Abstract
Postsynaptic density 93 (PSD-93) is a protein enriched at postsynaptic sites. As a key scaffolding protein, PSD-93 forms complexes with the clustering of various synaptic proteins to construct postsynaptic signaling networks and control synaptic transmission. Extracellular signal-regulated kinase (ERK) is a prototypic member of a serine/threonine protein kinase family known as mitogen-activated protein kinase (MAPK). This kinase, especially ERK2 isoform, noticeably resides in peripheral structures of neurons, such as dendritic spines and postsynaptic density areas, in addition to its distribution in the cytoplasm and nucleus, although little is known about specific substrates of ERK at synaptic sites. In this study, we found that synaptic PSD-93 is a direct target of ERK. This was demonstrated by direct protein-protein interactions between purified ERK2 and PSD-93 in vitro. The accurate ERK2-binding region seems to locate at an N-terminal region of PSD-93. In adult rat striatal neurons in vivo, native ERK from synaptosomal fractions also associated with PSD-93. In phosphorylation assays, active ERK2 phosphorylated PSD-93. An accurate phosphorylation site was identified at a serine site (S323). In striatal neurons, immunoprecipitated PSD-93 showed basal phosphorylation at an ERK-sensitive site. Our data provide evidence supporting PSD-93 as a new substrate of the synaptic species of ERK. ERK2 possesses the ability to interact with PSD-93 and mediate phosphorylation of PSD-93 at a specific site.
Keywords: PDZ, PSD-95, MAPK, JNK, synapse, signaling, striatum
1. Introduction
Postsynaptic density 93 (PSD-93) is a member of a superfamily of proteins containing PSD-95/DLG/Zo-1 (PDZ) domains. Proteins in this family, including PSD-95 and synapse-associated protein 97/102 (SAP97/102), usually have three tandem PDZ domains in their N-terminal regions, a Src homology (SH) domain in the middle, and a guanylate kinase (GK) domain at C-terminal ends (Fujita and Kurachi, 2000; Fitzjohn et al., 2006; Zheng et al., 2011). In neurons, PDZ proteins interact with a large number of cytoskeleton, receptor/transporter, and enzymatic proteins at synaptic sites. Through direct interactions, they support and maintain synaptic networks and participate in synaptic transmission (Kim and Sheng, 2004; Xu, 2011). To date, PSD-93 has been found to interact with a number of proteins, including NMDA receptor NR2A/2B subunits (Brenman et al., 1996; Kim et al., 1996), nitric oxide synthase (Kornau et al., 1995), K+ channels (Leyland and Dart, 2004; Ogawa et al., 2008), microtubule-associated protein 1A (Brenman et al., 1998), and ErbB-4 receptor tyrosine kinases (Garcia et al., 2000).
Interactions between protein kinases and PDZ proteins are thought to promote posttranslational modifications such as phosphorylation. In fact, PSD-95 is phosphorylated at serine 73 by Ca2+/calmodulin-dependent protein kinase II (CaMKII). This phosphorylation reduced synaptic expression of PSD-95 and inhibited spine growth and synaptic plasticity (Steiner et al., 2008). In addition, PSD-95 is subject to phosphorylation at serine 295 by c-Jun N-terminal kinase (JNK), which enhanced synaptic accumulation of modified PSD-95 and potentiated excitatory postsynaptic currents (Kim et al., 2007). Another PDZ protein SAP-97 is phosphorylated at serine 232 by CaMKII, which significantly impacted its binding to NR2A (Gardoni et al., 2003; Nikandrova et al., 2010). Compared to PSD-95 and SAP97, PSD-93 has been less investigated on its phosphorylation modification. Proteomic screen indicates that PSD-93 is phosphorylated at serine residues (Jaffe et al., 2004; DeGiorgis et al., 2005) and tyrosine sites (Nada et al., 2003), although responsible protein kinases catalyzing PSD-93 phosphorylation at specific site(s) are unknown.
Extracellular signal-regulated kinase (ERK) is a prototypic member of the mitogen-activated protein kinase (MAPK) family. This kinase, once activated, translocates from the cytoplasm to the nuclear envelop to phosphorylate discrete transcription factors such as Elk-1and cAMP response element-binding protein. It then modulates a distinct set of gene expression and transcriptionally regulates cellular and synaptic activities (Cagnol et al., 2010). Additionally, ERK resides in peripheral structures of neurons, including the PSD (Suzuki et al., 1995; 1999; Boggio et al., 2007). It is this subcellular (synaptic) pool of ERK that we know little about its interacting partners and phosphorylation substrates, while synaptic ERK is deemed to interact with and phosphorylate multiple targets at synaptic sites.
To search for a new interacting partner of ERK among prime synaptic proteins, we explored the possibility of PSD-93 in this study. We tested the existence of direct binding between purified ERK and PSD-93 proteins in vitro. We also assayed physical interactions of native ERK and endogenous PSD-93 in synaptosomal fractions from adult rat brains in vivo. Finally, we investigated phosphorylation responses of recombinant and native PSD-93 to ERK and identified an accurate phosphorylation site in PSD-93.
