Abstract
The sequence corresponding to the first 77 nucleotides of the L1Tc and NARTc non-LTR retrotransposons from Trypanosoma cruzi is an internal promoter (Pr77) that generates abundant, although poorly translatable, un-spliced transcripts. It has been recently described that L1TcRz, an HDV-like ribozyme, resides within the 5′-end of the RNA from the L1Tc and NARTc retrotransposons. Remarkably, the same first 77 nucleotides of L1Tc/NARTc elements comprise both the Pr77 internal promoter and the HDV-like L1TcRz. The L1TcRz cleaves on the 5′-side of the +1 nucleotide of the L1Tc element insuring that the promoter and the ribozyme functions travel with the transposon during retrotransposition. The ribozyme activity would prevent the mobilization of upstream sequences and insure the individuality of the L1Tc/NARTc copies transcribed from associated tandems. The Pr77/L1TcRz sequence is also found in other trypanosomatid’s non-LTR retrotransposons and degenerated retroposons. The possible conservation of the ribozyme activity in a widely degenerated retrotransposon, as the Leishmania SIDERs, could indicate that the presence of this element and the catalytic activity could play some favorable genetic regulation. The functional implications of the Pr77/L1TcRz dual system in the regulation of the L1Tc/NARTc retrotransposons and in the gene expression of trypanosomatids are also discussed in this paper.
Keywords: HDV-like ribozyme, L1Tc, LINE, SINE, Trypanosoma cruzi, genetic regulation, promoter, retrotransposition machinery, retrotransposons, trypanosomatid
Retrotransposons are DNA sequences able to mobilize their own copies within a host genome by transcription and reverse transcription of an intermediate RNA. The generation of this intermediate RNA is a crucial step for the mobilization mechanism. The LTR retrotransposons (with long terminal repeats at both ends) have an external RNA polymerase II promoter located upstream of the transcription initiation site. A complex and discontinuous reverse transcription process that requires two DNA strand transfers in addition to several enzymatic activities is necessary to regenerate the promoter1 (Fig. S1A). On the contrary, the retrotransposition mechanism described for non-LTR retrotransposons, called Target Primed Reverse Transcription (TPRT),2 consists of a single-step reverse transcription of the mRNA element and the synthesis of a cDNA strand to generate a new double-stranded DNA copy able to be inserted into a new position in the genome (Fig. S1B). Thus, the intermediate RNA of the non-LTR retrotransposons is likely to bear within its sequence the information required for its own transcription. Although internal promoters that satisfy these requirements have been described in a few elements,3-5 the way to generate the mRNAs by many others elements is still unknown. Eickbush, Lupták and coworkers recently described the system of the Drosophila R2 retrotransposon for the generation of mRNAs involving an HDV-like ribozyme coupled to an upstream host-promoter.6,7
Based on their coding capacity, the non-LTR retrotransposons are classified in two major groups: those that code for their own retrotransposition machinery or LINEs (long interspersed nuclear elements) and those that lack translation capability and are thought to mobilize using LINEs’ machinery in trans, like SINEs (short interspersed nuclear elements) and LINE truncated elements. A high copy number of these elements is present in the genome of the etiological agent of Chagas disease, the protozoan parasite Trypanosoma cruzi.8
The L1Tc (5 kb in length) is a non-LTR retrotransposon widely distributed along the T. cruzi genome. L1Tc is a LINE element that codes for the mobilization machinery including AP-endonuclease,9,10 reverse transcriptase,11 RNase H12 and nucleic acids chaperone activities.13,14 In addition, an active picornavirus-like 2A autoproteolytic motif resides at the N-terminal end of the encoded L1Tc polypeptide. The 2A autoproteolytic activity is expected to regulate the composition and abundance of the enzymatic machinery required for autonomous mobilization of L1Tc.15 NARTc (263 nt in length) is a non-autonomous element thought to be mobilized by the gen products of L1Tc.16 L1Tc and NARTc share 97% and 77% identity at their first 77 and last 30 nucleotides (including a short poly-A tail codified in the DNA), respectively. The conserved sequence located at their 3′-end is thought to be recognized by the reverse transcriptase encoded by the L1Tc element. The sequence corresponding to the first 77 nucleotides (Pr77) of L1Tc and NARTc has promoter activity and acts as an internal promoter that generates abundant, although poorly translatable, transcripts.17
HDV-Like Ribozymes Associated with Retrotransposons: L1TcRz and R2Rz. The Dual Promoter-Ribozyme System of L1Tc
We have recently described the L1TcRz, an hepatitis delta virus (HDV)-like ribozyme that resides within the 5′-end of the RNA of both the L1Tc and NARTc retrotransposons of T. cruzi (Fig. 1A).18 The ribozyme catalyzes the self-cleavage of the RNA sugar-phosphate backbone of its own molecule, leaving a 5′-hydroxyl end on the downstream product, and it is expected to leave a 2′,3′-clyclic phosphate end on the upstream cleavage product, similar to the HDV and other known HDV-like ribozymes.19 The cleavage position of L1TcRz is located immediately upstream of the +1 nucleotide of both L1Tc and NARTc. L1TcRz, then, is capable of determining the 5′-end of these two elements. The 5′-end is such that the ribozyme and the internal promoter sequence is present in the mature element RNA insuring their transmission to the new retrotransposon copy during TPRT process.
