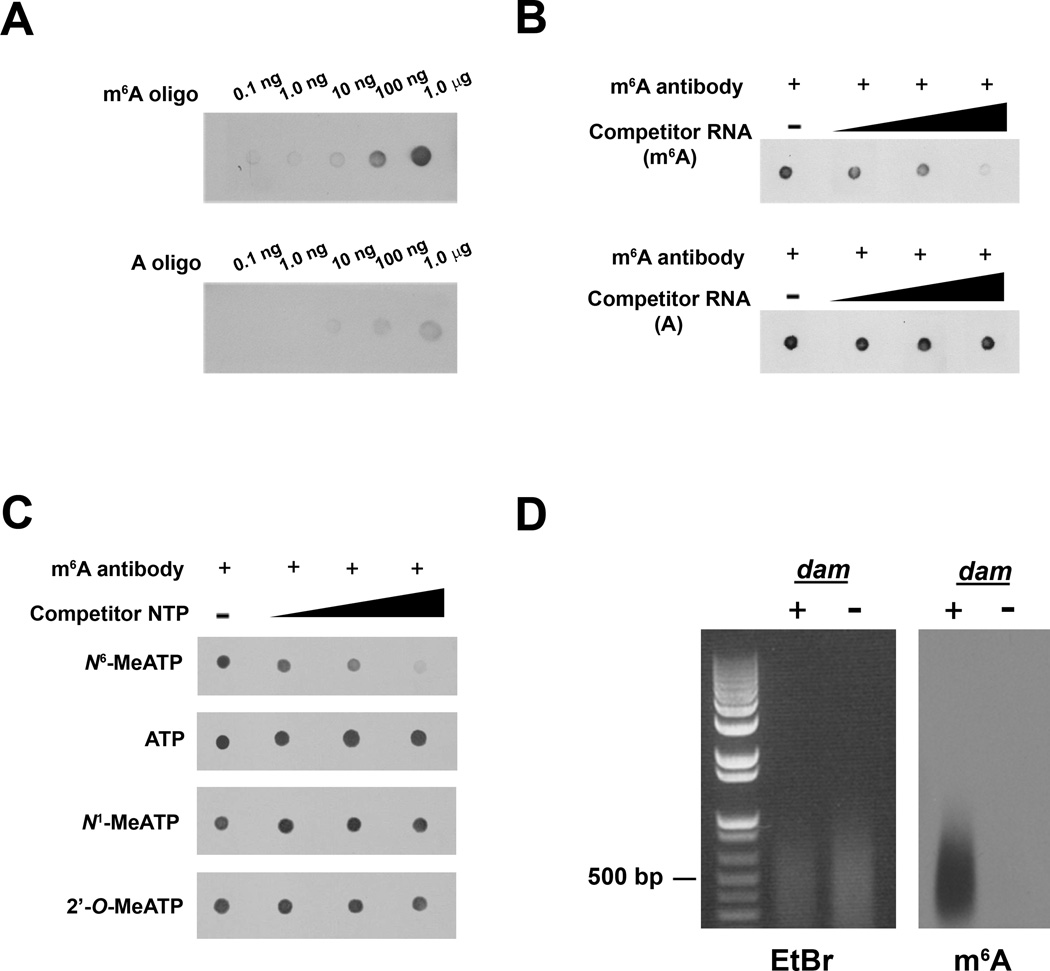
In order to simplify detection of m6A, we sought to develop an immunoblotting strategy. For these experiments, we used a previously described anti-m6A antibody (Bringmann and Luhrmann, 1987; Jia et al., 2011; Munns et al., 1977). To ensure the specificity of this antibody for m6A, we performed dot blots using modified oligonucleotides immobilized to a membrane. The m6A antibody selectively bound to oligonucleotides containing a single m6A residue, and exhibited negligible binding to oligonucleotides containing unmodified adenosine (Figure 1A). The binding was competed by incubating the antibody with increasing concentrations of an m6A-rich competitor RNA (Figure 1B). However, RNA containing unmodified adenosine did not compete for binding. Furthermore, binding was competed by N6-methyladenosine triphosphate, but not by ATP or other modified adenosine triphosphates including N1-methyladenosine and 2’-O-methyladenosine (Figure 1C). Finally, to examine the specificity of the antibody in the context of other nucleotide sequences, we took advantage of the fact that the enzyme encoded by the DNA adenine methylase (dam) gene in E. coli methylates the N6 position of adenosine in DNA. Upon subjecting digested DNA isolated from dam+ and dam− E. coli to immunoblotting using the m6A antibody, we found robust signals only in the DNA samples from the dam+ strain (Figure 1D). Collectively, these data demonstrate the high sensitivity and selectivity of this antibody for m6A, as well as its ability to detect m6A within cellular nucleotide pools.
Figure 1. Specificity and Sensitivity of m6A-Specific Antibody.
A. Dot blot analysis demonstrates antibody specificity for m6A. Increasing amounts of an oligonucleotide containing either m6A or unmodified adenosine were spotted onto a membrane and probed with the m6A antibody. While increased m6A immunoreactivity is observed in the presence of increasing concentrations of the m6A oligonucleotide (top), only background levels of immunoreactivity are observed at the highest concentrations of the A oligonucleotide (bottom). Blots shown are representative of results from three experiments.
B. Competition dot blot assays were performed on membranes spotted with 100 ng of m6A-containing oligonucleotide. Antibody binding to the m6A oligonucleotide is attenuated by pre-incubation with increasing amounts of m6A-containing competitor RNA (top), but not with RNA containing unmodified adenosine (bottom). Amount of competitor RNA used (left to right): 0 ng (0 nM), 10 ng (0.1 nM), 100 ng (1.1 nM), 1 µg (11.2 nM). Blots shown are representative of results from four experiments.
C. Competition dot blot assays were performed as in (B). Antibody was pre-incubated with increasing amounts of N6-methyladenosine triphosphate (N6-MeATP), adenosine triphosphate (ATP), N1-methyladenosine triphosphate (N1-MeATP), or 2’-O-methyladenosine triphosphate (2’-O-MeATP). Only N6-MeATP is able to compete with antibody binding. Concentration of competitor nucleotide used (left to right): 0 µM, 1 µM, 2 µM, 4 µM. Blots shown are representative of results from three experiments.
D. Detection of m6A in cellular DNA. Genomic DNA isolated from dam+ (containing m6A) or dam− (lacking m6A) E. coli was sheared and subjected to immunoblotting with the anti-m6A antibody. Although 1.5 times as much DNA from dam− E. coli was loaded (left panel), the antibody only recognizes the m6A present in DNA from dam+ E. coli (right panel). Blot shown is representative of results from three experiments.
See also Figure S2.