2. Results
2.1. ERK binds to PSD-93
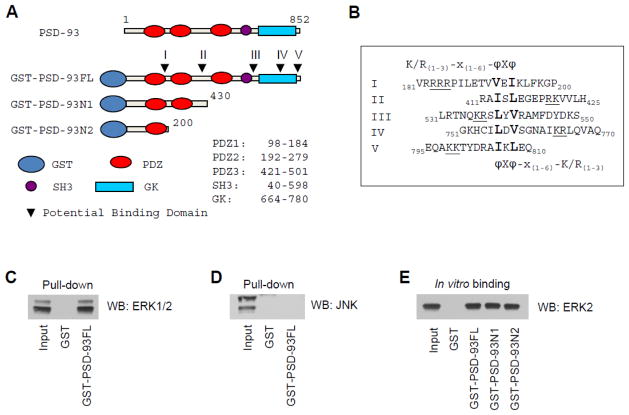
PSD-93 is enriched in the PSD and functions as a scaffolding protein. This protein has the ability to interact with a number of cytosolic molecules and membrane receptors; however, the relationship between PSD-93 with synaptic ERK has not been investigated. Here we attempted to explore whether ERK binds to PSD-93. A conserved ERK binding domain knowingly contains a group of amino acids: (K/R)1–3-X1–6-ϕ-X-ϕ. X represents any amino acid, while ϕ is a hydrophobic residue. Lysine (K) and arginine (R) can reside at either side. In amino acid sequence analysis of PSD-93, we found five regions potentially accommodating ERK binding (Fig. 1A and 1B). To determine the existence of ERK binding to PSD-93, we synthesized a GST-tagged full-length (FL) PSD-93 protein, GST-PSD-93FL(M1-L852). used this GST-fusion protein as bait in GST pull-down assays with adult rat striatal homogenates. We found that PSD-93 consistently pulled down ERK (Fig. 1C). -93 seemed to primarily pull down ERK2 isoform with a significantly less amount of ERK1. The selectivity of PSD-93 in pulling down ERK was confirmed by the lack of PSD-93 in precipitating JNK (Fig. 1D). To identify the region responsible for ERK binding and determine whether the ERK-PSD-93 interaction is direct, we produced two truncated GST-fusion proteins: GST-PSD-93N1(M1-G430) and GST-PSD-93N2(M1-P200) (Fig. 1A). These truncates were separately incubated with recombinant ERK2. As shown in Fig. 1E, GST-PSD-93FL and two truncated fragments all precipitated ERK2, while GST alone did not. Since ERK2 bound to PSD-93N2(M1-P200) to an extent comparable to PSD-93FL, we reasoned that a region in N-terminus of PSD-93(M1-P200) is responsible for ERK binding. In support of this, this region contains a well-conserved binding domain for ERK (183RRRPILETVVEI194).
Figure 1. Interactions between ERK and PSD-93.
(A) Structure domains of full length (FL) PSD-93. Arrowheads point to potential ERK binding domains in PSD-93. (B) Amino acid sequences of five possible ERK binding domains (I-V). K/R(1–3)-X(1–6)-ϕ-X-ϕ is the conserved sequence for ERK binding. (C and D) Pull-down assays showing that GST-PSD-93FL precipitated ERK1/2 (C), but not JNK (D), from rat striatal homogenates. (E) In vitro binding assays with immobilized GST-fusion proteins and purified ERK2. Note that GST-PSD-93FL and two fragments (N1 and N2) effectively precipitated ERK2, while GST alone did not. Proteins bound to GST-fusion proteins in pull-down and binding assays were visualized by Western blot (WB) with an antibody indicated.
2.2. ERK2 phosphorylates PSD-93
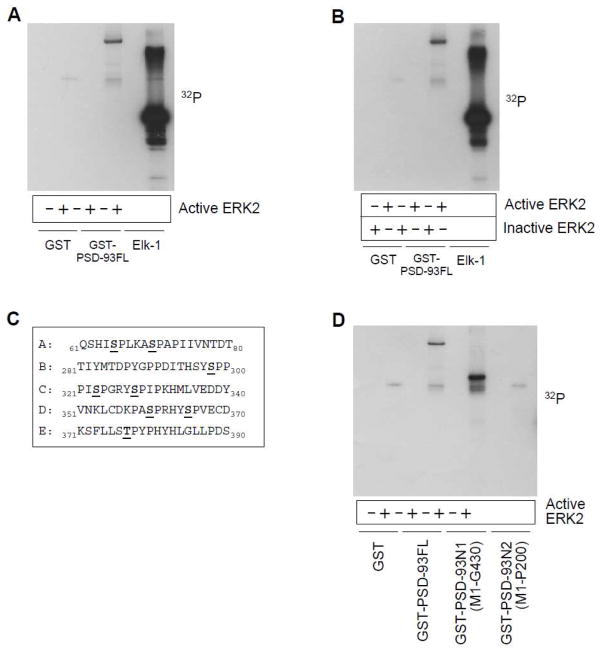
The direct binding of ERK2 to PSD-93 renders the opportunity for ERK2 to phosphorylate PSD-93. Sequence analysis shows that PSD-93 possesses eight possible phosphorylation sites specific for ERK (serine or threonine followed by proline, S/TP). To test ERK2-mediated phosphorylation of PSD-93, we investigated whether the free 32P isotope can be incorporated into PSD-93FL by active ERK2 in phosphorylation reactions carried out in vitro. GST-PSD-93 showed a clear phosphorylation band while GST alone did not (Fig. 2A). The transcriptional factor Elk-1, a well-known substrate of ERK2, also exhibited strong signals. Elk-1 is phosphorylated at multiple sites and served as a positive control in this experiment. These results support that PSD-93 is subject to the phosphorylation by ERK2. further confirm our observations, inactive ERK2 was used and compared with active ERK2 in the efficacy of phosphorylating PSD-93. As expected, no radioactive signals were detected in PSD-93 in the presence of inactive ERK2 (Fig. 2B).