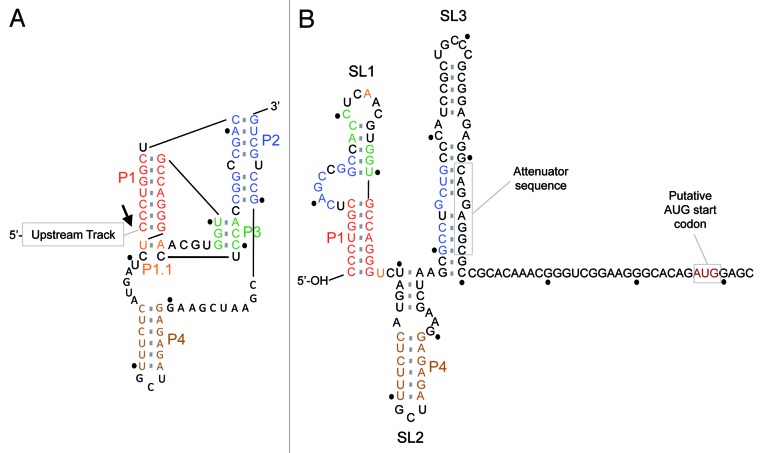
Figure 1. L1Tc RNA 5′-end region structural switching. The proposed folding of the L1TcRz is shown in (A). The three helixes P1, P2 and P4 and the two pseudoknots P1.1 and P3 are colored. The arrow points the ribozyme cleavage site. The proposed switching of the folding toward a supposed-IRES after elongation is shown in (B). RNAfold software proposes for the first 110 nt of the L1Tc RNA a structure involving three stem-loops called SL1, SL2 and SL3. P1 and P4 helixes are conserved after the folding switching but the two pseudoknots and the P2 helix are not maintained. A 5′-hydroxyl end is expected for SL1 to be occluded by the GC pair-rich P1 stem. Each ten nucleotides are marked by a dot.
Co-transcriptional cleavage assays were used to map the L1TcRz to the first 77nt (+1 to +77) of L1Tc/NARTc elements. This 77 nt sequence is predicted to fold into an HDV-like ribozyme, and can be fitted to the consensus HDV secondary structure with minimal divergences.6,20,21 Assays where L1TcRz sequence is preceded by various L1Tc target derived sequences indicate that these sequences upstream of the L1TcRz have the potential to modulate the L1TcRz activity. A similar modulation phenomenon has been described for other known HDV-like ribozymes.7,22 L1Tc sequences downstream of the L1TcRz have also proved to regulate the ribozyme activity.18 It has been shown that the 49 nt downstream of L1TcRz can induce an RNA structural change to a conformation not capable of L1TcRz activity (Fig. 1B).18 The sequence located downstream of L1TcRz in NARTc does not produce a conformational switch and consequently the ribozyme activity is preserved in NARTc RNA. It is possible that the downregulation effect of L1Tc downstream sequences may be an artificial consequence of the transcriptional activity of the T7 RNA polymerase used in the in vitro assays. T7 RNA polymerase may transcribe the downstream sequence so quickly, prior to ribozyme folding, allowing alternative folding pathways to occur. The inhibition of the ribozyme activity may not occur with an eukaryotic RNA polymerase, whose polymerase activity is slower than the T7 phage polymerase.
Other non-LTR retrotransposon has an HDV-like ribozyme in its 5′-end for the purpose of processing the retrotransposon RNA.6,7 L1Tc was the second reported case of a non-LTR retrotransposon to be associated to an HDV-like ribozyme, being the R2 the first one. R2, unlike L1Tc, is a site-specific element whose copies are always found at the same position in the redundant rDNA genes. The 5′-end of the R2 retrotransposon lacks a promoter relying on being co-transcribed with the rRNA. The R2 encoded HDV-like ribozyme is used to process the co-transcript, thus releasing the R2 mRNA. Like L1TcRz, the R2 encoded HDV-like ribozyme stays covalently attached to the R2 RNA.