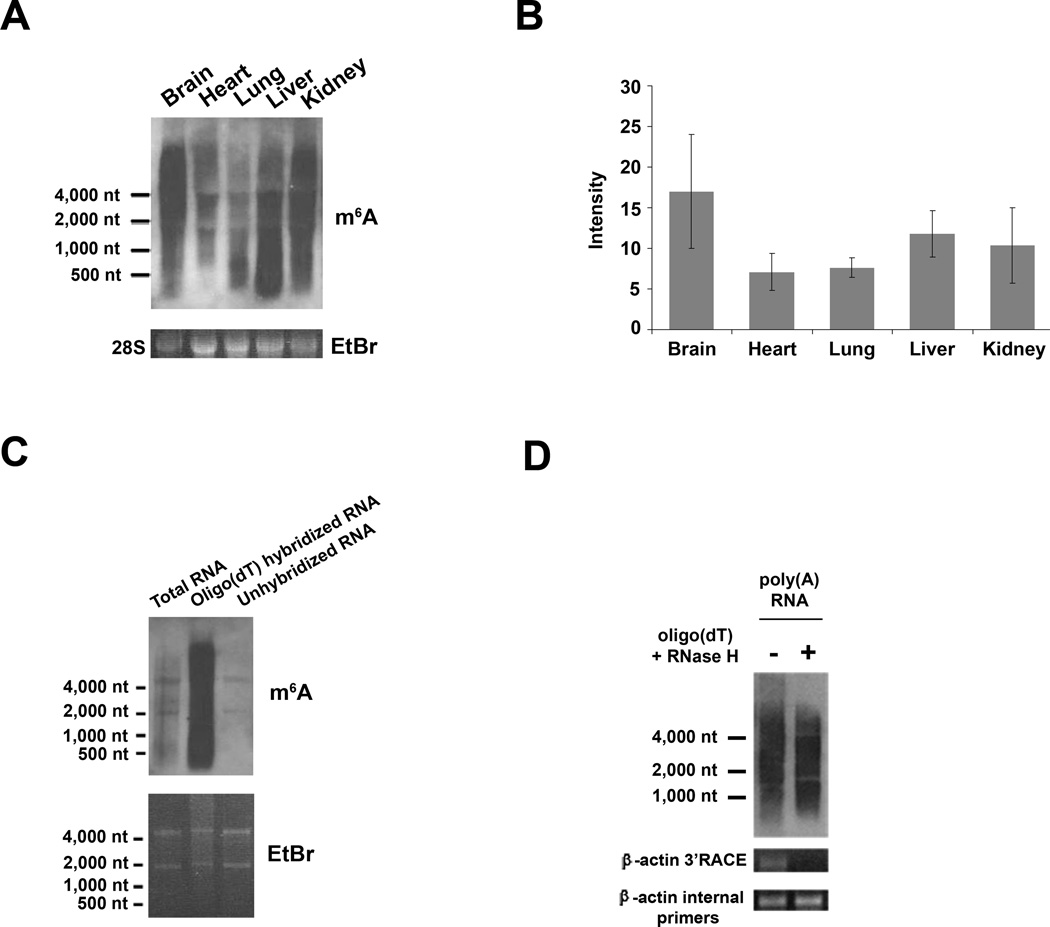
To explore the abundance of m6A within various RNA populations, we isolated RNA from several mouse tissues and subjected it to immunoblot analysis using the m6A antibody (Figure 2A). We found that m6A was present in all RNA samples tested, indicating that this modified nucleotide is widely distributed in many tissues, with particularly high enrichment in liver, kidney, and brain (Figure 2B). In addition, we observed large differences in the m6A content of various immortalized cell lines, including several cancer cell lines, which further indicates that large differences in m6A levels exist in different cell populations (Figure S1A).
Figure 2. Distribution and Dynamic Cellular Regulation of m6A in RNA.
A. Widespread distribution of m6A levels in a variety of tissues. Total RNA isolated from mouse brain, heart, lung, liver, and kidney (top) was subjected to m6A immunoblot analysis. Ethidium bromide staining of the 28S rRNA is shown as a loading control (bottom).
B. Quantification of m6A abundance within various tissues. Quantification of m6A immunoreactivity in (A) was measured by densitometry and normalized to the intensity of the corresponding 28S rRNA band for each tissue (n = 3; data are presented as mean ± SEM).
C. m6A is enriched within mRNAs. Oligo(dT) Dynabeads were used to isolate poly(A) RNA from total mouse brain RNA, and the unbound “flow-through” RNA was saved as the poly(A)-depleted fraction. Equal amounts of total RNA, poly(A) RNA, and poly(A)-depleted RNA were then subjected to m6A immunoblot analysis (top). Ethidium bromide staining of 28S rRNA is shown as a loading control (bottom). Intense m6A immunoreactivity is observed in the poly(A) RNA fraction, consistent with high levels of m6A within mRNAs.
D. Depletion of poly(A) tails from mRNA does not reduce levels of m6A in mRNA. Poly(A) RNA was isolated from total mouse brain RNA using oligo(dT) Dynabeads. Half the sample was then subjected to poly(A) tail depletion by hybridizing to oligo(dT) primers and digestion with RNase H. Immunoblot analysis with the m6A antibody (top panel) shows that levels of m6A in poly(A) RNA (left) and poly(A) tail-depleted RNA (right) are comparable. Removal of poly(A) tails was confirmed using 3’RACE and RTPCR to detect β-actin; no product is detected in the tail-depleted sample when oligo(dT) primers are used for cDNA synthesis (middle panel). As a control, use of random hexamers successfully generates a product in both samples (bottom panel).
See also Figure S1.
m6A immunoreactivity was detected in bands throughout the molecular weight range of the blot (~0.2 kb to ~ 10 kb), consistent with incorporation of m6A in mRNA. Indeed, fractionation of whole cellular RNA into polyadenylated and nonpolyadenylated RNAs indicates that m6A immunoreactivity is enriched in the polyadenylated RNA pool, which further suggests that m6A in cellular RNA is localized to mature mRNA (Figure 2C). To determine whether m6A is present in poly(A) tails, we selectively removed the poly(A) tail from cellular mRNA using oligo(dT) hybridization and RNase H treatment.
Transcripts depleted of the poly(A) tail did not exhibit an appreciable reduction in m6A levels (Figure 2D). In addition, immunoblotting poly(A) tails alone showed minimal m6A immunoreactivity (Figure S2C-E). Together, these data demonstrate that m6A is primarily an internal modification which is largely absent from the poly(A) tail.
Dynamic Regulation of m6A
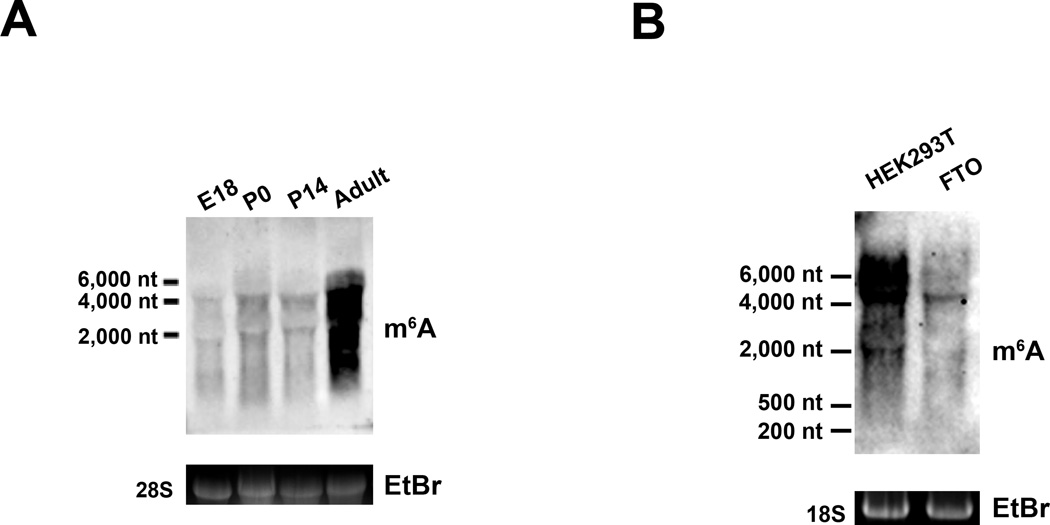
Our observation that m6A is highly enriched within the brain prompted us to investigate the temporal dynamics of m6A levels during different stages of neural development. Immunoblotting RNA samples with the m6A antibody indicates that m6A is present in mRNA at low levels throughout embryogenesis but increases dramatically by adulthood (Figure 3A). A similar increase in m6A levels is also observed in RNA isolated from embryonic and postnatal rat brain cultured neurons (Figure S1B), which suggests that upregulation of m6A levels accompanies neuronal maturation.