Figure 2. ERK2-mediated phosphorylation of PSD-93 in vitro.
(A) Active ERK2 phosphorylated PSD-93 and Elk-1. (B) Active but not inactive ERK2 phosphorylated PSD-93 and Elk-1. (C) PSD-93 fragments containing potential phosphorylation sites. (D) Phosphorylation of PSD-93FL and fragments (N1 and N2) by active ERK2. Note that PSD-93FL and PSD-93N1 were phosphorylated, while PSD-93N2 was not. Phosphorylation reactions were carried out at 30°C for 30 min with [γ-32P]ATP. The reactions were then subjected to gel electrophoresis followed by autoradiograph.
The eight possible phosphorylated sites are distributed in N-terminal 1–390 amino acids of PSD-93 (Fig. 2C). To identify accurate site(s) of phosphorylation, we tested ERK2-mediated phosphorylation of truncated PSD-93 proteins. As can be seen in Fig. 2D, while PSD-93FL and the N1 fragment (M1-G430) were phosphorylated by ERK2, the N2 fragment (M1-P200) was not. Thus, a region from 200 to 390 amino acids seems to harbor an ERK-sensitive phosphorylation site.
2.3. ERK2 phosphorylates PSD-93 at S323
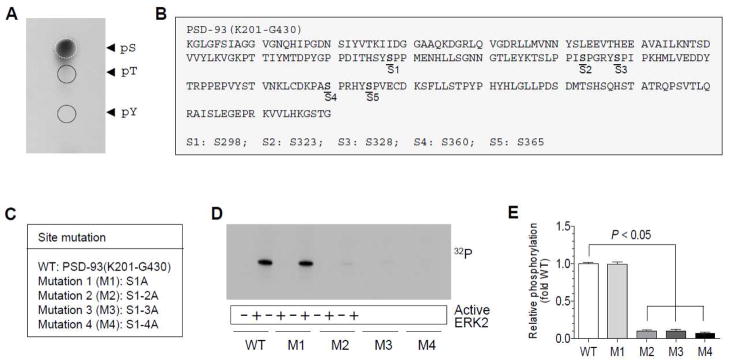
Above results indicate that ERK2-mediated phosphorylation of PSD-93 occurs in a confined region. We next wanted to identify accurate site(s) in this region for phosphorylation. To determine whether serine or threonine is phosphorylated by the serine/threonine protein kinase ERK2, we performed phosphoamino acid analysis (PAA) with the phosphorylation-positive fragment of PSD-93, i.e., PSD-93(K201-G430). We found that serine was strongly phosphorylated by ERK2 (Fig. 3A). In contrast, threonine and tyrosine phosphorylation was nearly undetectable. Thus, serine residues rather than threonine and tyrosine sites are phosphorylated in PSD-93. It is noticeable that the K201-G430 fragment contains five serine sites (S298, S323, S328, S360, and S365) (Fig. 3B). To identify exact serine site(s) responsible for phosphorylation, we carried out phosphorylation assays with site-directed mutants. Upon five serine sites, we generated four mutants (M1-M4) (Fig. 3C) and subsequently tested their phosphorylation responses to ERK2. The wild type (WT) and M1 mutant were similarly phosphorylated by ERK2 (Fig. 3D and 3E)., M2 mutants showed a deficit of phosphorylation and so did M3 and M4 mutants (Fig. 3D and 3E). M2 contains an additional mutation at S323 compared to M1. The mutation of this site seems to cause a nearly complete loss of phosphorylation signals. Therefore, we reasoned that S323 is a primary phosphorylation site in PSD-93 in response to ERK2.
Figure 3. ERK2-mediated phosphorylation of PSD-93 at S323.
(A) PAA of PSD-93 after phosphorylation by ERK2. Note that strong phosphorylation signals were shown at serine (pS), but not at threonine (pT) and tyrosine (pY), residues. (B) Potential serine phosphorylation sites within the PSD-93(K201-G430) region. (C) Wild type (WT) and four mutants derived from the PSD-93(K201-G430) fragment. Serines (S) were mutated to alanines (A) in a stepwise manner (M1: S298A; M2: S298/323A; M3: S298/323/328A; and M4: S298/323/328/360A). (D) Phosphorylation of WT and M1–4 mutants in phosphorylation reactions containing active ERK2. Note that M1 was phosphorylated to a level comparable to WT, while M2, M3, or M4 was insensitive in phosphorylation responses to ERK2. (E) A graph showing relative phosphorylation levels of WT and M1–4 mutants in response to ERK2. Data are presented as means ± SEM (n = 3 per group).
2.4. S323 phosphorylation detected by a motif antibody
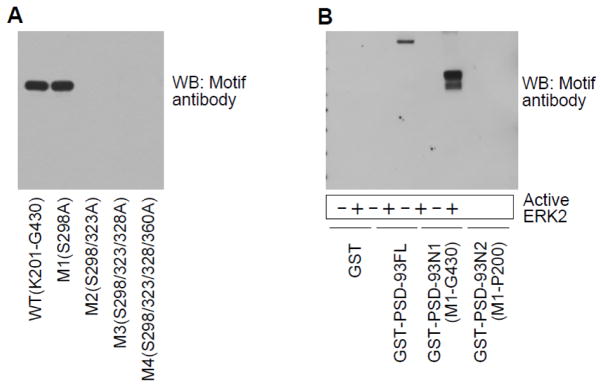
Amino acid residues flanking S323 (321PIpSPGR326) match with the common ERK phosphorylation motif, PXpSP or pSPXR/K, X representing any amino acid. We then used a motif antibody against PXpSP/pSPXR/K to probe phosphorylation of PSD-93. As we can see in Fig. 4A, PSD-93(K201-G430) WT and M1 proteins showed comparable phosphorylation signals. M2, M3, and M4 mutants, however, had no signals detected by the motif antibody. We also noticed that M2 and M3 actually contain a sequence of PASP (S360). If phosphorylation occurred at this site, the motif antibody may visualize phosphorylation signals accordingly. However, the results showed that no signals were detected in M2 and M3 proteins (Fig. 4A), indicating that S360 was not phosphorylated. These data are consistent with those observed from above 32P autoradiographic experiments and thus solidify the notion that ERK2 phosphorylates PSD-93 at S323. Furthermore, the motif antibody was demonstrated to be sensitive enough to assess ERK2-mediated phosphorylation in PSD-93FL and PSD-93N1 (Fig. 4B). PSD-93N2, even though it contains a sequence of SPLK (S60), did not exhibit detectable phosphorylation signals.
Figure 4. PSD-93 S323 phosphorylation detected by a motif antibody.
(A) A representative Western blot (WB) showing phosphorylation of PSD-93(K201-G430) WT and corresponding mutants detected by the motif antibody. Note that only WT and M1 showed strong phosphorylation signals, while M2, M3 and M4 did not. (B) A representative WB showing phosphorylation of PSD-93FL and fragments (N1 and N2). Only WT and N1 fragments that contain S323 were phosphorylated by ERK2.
2.5. Interactions of ERK with PSD-93 in vivo
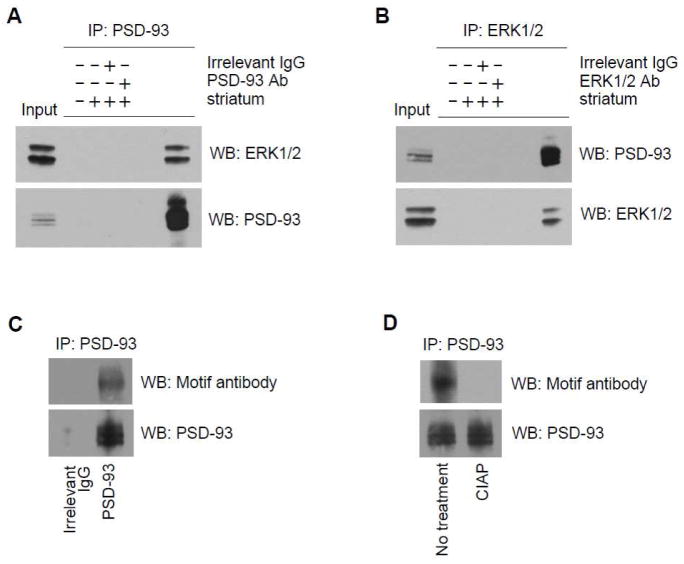
To determine whether native ERK interacts with endogenous PSD-93 in neurons in vivo, we carried out coimmunoprecipitation to examine the interaction between two proteins in adult rat striatal synaptosomal fractions. In coimmunoprecipitation assays with the PSD-93 antibody, we found that ERK1/2 coimmunoprecipitated with PSD-93 (Fig. 5A), demonstrating that these proteins are associated with each other in striatal neurons. In reverse coimmunoprecipitation assays with the ERK1/2 antibody, PSD-93 was found to coimmunoprecipitate with ERK1/2 (Fig. 5B). To explore whether ERK-mediated S323 phosphorylation occurs in native PSD-93, we used the motif antibody to probe phosphorylation of immunoprecipitated PSD-93. A clear signal band was detected in immunoprecipitated PSD-93 proteins (Fig. 5C)., in precipitated PSD-93 proteins pretreated with calf intestine alkaline phosphatase (CIAP), no phosphorylation signals were detected by the motif antibody (Fig. 5D). These results provide evidence that ERK interacts with PSD-93 and phosphorylates PSD-93 in neurons in vivo.