Figure 3. Regulation of m6A Levels in Cells and During Development.
A. Ontogeny of m6A abundance throughout brain development. Total RNA was isolated from mouse brain at embryonic day 18 (E18), postnatal day 0 (P0), postnatal day 14 (P14), and adulthood, then subjected to immunoblot analysis to detect m6A-containing transcripts. Ethidium bromide staining of 28S rRNA bands is shown as a loading control.
B. FTO demethylates a wide range of cellular transcripts. FTO was expressed in HEK293T cells for 48h, and cellular RNA was subjected to immunoblot analysis to detect m6A.
See also Figure S1.
We next asked if adenosine methylation is a dynamically regulated post-transcriptional modification and if its levels can be regulated by specific demethylating enzymes. In our search for potential demethylating enzymes that act to remove the methyl group from m6A, we focused on members of the family of Fe(II)- and 2-oxoglutarate-dependent oxygenases, several of which have previously been shown to demethylate both DNA and RNA (Falnes et al., 2007; Gerken et al., 2007). Consistent with the findings of Jia et al (2011), we observed that FTO decreased m6A levels when overexpressed in mammalian cells (Figure 3B). Furthermore, we find that overexpression of FTO resulted in a broad size range of RNAs that exhibit reduced m6A immunoreactivity (Figure 3B).
MeRIP-Seq Identifies m6A-Containing RNAs Throughout the Transcriptome
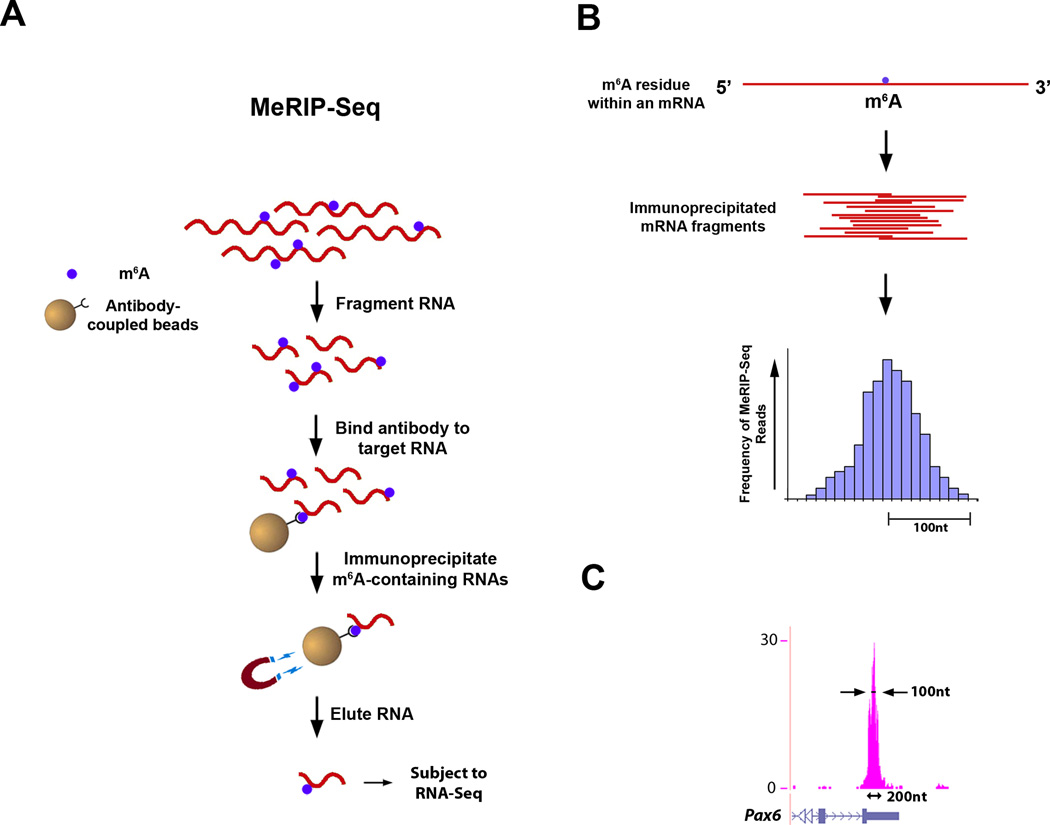
In order to obtain insight into potential roles for m6A, we sought to characterize its distribution throughout the transcriptome. To do this, we first determined whether the m6A antibody could be used to enrich m6A-containing RNAs. In vitro immunoprecipitation experiments showed that a single round of MeRIP produces ~70-fold enrichment, and two rounds produce over 130-fold enrichment for m6A-containing targets (Figure S3). To identify m6A sites throughout the transcriptome, we developed a method that combines m6A-specific methylated RNA immunoprecipitation (MeRIP) with next-generation sequencing (RNA-Seq). The procedure for MeRIP-Seq (outlined in Figure 4A) involves randomly fragmenting the RNA to approximately 100 nt-sized fragments prior to immunoprecipitation. Because an m6A site could lie anywhere along the length of a given immunoprecipitated 100 nt fragment, sequencing reads are expected to map to a region which contains the m6A site near its center. At its extremes, this region would be predicted to be roughly 200 nt wide (100 nt up- and downstream from the m6A site) (Figure 4B,C).
Figure 4. Outline of MeRIP-Seq Protocol and Distribution of Sequencing Reads.
A. Schematic representation of MeRIP-Seq. Total RNA is subjected to RiboMinus treatment to remove rRNA species. RNAs containing m6A are then immunoprecipitated by mixing the RNA with m6A antibody-coupled Dynabeads. m6A-containing RNAs are then eluted from the antibody-coupled beads and subjected to a second round of m6A immunoprecipitation. The resulting RNA pool, which is highly enriched for m6A-containing RNAs, is then subjected to next-generation sequencing.
B. Schematic of sequencing reads and their alignment to locations in the genome surrounding an m6A site. Top: an mRNA that contains a single m6A residue along its length. Middle: individual 100 nt-wide mRNA fragments which are isolated following m6A immunoprecipitation, each of which contains the same m6A residue from the mRNA depicted above. Bottom: histogram showing predicted frequency of MeRIP-Seq reads obtained by sequencing individual immunoprecipitated fragments. Read frequency is predicted to increase with closer proximity to the m6A site, forming a “peak” which is roughly 200 nt wide at its base and 100 nt wide at its midpoint.
C. Sequencing reads from MeRIP-Seq converge over m6A sites. Representative UCSC Genome Browser plot from MeRIP-Seq data which demonstrates typical read frequency peak formation surrounding a site of m6A (shown here is the 3’ UTR of Pax6). Peak height is displayed as reads per base per million mapped reads (BPM).
See also Figures S2, S3.
We next utilized MeRIP-Seq to identify m6A sites in total mouse brain RNA. Reads from the MeRIP sample frequently mapped to mRNAs and clustered as distinct peaks. As predicted, these peaks frequently converged to approximately 100 nt-wide regions near their midpoint (Figure 4C). Furthermore, enrichment of reads in these regions was not observed in the non-IP control sample, which was composed of the input RNA prior to m6A immunoprecipitation, demonstrating the specificity of these peaks (Figure S3).
To determine the location of these peaks throughout the transcriptome, and thus characterize the regions of m6A localization, we developed an algorithm for identifying m6A peaks (see Extended Experimental Procedures). Additionally, we performed replicate MeRIP-Seq experiments in which we utilized (1) a different sequencing platform (Illumina’s GAIIx vs HiSeq2000), (2) independently prepared RNA samples from different animals, and (3) an unrelated m6A antibody (Kong et al., 2000), which exhibited similarly high specificity for m6A (Figure S4). We employed our algorithm to identify m6A peaks that met a minimum p-value (p ≤ 0.05, Benjamini and Hochberg corrected) within each individual sample. From the three samples, we identified a total of 41,072 distinct peaks in the RNAs of 8,843 genes, which we call our “filtered” set of m6A peaks (Table S1). Of these peaks, 80% were detected in at least two different replicates. The high concordance between these samples indicates that MeRIP-Seq is highly reproducible across different sequencing platforms and using different m6A antibodies. For subsequent bioinformatic analyses, we used the list of 13,471 m6A peaks in RNAs from 4,654 genes which were detected in all three replicates (our “high-confidence” list; Table S2A). This list demonstrates the presence of m6A in a substantial fraction of the transcriptome and indicates that m6A is a common feature of mammalian mRNAs.
m6A is Detected in Non-Coding RNAs
The majority of our high-confidence m6A peaks (94.5%) are found within mRNAs. However, we also observed that 236 (1.1%) of our peaks mapped to non-coding RNAs (ncRNAs) that were annotated in the RefSeq database (Table S2A). In addition, 588 m6A peaks did not map to a known RefSeq mRNA or ncRNA. To determine whether these unannotated peaks localize to ncRNAs predicted in other databases, we aligned them to genomic regions of a set of 32,211 ncRNAs from the RIKEN functional annotation of mouse (FANTOM3) dataset that we obtained from the mammalian noncoding RNA database (RNAdb; Pang et al., 2005). We found that 216 of these peaks mapped to a FANTOM3 ncRNA (Table S2B). All of these ncRNAs were greater than 200 nt in length, indicating that long ncRNAs are substrates for adenosine methylation. Additionally, when we interrogated a set of conserved human lincRNAs (Cabili et al., 2011) for overlaps with m6A peaks, we found nine additional peaks that overlapped with these lincRNAs (Table S2C). Collectively, these data identify several classes of ncRNAs as targets of adenosine methylation.
Biochemical Validation of m6A-Containing Transcripts
We next sought to validate the presence of m6A in mRNAs identified with MeRIP-Seq. To do this, we used RNA pull-down assays to isolate individual mRNAs from total mouse brain RNA by hybridization to target-specific probes. Isolated mRNAs were then subjected to immunoblot analysis using the m6A antibody to detect the presence of m6A. Using this method, we validated the presence of m6A within low density lipoprotein receptor (Ldlr) (Figure 5A,B), metabotropic glutamate receptor 1 (Grm1), and dopamine receptor D1A (Drd1a) (Figure S5A-D). These mRNAs were chosen to demonstrate our ability to validate m6A presence in transcripts with multiple methylation sites (Grm1 and Drd1a) as well as those with single m6A peaks (Ldlr). To further demonstrate that MeRIP-Seq selectively enriches for these endogenous methylated targets, we performed qRT-PCR on the unbound fractions after RNA precipitation with the m6A antibody. As expected, we observed substantial immunodepletion of Grm1, Drd1a, and other methylated targets in the unbound fraction. In contrast, transcripts which lack m6A peaks, such as Rps21 and Ndel1, were detectable at high levels in the unbound fraction (Figure S5E).
Figure 5. Validation of m6A Targets and Characteristics of m6A Localization.
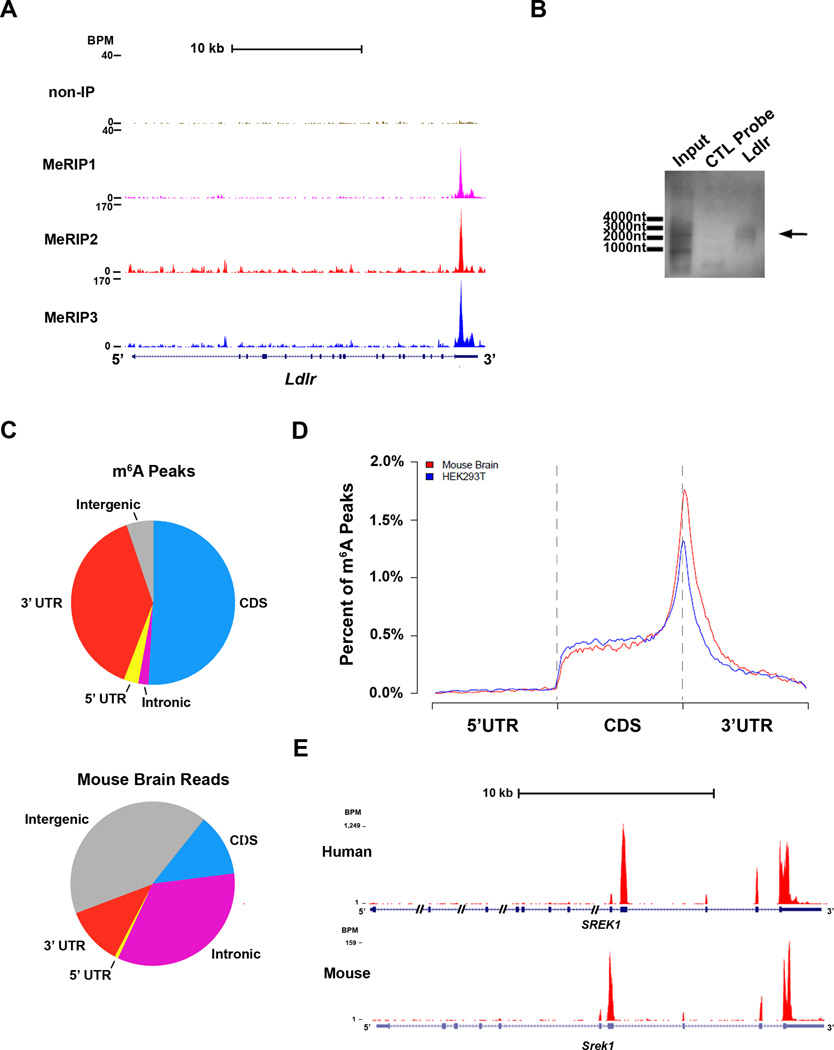
A. Different sequencing platforms and antibodies result in similar m6A profiles. UCSC Genome Browser tracks displaying read clusters from three MeRIP-Seq replicates (MeRIP1, MeRIP2, and MeRIP3) are shown along the length of the Ldlr transcript. The upper-most track (non-IP) represents the non-immunoprecipitated control sample.
B. Validation of m6A-containing mRNA identified with MeRIP-Seq. Hybridization-based RNA pulldown was used to isolate Ldlr mRNA from total brain RNA, followed by confirmation of m6A presence (arrow) by immunoblot analysis with anti-m6A. A control sample using a non-specific probe of equal size (Control Probe) was run in parallel. Total mouse brain RNA (Input RNA) is shown as a reference for m6A labeling.