Figure 5. ERK associates with PSD-93 in striatal neurons in vivo.
(A) Immunoprecipitation (IP) with the PSD-93 antibody (Ab) showing coimmunoprecipitation of ERK1/2 and PSD-93. (B) IP with the ERK1/2 antibody showing coimmunoprecipitation of PSD-93 and ERK1/2. (C) Phosphorylation of immunoprecipitated PSD-93 from the rat striatum detected by the ERK motif antibody. (D) Dephosphorylation of immunoprecipitated PSD-93 by CIAP. Immunoprecipitated proteins were visualized by Western blot (WB) with an antibody indicated.
3. Discussion
ERK, especially ERK2, and PSD-93 are both locsated in the PSD (Suzuki et al., 1995; 1999). This spatial proximity provides the opportunity for them to interact with each other. In this study, we demonstrated that ERK directly binds to the N-terminal region of PSD-93 in vitro. binding may support and promote phosphorylation of PSD-93 by ERK. Indeed, active ERK2 was found to effectively phosphorylate PSD-93. The precise phosphorylation site seems to be S323 according to phosphorylation assays with site-directed mutant proteins. Native ERK also associates with PSD-93 in striatal neurons in vivo. The ERK phosphorylation motif antibody was able to detect phosphorylation signals in immunoprecipitation-purified PSD-93 proteins from striatal neurons. Together, our data demonstrate PSD-93 as a new substrate of ERK. Both in vitro and in vivo results support that ERK directly binds to PSD-93 and phosphorylates PSD-93 at a specific site.
Proteins in the PDZ family are characterized to contain multiple domains for protein-protein interactions. Through interactions, PDZ proteins organize a set of synaptic proteins into macromolecules to stabilize them at a specific subsynaptic microdomain and/or facilitate discrete synaptic signaling. Like other PDZ proteins, PSD-93 is believed to interact with a large number of synaptic proteins. Among various interacting proteins, protein kinases are particularly interesting. This is because a protein kinase may exert a phosphorylation-dependent regulation of PSD-93 via its interaction with PSD-93. In this study, we discovered that a MAPK family member, ERK, binds to PSD-93. This indicates that ERK is among kinases that can modulate PSD-93 via a direct interaction. It is noticeable that ERK2 binds to the N-terminal region likely flanking 183RRRPILETVVEI194, whereas it phosphorylates PSD-93 at a different site (S323). This is consistent with a traditional model that binding and phosphorylation sites are usually separate in most, in not all, substrates of ERK (Songyang et al., 1996).
We were interested in ERK-PSD-93 interactions for the following reasons. First, PSD-93 contains multiple well-conserved ERK binding domains, indicating that PSD-93 could be a potential interacting partner of ERK. Many known substrates of ERK, including Elk-1 and MEK1 (MAPK kinase 1), are directly interacted by ERK and are thereby efficiently phosphorylated by the kinase (Yang et al., 1998; Xu et al., 1999). Second, PSD-93 possesses eight proline-directed ERK phosphorylation motifs (S/TP), suggesting that PSD-93 could be a phosphorylation substrate of ERK. Finally, these two proteins are colocalized in the PSD (Suzuki et al., 1995; 1999). The spatial vicinity supports the intimate interaction between them. Indeed, our results support the existence of ERK-PSD-93 coupling. This coupling results in ERK2-mediated phosphorylation of PSD-93 at a serine site (S323). Of note, S323 lies in the center of a sequence (PISPGR) flanking a consensus ERK phosphorylation motif (SP). S323 phosphorylation seems to also occur in striatal neurons in vivo since phosphorylation of PSD-93 immunoprecipitated from the striatum was detected by a motif antibody against PXpSP/pSPXR/K. PSD-93 in the purified PSD fraction from adult rat brains has been reported to be phosphorylated at S365 by an unknown kinase in mass spectrometric analyses (Jaffe et al, 2004). this study, we did not observe reliable phosphorylation at S365 in site-directed mutation assays., phosphorylation of PSD-93 at S365 is catalyzed by a different proline-directed kinase.