C. Transcriptome-wide distribution of m6A peaks. Pie charts showing the percentage of m6A peaks (top) and non-IP sample reads (bottom) within distinct RNA sequence types. m6A is highly enriched in 3’ UTRs and CDSs compared to the distribution of reads in the non-IP samples.
D. Distribution of m6A peaks across the length of mRNA transcripts. 5’ UTRs, CDSs, and 3’ UTRs of RefSeq mRNAs were individually binned into regions spanning 1% of their total length, and the percentage of m6A peaks that fall within each bin was determined. The moving averages of mouse brain peaks percentage (red) and HEK293T peak percentage (blue) are shown.
E. Highly similar m6A peak distribution is observed within many human and mouse transcripts. UCSC Genome Browser plots showing MeRIP-Seq read clusters in the representative transcript SREK1. MeRIP-Seq reads cluster at the same distinct regions of SREK1 in both HEK293T cell RNA (top) and mouse brain RNA (bottom).
See also Figures S4 – S7, Tables S1-S6.
m6A-Containing mRNAs are Involved in Important Biological Pathways
To predict potential signaling pathways and cellular processes that involve m6A, we used the DAVID bioinformatics database to identify the gene ontology (GO) terms that are enriched for m6A-containing transcripts. We found that genes encoding m6A-containing RNAs are involved in a variety of cellular functions, including transcriptional regulation, RNA metabolism, and intracellular signaling cascades (Table S5). In addition, we observed that m6A peaks mapped to many genes linked to neurodevelopmental and neurological disorders, such as Bdnf, Dscam, Lis1, and Ube3a, as well as the neurexins and several neuroligins. Collectively, these data demonstrate that m6A-containing RNAs are involved in a variety of biological pathways relevant to cellular signaling and disease.
Since m6A is a physiological target of FTO, we sought to determine whether mRNAs whose levels have previously been shown to be influenced by FTO activity contain m6A. We examined a list of 77 mRNAs whose levels are either up- or downregulated in the liver, skeletal muscle, or white adipose tissue of mice homozygous for a nonsynonymous FTO point mutation (Church et al., 2009). mRNAs from seven genes which were significantly upregulated in FTO mutants (Acaca, Atf6, Bip, Gcdh, Irs1, Perk, and Xbp1) also contain m6A peaks. Intriguingly, some of these genes are involved in important metabolic pathways, raising the possibility that demethylation of the transcripts of these genes may contribute to the mechanism by which FTO regulates metabolism and energy homeostasis.
Diverse Patterns of m6A Localization within Transcripts
We next characterized the pattern of adenosine methylation in mRNAs. mRNAs from many genes (46.0%) exhibit a single m6A peak, consistent with a single m6A site or a cluster of adjacent m6A residues. However, 37.3% contain two m6A peaks, 11.2% contain three peaks, and 5.5% contain four or more peaks. Several genes contain ten or more m6A peaks, suggesting the existence of multiple m6A residues along their length. Indeed, of the twenty genes that exhibited the largest number of m6A peaks, all had 15 or more m6A peaks along their length (Table S3). Additionally, of the genes that contain more than one m6A peak, 90.1% contain two or more contiguous m6A peaks, suggesting that m6A sites are frequently clustered in adjacent regions along the transcript. Indeed, 32.8% of m6A peaks are part of clusters that contain three or more adjacent m6A peaks, suggesting that m6A clustering is a common feature in methylated transcripts (Figure S6A). We also identified 68 genes that have long (≥ 1 kb) stretches of contiguous m6A peaks (Table S4), which likely indicates the presence of multiple m6A residues throughout these regions.
We next determined which mRNAs contain m6A sites with the highest degree of methylation. To do this, we developed a method of calculating the level of m6A enrichment at individual m6A peaks which normalized the number of MeRIP sample reads within each peak to the abundance of the individual transcript in which the peak resides (see Extended Experimental Procedures). The genes which contain the most enriched m6A peaks are listed in Table 1. Importantly, because MeRIP-Seq identifies m6A sites at a resolution of 200 nt, there could be multiple individual m6A residues within the area covered by each peak. Therefore, the peaks with the highest levels of local m6A enrichment may represent a single adenosine residue which exhibits a high degree of methylation, or multiple adjacent m6A residues with a lower stoichiometry of methylation. In either case, however, the high levels of methylation observed at these sites likely indicate transcripts that are most influenced by m6A-dependent regulatory processes.
Table 1.
Genes Encoding Transcripts with the Highest Degree of m6A Enrichment
| Chr | Peak Start | Peak End | RefSeq Accession | Gene Symbol |
Enrichment Score |
|---|---|---|---|---|---|
| chr6 | 58856032 | 58856214 | NM_021432 | Nap1l5 | 3.859 |
| chr3 | 30717625 | 30717825 | NM_027016 | Sec62 | 3.693 |
| chr3 | 88566067 | 88566234 | NM_018804 | Syt11 | 3.452 |
| chr6 | 58855850 | 58856032 | NM_021432 | Nap1l5 | 3.327 |
| chr19 | 5801367 | 5801534 | NR_002847 | Malat1 | 3.322 |
| chr12 | 110898950 | 110899150 | NR_028261 | Rian | 3.222 |
| chr10 | 34018834 | 34018993 | NM_030203 | Tspyl4 | 3.162 |
| chr17 | 6138575 | 6138775 | NM_054040 | Tulp4 | 2.984 |
| chr2 | 34631357 | 34631514 | NM_001163434 | Hspa5 | 2.935 |
| chr2 | 102630175 | 102630350 | NM_001077514 | Slc1a2 | 2.933 |
| chr11 | 7072225 | 7072425 | NM_009622 | Adcy1 | 2.865 |
| chr2 | 158211200 | 158211350 | NM_175692 | Snhg11 | 2.788 |
| chr15 | 37326800 | 37327000 | NM_134094 | Ncald | 2.685 |
| chr2 | 102629600 | 102629788 | NM_001077514 | Slc1a2 | 2.672 |
| chr15 | 37327000 | 37327200 | NM_134094 | Ncald | 2.656 |
| chr10 | 80644950 | 80645125 | NM_007907 | Eef2 | 2.451 |
| chr2 | 102629788 | 102629975 | NM_001077514 | Slc1a2 | 2.392 |
| chr11 | 59251613 | 59251750 | NM_144521 | Snap47 | 2.306 |
| chr15 | 74581075 | 74581267 | NM_011838 | Lynx1 | 2.258 |
| chr2 | 158211050 | 158211200 | NM_175692 | Snhg11 | 2.102 |
Shown are the top twenty genes which contain m6A peaks with the highest levels of enrichment.
m6A is Enriched Near Stop Codons and in 3’ UTRs of mRNAs
We next examined the distribution of m6A peaks within regions of the transcriptome in our high confidence set. The majority (94.8%) of m6A peaks occur within intragenic regions (Figure 5C). These m6A peaks are abundant in coding sequences (CDS; 50.9%), and untranslated regions (UTRs; 41.9%), with relatively few in intronic regions (2.0%) (Figure 5C). Additionally, m6A peaks are less abundant in the 5’ UTR (7.0% of UTR peaks) than in the 3’ UTR (93.0% of UTR peaks) (Figure 5C). This distribution deviates substantially from the distribution of reads in the non-IP sample, indicating the high degree of enrichment of m6A peaks in the CDS and UTRs (Figure 5C). Although a low percentage of m6A peaks was observed in intronic regions, because our samples were not enriched for unspliced pre-mRNAs, it is possible that additional methylated intronic sequences exist.
We next sought to determine if m6A peaks are preferentially found in certain portions of transcripts. To do this, we assigned each m6A peak to either a 5’ UTR, CDS, or 3’ UTR category, and assigned it to one of 100 bins a based on its location along the 5’ UTR, CDS, or 3’ UTR. These data show that m6A occurs at low levels in the 5’ UTR and the 5’ end of the CDS. In the CDS, the percentage of m6A peaks increases steadily along transcript length and is on average 5–6 fold higher at the end of the CDS than at the beginning (Figure 5D). In the 3’ UTR, the peaks are enriched near the stop codon and decrease in abundance along the length of the 3’ UTR. Indeed, 61% of m6A peaks are in the first quarter of the 3’ UTR and a quarter of all m6A peaks across the entire transcriptome are found within the first 26% of the 3’ UTR (Figure 5D). Mapping the number of m6A peaks 1 kb up and downstream of CDS end sites further demonstrated the high levels of methylation in the vicinity of the stop codon (Figure S7A,B). Collectively, these data indicate that m6A peaks are highly clustered in the vicinity of the stop codon in mRNAs.
m6A Occurs in Highly Conserved Regions within Unique Sequence Motifs
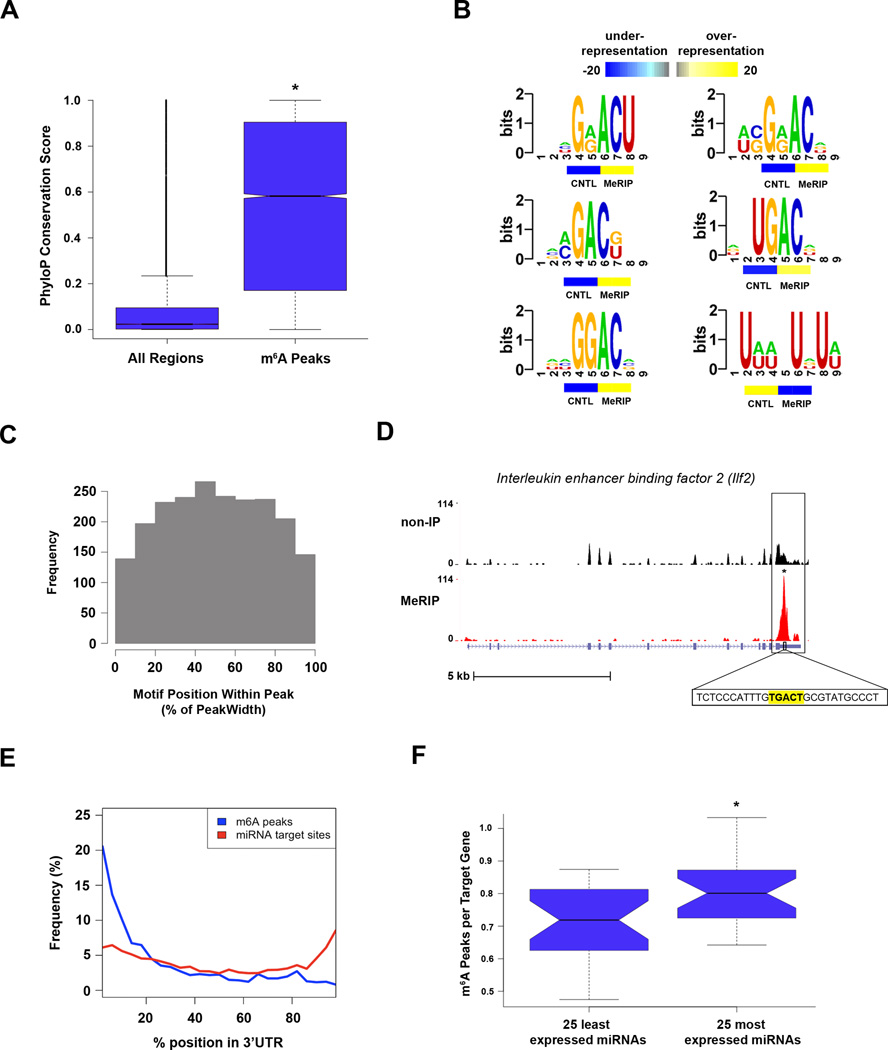
We next asked if m6A sites are conserved across species. We compared PhyloP scores across 30 vertebrates (Pollard et al., 2010) of m6A peak regions to those of random regions of the same size in gene exons. We found that the distribution of conservation scores of the m6A peaks was significantly different from that of the random regions (p ≤ 2.2×10−16, Kolmogorov-Smirnov (K-S) test, Figure 6A) and that m6A peaks’ median conservation score (0.578) was much higher than that of the random regions (0.023). The fact that m6A frequently occurs in evolutionarily conserved sequences suggests that m6A-containing regions are maintained through selection pressure.
Figure 6. MeRIP-Seq Reveals Features of m6A in mRNA.
A. Phylogenetic conservation of m6A peaks. PhyloP scores of m6A peak regions were compared to those of randomly shuffled regions throughout gene exons. There was a significantly higher median conservation score (K-S test, * p ≤ 2.2e−16) in m6A peaks (0.578) than in the random regions (0.023).
B. Sequence motifs identified within m6A peaks. The motif G[AG]ACU and variants thereof ([AC]GAC[GU], GGAC, [AU][CG]G[AG]AC, and UGAC) was highly enriched in m6A peaks. Additionally, one U-rich motif (bottom right) was identified as being significantly underrepresented within m6A peaks. Color bars under each motif indicate the degree of underrepresentation (blue) or overrepresentation (yellow) within regions of m6A peaks in the non-IP control sample (CNTL) and the MeRIP sample (MeRIP).
C. m6A motif sequences frequently lie near the center of m6A peaks. Shown is a plot of the cumulative distribution of m6A motif positions within m6A peaks containing a single motif. Motifs cluster in the center of peaks, suggesting that the methylated adenosines in these motifs account for the m6A peaks identified in MeRIP-Seq.
D. Example of a m6A motif sequence near the center of a peak. UCSC Genome Browser plot containing tracks for MeRIP-Seq reads (red) and non-IP control reads (black) at the Ilf2 locus. The m6A peak within the Ilf2 3’ UTR contains a single m6A motif identified in (B). The sequence of this motif (highlighted in yellow) is located at the center of the m6A peak.
E. Distribution of m6A peaks and miRNA target sites within 3’ UTRs. The frequency of m6A peaks (blue) and miRNA target sites (red) along the length of 3’ UTRs is shown.
F. Association between 3’ UTR methylation and miRNA abundance. The 25 most abundant miRNAs in brain have a significantly greater percentage of m6A peaks within their target mRNA 3’ UTRs than do the 25 most weakly expressed brain miRNAs (*p<0.05, Wilcoxon test). The error bars in A and F indicate the highest and lowest values, and the box boundaries denote the 1st quartile, median, and 3rd quartile.
See also Figures S3, S6, and S7.
Because the tools for transcriptome-wide localization of m6A sites have until now been unavailable, only a few studies to date have examined the sequence contexts of m6A formation (Canaani et al., 1979; Dimock and Stoltzfus, 1977; Wei et al., 1976). Using methods such as RNase T1 fingerprinting of radiolabeled RNA followed by separation by thin-layer chromatography, these studies reported that m6A exists within two unique sequence contexts: GAC and AAC (underlined adenosines indicate m6A). Subsequently, an extended m6A consensus sequence was identified: PuPuACX (Pu= purine; X= A, C, or U). However, since the methods used in these studies are not practical for use in a high-throughput manner, it is unclear whether these motifs are relevant to the transcriptome-wide m6A sites identified by MeRIP-Seq.