It is currently unclear whether the ERK-mediated PSD-93 phosphorylation is a regulated event. is a well-known kinase highly sensitive to a variety of extracellular stimuli and changing synaptic inputs (Volmat and Pouyssegur 2001; Wang et al., 2007). Activity-dependent activation of ERK increases phosphorylation of its downstream substrates, leading to a significant modification of expression, location, and/or function of phosphorylated proteins. It is thought that the PSD-93 phosphorylation by ERK is linked to cellular and synaptic activity levels. ERK-dependent phosphorylation and regulation of PSD-93 join posttranslational modifications of other synaptic proteins to modulate efficacy and strength of synaptic transmission. These interesting questions will be investigated in future.
4. Experimental procedures
4.1. Animals
Adult male Wistar rats (225–275 g) from Charles River (New York, NY) were individually housed in a controlled environment at a constant temperature of 23°C and humidity of 50 ± 10% with food and water available ad libitum. The animal room was on a 12-h/12-h light/dark cycle. All animal use and procedures were in strict accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee.
4.2. Cloning, expression, and purification of GST-fusion proteins
The cDNA fragments for PSD-93FL(M1-L852), PSD-93N1(M1-G430), PSD-93N2(M1-P200), and PSD-93(K201-G420) were generated through polymerase chain reaction (PCR) using PCR kit with high fidelity (Invitrogen, Carsbad, CA). The forward or reverse primer was added with a nucleotide fragment in its 5′ end which can be digested by BamHI or EcoRI, respectively. Both PCR products and pGEX4T-3 plasmid (Amersham Biosciences, Arlington Heights, IL) were double digested by BamHI and EcoRI in 37°C for 3 h, and after purification (quick gel purification kit, Invitrogen), they were put together for ligation in the presence with T4 ligase (1 unit, Invitrogen) at 14°C with overnight incubation. The ligation products were transformed into fresh DH5α competent cells for clone growth. Individual clone was selected and cultured for extracting the recombinant GST plasmids (HQ mini-plasmid extraction kit, Invitrogen). These recombinant plasmids were transformed into BL21 prokaryotic cells for producing GST-fusion proteins according to standard protocol provided by the manufacturer.
4.3. Affinity purification (pull-down) assay
Solubilized striatal proteins (500 – 1000 μg) were diluted with PBS/1% Triton X-100 and incubated with 50% (v/v) slurry of glutathione–Sepharose 4B beads (Amersham) saturated with GST alone or an indicated GST-fusion protein (5 – 10 μg) for 2 – 3 h at 4°C. Beads were washed four times with PBS/1% Triton X-100. Bound proteins were eluted with 2X lithium dodecyl sulfate (LDS) sample buffer, resolved by SDS–PAGE, and immunoblotted with indicated antibodies.
4.4. In vitro binding assay
As described previously (Guo et al., 2010), his-tagged, purified ERK2 (Signal Chem, Richmond, Canada) were equilibrated to binding buffer (200 mM NaCl, 0.2% Triton X-100, 0.1 mg/ml BSA, and 50 mM Tris, pH 7.5). Binding reactions were initiated by adding purified GST or GST-fusion proteins and were maintained at 4°C for 2 – 3 h with occasionally gentle rotation. GST-fusion proteins were precipitated using 100 μl of 50% glutathione–Sepharose. The precipitate was washed three times with binding buffer. Bound proteins were eluted with 2X LDS loading buffer, resolved by SDS–PAGE, and immunoblotted with an indicated antibody.
4.5. Phosphorylation and dephosphorylation assays in vitro
GST and GST-fusion proteins were incubated with active ERK2 (Signal Chem, 25 ng) or inactive ERK2 (Millipore, Billerica, MA, 25 ng) for 30 min at 30°C in a volume of 25 μl of the reaction buffer containing 10 mM HEPES, pH 7.4, 10 mM MgCl2, 1 mM Na3VO4, 1 mM DTT, 50 μM ATP, and 2.5 μCi/tube [γ-32P]ATP (~3000 Ci/mmol, PerkinElmer, Waltham, MA). Reactions were stopped by adding LDS sample buffer (10 μl) and boiling for 3 min. Phosphorylated proteins in one-third reaction volume were resolved by SDS-PAGE, transferred to polyvinylidene fluoride membrane, and visualized by autoradiography. For dephosphorylation, rat striatal P2 samples were incubated with calf intestine alkaline phosphatase (5 units, Invitrogen) for 1 h at 37°C in reaction buffer containing 50 mM Tris-HCl, pH 8.5, 0.1 mM EDTA, 1 mM MaCl2, and 0.1 mM ZnCl2. The reaction was stopped by adding LDS sample buffer. Samples were used in the following Western blot analysis.