We therefore sought to identify sequence motifs that are enriched within m6A peaks. To do this, we used FIRE, a sensitive and unbiased tool for discovering RNA regulatory elements (Elemento et al., 2007). Remarkably, FIRE independently identified the GAC and AAC motif, G[AG]ACU, and related variants ([AC]GAC[GU], GGAC, [AU][CG]G[AG]AC, and UGAC) as being highly enriched in m6A peaks (Figure 6B). For example, the G[AG]ACU motif occurs in 42% of all m6A mRNA peaks and in a much lower fraction (21%) of non-m6A control peaks from the same mRNAs (p<1.0 × 10−124, chi-square test). Altogether, we found that >90% of all m6A peaks contain at least one of the motifs identified by FIRE.
We next examined the position of the motifs within m6A peaks. Nearly 30% of m6A peaks have only one motif (Figure S6B), indicating that these peaks are likely to contain only a single methylated residue. Motifs are also preferentially found in the center of m6A peaks (Figure 6C, D), suggesting that these peaks derive from a centrally located methylated adenosine residue. Notably, other RNA regulatory elements, such as AU-rich elements, poly(A) signals, or binding sites for known RNA-binding proteins, were not identified by FIRE, suggesting that m6A is unlikely to primarily function by modifying these known regulatory elements.
Relationship Between m6A Sites and Polyadenylation Signals in 3’ UTRs
FIRE did not identify an enrichment of poly(A) signals (PASs), which are involved in 3’ UTR end processing, in m6A peaks. However, PASs exhibit considerable sequence heterogeneity beyond the canonical AAUAAA consensus (Tian et al., 2005). This sequence heterogeneity might allow these PASs to evade detection by FIRE, despite being enriched in m6A peaks. Therefore, we sought to further investigate whether m6A peaks within 3’ UTRs are enriched at PASs. We obtained a high-confidence list (Brockman et al., 2005) of poly(A) cleavage sites (the site downstream of a PAS where the mRNA is actually cleaved and polyadenylated) for the mRNAs that contain m6A peaks within their 3’ UTRs. We then examined whether m6A peaks were enriched near these sites by determining the number of 3’ UTR m6A peaks that fell within 50 nt upstream of each cleavage site. Since a PAS is located approximately 10–30 nt upstream of an actual mRNA cleavage site (reviewed in Proudfoot, 1991), these 50 nt-long regions are expected to contain the PAS. Of the 6,288 m6A peaks found within 3’ UTRs, 1,042 (16.6%) overlapped with the 50 nt-long regions upstream of poly(A) cleavage sites, compared to 1,070 (17.0%) control peaks, which were generated from random nonoverlapping regions of the same 3’ UTRs. Thus, these data suggest that m6A do not have a significant association with known PASs (p = 0.39; chi-square test).
m6A is Not Enriched at Splice Junctions
Prior studies that used nonspecific methylation inhibitors to explore possible functions for m6A revealed impaired splicing in a small number of RNAs (Carroll et al., 1990; Stoltzfus and Dane, 1982). We therefore asked whether the localization of m6A peaks is compatible with a role for influencing the binding of splicing factors. However, only 80 splice junctions were found in regions contiguous with m6A peaks, significantly less than the overlap seen with a set of randomly-generated peaks (9,531; p=0.0; chi-square test). Thus, unlike CLIP-Seq tag clusters from RNA-binding proteins that influence splicing (Licatalosi et al., 2008), m6A peaks did not significantly coincide with exon-exon junctions, suggesting that m6A is unlikely to primarily function to directly influence the binding of splicing factors.
Relationship Between m6A and MicroRNA Binding Sites Within 3’ UTRs
The strong enrichment of m6A peaks in 3’ UTRs prompted us to investigate whether m6A peaks are found near microRNA (miRNA) binding sites, which are also frequently observed within 3’ UTRs. We found that 67% of 3’ UTRs that contain m6A peaks also contain at least one TargetScan-predicted miRNA binding site. Since ~30% of genes have miRNA binding sites in their 3’ UTRs (Lewis et al., 2005), this is a significantly greater association than what would be expected by chance alone. Intriguingly, we also found that in 3’ UTRs with both m6A peaks and miRNA binding sites, the m6A peaks precede miRNA binding sites 62% of the time. Moreover, we found that the overall distribution of m6A peaks and miRNA binding sites within 3’ UTRs are anti-correlated; while m6A peaks are most abundant near the stop codon and generally decrease in frequency along 3’ UTR length, miRNA target sites are more enriched near the 3’ end of 3’ UTRs (Figure 6E). The reason for this inverse localization pattern is unknown, although it could indicate that a certain spatial separation is necessary for m6A to influence the function of a downstream bound miRNA or vice versa.
We next sought to determine whether miRNA-targeted transcripts in the brain are more likely to contain m6A. To test this, we used TargetScan to identify the target transcripts of the 25 most highly expressed and 25 least highly expressed miRNAs within the brain. Intriguingly, we observed that the most highly expressed miRNAs have a significantly greater percentage of target transcripts that contain m6A (p<0.05, Wilcoxon test; Figure 6F). These data suggest that miRNA levels may control methylation of their target transcripts.
Prominent Features of m6A Distribution are Conserved in the Human Transcriptome
We next asked whether the enrichment of m6A in the 3’ UTR is also observed in other species. We therefore profiled m6A in HEK293T cells, a human cell line with high levels of adenosine methylation (Figure 3B). We generated a high-confidence list of m6A peaks using three MeRIP-Seq biological replicates and confirmed by both m6A antibodies. We found that the distribution of m6A peaks in HEK293T cells closely mirrored the distribution in mouse brain, with 31% and 53% of m6A peaks falling within the 3’ UTR and the CDS, respectively (Figure 5D, Figure S7D). As with the pattern of m6A distribution in the mouse brain transcriptome, HEK293T m6A peaks were predominantly localized near stop codons (Figure 5D and Figure S7C).
In total, we identified 18,756 peaks in RNAs encoded by 5,768 genes in HEK293T cells (Table S6). Additionally, we found that transcripts from 2,145 and 3,259 genes were methylated only in the mouse brain and HEK293T datasets, respectively, and that transcripts from 2,509 genes were methylated in both datasets (Table S6B). Interestingly, among the transcripts methylated in both tissues, m6A peaks were often localized to the same distinct regions of both orthologs (Figure 5E). Collectively, these data indicate that m6A peaks are enriched near the stop codon in human transcripts and that many sites of methylation are conserved in mouse and human transcripts.
DISCUSSION
Unlike DNA, which undergoes cytosine methylation and hydroxymethylation, dynamic internal modifications of mRNA other than RNA editing have not been established. Recent evidence that the obesity risk gene, FTO, is a physiologic m6A demethylase suggests that m6A has central roles in cellular function. Here we use MeRIP-Seq to provide the first transcriptome-wide characterization of m6A. We show that m6A is a reversible and widespread modification which is primarily located in evolutionarily conserved regions and is particularly enriched near the stop codon. We also find that many features of m6A localization are conserved between the human and mouse transcriptomes, and we uncover a previously unidentified link between m6A and miRNA signaling. Collectively, these studies reveal that m6A is a widespread and dynamically regulated base modification in mRNA, and they identify mRNAs which are most likely to be influenced by signaling pathways that influence m6A levels.
One of the most striking features of m6A localization is its prevalence within 3’ UTRs. The 3’ UTR is an important region for RNA regulation, as it can influence RNA stability, subcellular localization, and translation regulation. Several of these events are regulated by RNA binding proteins (RBPs) that bind to cis-acting structural motifs or consensus sequences within the 3’ UTR and act to coordinate RNA processing. Conceivably, m6A may influence the affinity of specific RBPs for their target mRNAs, analogous to the recruitment of methyl-CpG binding protein 2 (MeCP2) to methylated cytosine residues in DNA (Lewis et al., 1992). Given the abundance of m6A throughout the transcriptome and its widespread localization, such a role for m6A would be likely to have important consequences for the regulation of numerous mRNAs.
Our profiling of m6A in HEK293T cells revealed thousands of transcripts that are also methylated in the mouse brain. In many cases, the patterns of m6A localization within these transcripts are nearly identical, suggesting that some RNAs possess highly conserved methylation profiles. However, we also uncovered many transcripts that exhibit distinct cell type-specific methylation patterns, demonstrating that m6A is also capable of being differentially regulated within unique cellular environments.
Our finding that a large proportion of 3’ UTRs that contain m6A peaks also contain miRNA binding sites is highly suggestive of an association between m6A and miRNA function. Additionally, our analysis also indicated an inverse localization of m6A peaks and miRNA binding sites within 3’ UTRs, with m6A sites typically preceding, but not overlapping, the miRNA sites in the 3’ UTRs. Although miRNAs can inhibit their target mRNAs by promoting either transcript degradation or translational repression (Guo et al., 2010; Hendrickson et al., 2009), the factors that determine which fate predominates are not well understood. Conceivably, the proximity of m6A to a miRNA binding site could influence the mechanism of miRNA-mediated transcript inhibition. Additionally, it is possible that miRNA binding influences m6A levels within 3’ UTRs. Indeed, our finding that abundant miRNAs are more significantly enriched in m6A peaks than weakly expressed miRNAs raises the possibility that miRNAs regulate methylation status.
A surprising result of these studies is the finding that m6A is highly enriched near stop codons. This recurrent localization within transcripts suggests that adenosine methylation in the vicinity of the stop codon may be of functional importance. Interestingly, the consensus for adenosine methylation is relatively short, and sequences that match the consensus are found throughout the transcriptome. However, despite the frequency of m6A consensus sites, methylation occurs primarily near stop codons. It will be important to determine how this specificity is achieved and whether cellular mechanisms that involve recognition of the stop codon or the beginning of the 3’ UTR are involved in providing specificity to adenosine methylation.
The finding that FTO demethylates m6A suggests that misregulation of pathways controlled by adenosine methylation ultimately affect physiologic processes in humans. Although m6A is found in many classes of RNA, it is intriguing to speculate that FTO mutations mediate their effects by affected m6A in mRNA. Indeed, our finding that FTO can demethylate diverse mRNAs is consistent with this model. Direct characterization of m6A profiles in patients with FTO mutations will be useful to establish the mechanisms by which this mutation leads to disease.
In summary, our study demonstrates that m6A is a widespread modification found in a large fraction of cellular mRNA. The pervasive nature of this modification suggests that adenosine methylation has important roles in RNA biology. Much how cytosine methylation and hydroxymethylation in DNA are important epigenetic regulators of the genome, our data demonstrate that adenosine methylation in RNA is a reversible modification that is likely to influence a wide variety of biological pathways and physiological processes.
EXPERIMENTAL PROCEDURES
MeRIP-Seq
Total mouse brain RNA was subjected to RiboMinus treatment to reduce rRNA content as per the manufacturer’s instructions (Invitrogen). RNA was then fragmented to 100 nt-sized fragments using Illumina Fragmentation Buffer according to the manufacturer’s instructions, and subjected to two rounds of m6A immunoprecipitation. Sequencing libraries were prepared using the Illumina protocol for mRNA samples, and sequencing was performed on an Illumina GAIIx or an Illumina HiSeq2000 as indicated. Genomic alignment (mm9 from UCSC genome browser) was done using the Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2010) at default settings. We analyzed only those reads which (1) uniquely mapped to the genome and (2) had a Phred quality score greater or equal to 20.
Additional methods are detailed in Extended Experimental Procedures.
Supplementary Material
Highlights.
m6A is a widespread RNA modification in many tissues with high levels in the brain
MeRIP-Seq identifies m6A in 7,913 genes encoding both coding and noncoding RNAs
m6A is enriched near stop codons and within 3' UTRs in both mouse and human mRNAs
The transcriptome-wide landscape of m6A provides important insights into m6A function
ACKNOWLEDGMENTS
We thank Dr. Richard Roberts and New England Biolabs for generously providing the m6A (NEB) antibody. We also thank members of the Jaffrey and Mason laboratories for helpful comments and suggestions. This work was supported by NIH grants T32CA062948 (K.D.M.), 1F32MH095353-01 (K.D.M.), 1R01NS076465-01 (C.E.M.) and MH080420 (S.R.J.), NSF CAREER grant 1054964 (O.E.), and the Starr Cancer Consortium (I4-A411; C.E.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
MeRIP-Seq data have been deposited in the GEO database under accession number GSE29714.
REFERENCES
- Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113:165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann P, Luhrmann R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987;213:309–315. doi: 10.1016/0014-5793(87)81512-0. [DOI] [PubMed] [Google Scholar]
- Brockman JM, Singh P, Liu D, Quinlan S, Salisbury J, Graber JH. PACdb: PolyA Cleavage Site and 3'-UTR Database. Bioinformatics. 2005;21:3691–3693. doi: 10.1093/bioinformatics/bti589. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani D, Kahana C, Lavi S, Groner Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979;6:2879–2899. doi: 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Lee S, Bagg EA, McTaggart JS, Deacon R, Gerken T, Lee A, Moir L, Mecinovic J, Quwailid MM, et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 2009;5:e1000599. doi: 10.1371/journal.pgen.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Bruning JC, Nolan PM, Ashcroft FM, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csepany T, Lin A, Baldick CJ, Jr, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem. 1990;265:20117–20122. [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977;16:471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Klungland A, Alseth I. Repair of methyl lesions in DNA and RNA by oxidative demethylation. Neuroscience. 2007;145:1222–1232. doi: 10.1016/j.neuroscience.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, Ruther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami Y, Brown GM. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch Biochem Biophys. 1968;126:8–15. doi: 10.1016/0003-9861(68)90553-5. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011 doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagsbrun M. An evolutionary study of the methylation of transfer and ribosomal ribonucleic acid in prokaryote and eukaryote organisms. J Biol Chem. 1973;248:2612–2620. [PubMed] [Google Scholar]
- Kong H, Lin LF, Porter N, Stickel S, Byrd D, Posfai J, Roberts RJ. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 2000;28:3216–3223. doi: 10.1093/nar/28.17.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns TW, Liszewski MK, Sims HF. Characterization of antibodies specific for N6-methyladenosine and for 7-methylguanosine. Biochemistry. 1977;16:2163–2168. doi: 10.1021/bi00629a019. [DOI] [PubMed] [Google Scholar]
- Narayan P, Rottman FM. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Engstrom PG, Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS. RNAdb--a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus. Cell. 1975;4:387–394. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Saneyoshi M, Harada F, Nishimura S. Isolation and characterization of N6-methyladenosine from Escherichia coli valine transfer RNA. Biochim Biophys Acta. 1969;190:264–273. doi: 10.1016/0005-2787(69)90078-1. [DOI] [PubMed] [Google Scholar]
- Stoltzfus CM, Dane RW. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J Virol. 1982;42:918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. 5'-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15:397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.