4.6. Phosphoamino acid analysis
Phosphorylated proteins (32P-incorporated proteins) were separated on an SDS-PAGE gel. These proteins were then transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was stained and the targeted band containing a protein of interest was cut for subsequent PAA. PAA was carried out by hydrolyzing the 32P-labeled phosphoprotein and separating the hydrolysate by one-dimensional thin-layer electrophoresis. Briefly, samples containing 32P-labeled proteins were hydrolyzed into individual amino acids in 6 N HCl (110°C, 2 h) and concentrated by drying samples in a Savant DNA120 SpeedVac (Thermo Scientific). Resuspended samples were then spotted onto glass-backed thin-layer chromatography (TLC) cellulose plates (Merck), along with phosphoserine, phoshothreonine, and phosphotyrosine standards. The phosphoamino acids were separated by electrophoresis in pH 3.5 buffer (5% v/v acetic acid, 0.5% v/v pyridine, 100 ml) in a flatbed Multiphor II electrophoresis apparatus (GE Healthcare) at 1000 V (30 mA) for 45 min. The phosphoamino acid standards were visualized with ninhydrin. The TLC plate was exposed to X-ray film to visualize the 32P-labeled amino acids.
4.7. Coimmunoprecipitation
Rats were anesthetized with Equithesin (5 ml/kg, i.p.) and decapitated. Brains were quickly removed. The dissected striatum was homogenized using a motor-driven glass-teflon homogenizer (10–15 strokes) in ice-cold homogenization buffer containing 0.32 M sucrose, 20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, 5 mM dithioerythritol, a protease inhibitor cocktail (Thermo Scientific), and a phosphatase inhibitor cocktail (Thermo Scientific). Samples were centrifuged at 1,000 g for 15 min at 4°C. Supernatants were centrifuged again at 12,000 g for 30 min at 4°C. The P2 pellets were resuspended and solubilized in a buffer containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 5 mM dithioerythritol, 1% NP-40, 1% sodium deoxycholate, a protease inhibitor cocktail, and a phosphatase inhibitor cocktail. They were stored on ice for 1 h incubation with occasional mixing. Resuspended pellets were centrifuged at 12,000 g for 30 min at 4°C. Supernatants were collected for coimmunoprecipitation. Samples were incubated with a rabbit antibody against PSD-93 (Invitrogen) or ERK1/2 (Cell Signaling, Danvers, MA) at 4°C for overnight. The complex was then incubated with 50% protein A or G agarose/sepharose bead slurry at 4°C for 2–3 h (Amersham). After centrifugation, agarose beads and protein complex were washed 3 times using sample buffer. The beads were added with 20 μl SDS-loading buffers and boiled in 94°C for 10 min for eluting proteins. Detached proteins were separated on Novex 4–12% gels and probed with indicated antibodies.
4.8. Western blot analysis
The equal amount of proteins was separated on SDS NuPAGE Novex 4–12% gels (Invitrogen). Proteins were transferred to the PVDF membrane (Millipore) and blocked in a blocking buffer (5% nonfat dry milk in phosphate-buffered saline and 0.1% Tween 20) for 1 h. The blots were washed and incubated in the blocking buffer containing a primary rabbit antibody against PSD-93 (Invitrogen), ERK1/2 (Cell Signaling), or a phospho-MAPK motif (PXpSP or pSPXR/K) (Cell Signaling, Cat. #:2325S). The incubation was carried out usually at 1:1000 overnight at 4°C. This was followed by 1 h incubation in a horseradish peroxidase-linked secondary antibody against rabbit 1:5000. were developed with the enhanced chemiluminescence reagents (ECL; Amersham). Kaleidoscope-prestained standards (Bio-Rad, Hercules, CA) or MagicMark XP Western protein standards (Invitrogen) were used for protein size determination.
4.9. Statistics
The results are presented as means ± S.E.M., and were evaluated using a one-way analysis of variance followed by a Bonferroni (Dunn) comparison of groups using least squares-adjusted means. Probability levels of p< 0.05 were considered statistically significant.
Highlights.
Purified ERK2 protein binds to PSD-93 at its N-terminal region in vitro
Active ERK2 phosphorylates PSD-93 at S323 site in vitro
Native ERK from synaptosomal fractions associates with PSD-93
In striatal neurons, immunoprecipitated PSD-93 shows basal phosphorylation at an ERK-sensitive site
Acknowledgments
This work was supported by NIH grants R01DA10355 (JQW) and R01MH61469 (JQW).
Footnotes
Conflict of interest: there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brenman JE, Christopherson KS, Craver SE, Mcgee AW, Bredt DS. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein. J Neurosci. 1996;16:7407–7415. doi: 10.1523/JNEUROSCI.16-23-07407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenman JE, Topinka JR, Cooper EC, Mcgee AW, Rosen J, Mirroy T, Ralston HJ, Bredt DS. Localization of postsynaptic density-93 to dendritic microtubules and interaction with microtubule-associated protein 1A. J Neurosci. 1998;18:8805–8813. doi: 10.1523/JNEUROSCI.18-21-08805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio EM, Putignano E, Sassoe-Pognetto M, Pizzorusso T, Glustetto M. Visual stimulation activates ERK in synaptic and somatic compartments of rat cortical neurons with parallel kinetics. PLoS ONE. 2007;2(7):e604. doi: 10.1371/journal.pone.0000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death-apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- Degiorgis JA, Jaffe H, Moreira JE, Carlotti CG, Jr, Leite JP, Dosemeci A. Phosphoproteomic analysis of synaptosomes from human cerebral cortex. J Proteome Res. 2005;4:306–315. doi: 10.1021/pr0498436. [DOI] [PubMed] [Google Scholar]
- Fitzjohn SM, Doherty AJ, Collingridge GL. Promiscuous interaction between AMPA-Rs and MAGUKs. Neuron. 2006;52:222–224. doi: 10.1016/j.neuron.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Fujita A, Karachi Y. SAP family proteins. Biochem Biophys Res Commun. 2000;269:1–6. doi: 10.1006/bbrc.1999.1893. [DOI] [PubMed] [Google Scholar]
- Gardoni F, Mauceri D, Fionrentini C, Bellone C, Missale C, Cattabeni F, Luca MD. CaMKII-dependent phosphorylation regulates SAP97/NR2A interaction. J Biol Chem. 2003;45:44745–44752. doi: 10.1074/jbc.M303576200. [DOI] [PubMed] [Google Scholar]
- Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Pro Natl Acad Sci USA. 2000;97:3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ML, Fibuch EE, Liu XY, Choe ES, Buch S, Mao LM, Wang JQ. CaMKIIα interacts with M4 muscarinic receptors to control receptor and psychomotor function. EMBO J. 2010;29:2070–2081. doi: 10.1038/emboj.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe H, Vinade L, Dosemeci A. Identification of novel phosphorylation sites on postsynaptic density proteins. Biochem Biophys Res Commun. 2004;321:210–218. doi: 10.1016/j.bbrc.2004.06.122. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Futai K, Jo J, Hayashi Y, Cho K, Sheng M. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron. 2007;56:488–502. doi: 10.1016/j.neuron.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Eennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Leyland ML, Dart C. An alternatively spliced isoform of PSD-93/chapsyn 110 binds to the inwardly rectifying potassium channel, Kir2.1. J Biol Chem. 2004;279:43427–43436. doi: 10.1074/jbc.M407575200. [DOI] [PubMed] [Google Scholar]
- Nada S, Shima T, Yanai H, Husi H, Grant SGN, Okada M, Akiyama T. Identification of PSD-93 as a substrate for the Src family tyrosine kinase Fyn. J Biol Chem. 2003;48:47610–47621. doi: 10.1074/jbc.M303873200. [DOI] [PubMed] [Google Scholar]
- Nikandrova YA, Jiao Y, Baucum AJ, Tavalin SJ, Colbran RJ. Ca2+/calmodulin-dependent protein kinase II binds to and phosphorylates a specific SAP97 splice variant to disrupt association with AKAP79/150 and modulate alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid –type glutamate receptor (AMPAR) activity. J Biol Chem. 2010;285:10376–10384. doi: 10.1074/jbc.M109.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Horrshe I, Trimmer JS, Bredt DS, Peles E, Rasband MN. Postsynaptic density-93 clusters Kv1 channels at axon initial segments independently of Caspr2. J Neurosci. 2008;28:5737–5739. doi: 10.1523/JNEUROSCI.4431-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T, Cantley LC. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference for casein kinase I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner P, Higley MJ, Xu W, Czervionke BL, Malenka RC, Sabatini BL. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron. 2008;60:788–802. doi: 10.1016/j.neuron.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Mitake S, Murata S. Presence of up-stream and downstream components of a mitogen-activated protein kinase pathway in the PSD of the rat forebrain. Brain Res. 1999;840:36–44. doi: 10.1016/s0006-8993(99)01762-x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Okumura-Noji K, Nishida E. ERK2-type mitogen-activated protein kinase (MAPK) and its substrates in postsynaptic density fractions from the rat brain. Neurosci Res. 1995;22:277–285. doi: 10.1016/0168-0102(95)00902-6. [DOI] [PubMed] [Google Scholar]
- Volmat V, Pouyssegur J. Spatiotemporal regulation of the p42/p44 MAPK pathway. Biol Cell. 2001;93:71–79. doi: 10.1016/s0248-4900(01)01129-7. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Fibuch EE, Mao LM. Regulation of mitogen-activated protein kinases by glutamate receptors. J Neurochem. 2007;100:1–11. doi: 10.1111/j.1471-4159.2006.04208.x. [DOI] [PubMed] [Google Scholar]
- Xu B, Wilsbacher JL, Collisson T, Cobb MH. The N-terminal ERK-binding site of MEK1 is required for efficient feedback phosphorylation by ERK2 in vitro and ERK activation in vivo. J Biol Chem. 1999;274:34029–34035. doi: 10.1074/jbc.274.48.34029. [DOI] [PubMed] [Google Scholar]
- Xu W. PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) and synaptic plasticity. Curr Opin Neurobiol. 2011;21:306–312. doi: 10.1016/j.conb.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Seabold GK, Horak M, Petralia RS. MAGIKs, synaptic development, and synaptic plasticity. Neuroscientist. 2011;17:493–512. doi: 10.1177/1073858410386384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]