Non-LTR retrotransposons that lack a promoter, like R2, necessarily rely on host promoters and co-transcription to replicate. Target site specificity would ensure that a promoter-less element like R2 would always insert downstream of a host promoter. An RNA endonuclease system, like an element encoded ribozyme or a signal recognized by a host RNA endonuclease, is required to process the element RNA away from the co-transcript. Conversely, non-LTR retrotransposons that contain internal promoter may become more permissive for their insertion site and may, by the virtue of the internal promoter, generate unit length element RNA without the need for processing by a ribozyme. L1Tc is the first described retrotransposon that carries both internal promoter function (Pr77) as well as a ribozyme (L1TcRz) function, combining features expected for non-specific and site-unspecific elements, respectively. The L1Tc element is widely distributed throughout the T. cruzi genome but there is some degree of conservation of the sequence upstream the insertion sites that flanks the L1Tc element copies.23 A similar consensus sequence has been detected flanking other retrotransposons belonging to the ingi/L1Tc clade.24-26 Since the conservation of these nucleotides is considered to be a trace of the insertion site selection, it may be possible to hypothesize which retrotransposition machinery of an autonomous element is used by each non-autonomous element.26 However, this hypothesis is only supported by bioinformatic data as the function of the upstream sequence motifs remains unknown. The implications of the Pr77/L1TcRz dual function in trypanosomes are explored in the next section.
Functional Implications of Pr77/L1TcRz Dual System in the Regulation of L1Tc and Trypanosoma Cruzi Gene Expression
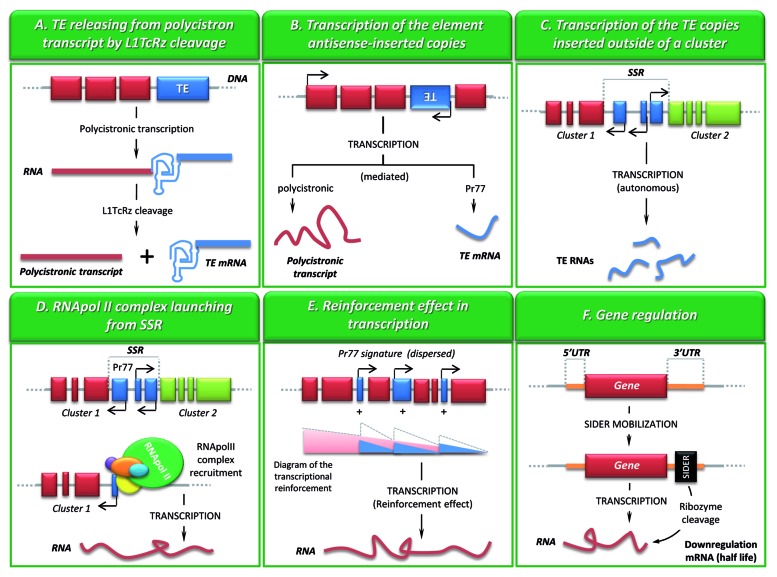
The coexistence of both, ribozyme and internal promoter systems within L1Tc and NARTc elements may be related to the genetic regulation of the host. Trypanosomatid genomes are organized in large directional polycistronic clusters that are transcribed by RNA polymerase II27 and separated by the so-called strand switch regions (SSRs). The main cluster transcription is initiated at the SSRs located between diverging clusters.28 Monocistronic mRNAs are produced through trans-splicing of capped spliced leader (SL) sequence onto the 5′-end of the individual coding sequences. Most likely, the L1TcRz ensures the proper release of any L1Tc (or NARTc) elements that were co-transcribed as part of a polycistronic RNA (Fig. 2A).
Figure 2. Functional roles of the ingi/L1Tc clade 77–79 bp signature: promoter-ribozyme duality. (A) Transposable element mRNA is released from policystronic transcripts by L1TcRz cleavage when the copy is located in sense orientation within a cluster. (B) Pr77-mediated transcription of antisense-inserted copies of the mobile element. (C) Autonomous Pr77-mediated transcription of the mobile elements inserted outside the clusters. (D) The bi-directional launching of the RNA polymerase II (pol II) for transcription of the large polycistronic clusters may be mediated by the Pr77 promoter from the mobile elements accumulated within the SSRs. (E) Pr77- mediated transcription of sense-inserted copies located within a cluster could prevent the transcriptional decay level of genes located far from the cluster transcription start site. (F) Mobile element copies inserted in sense at the 3′-UTRs of somatic genes could be downregulating the mRNA level reducing the RNA stability after L1TcRz cleavage (as the SIDER-dependent downregulation described in Leishmania spp). Code: TE, transposable element; same color boxes (red or green), somatic genes within the same cluster; blue boxes, copies of transposable elements; black arrows, transcription start site and orientation; black box, SIDER copy. Code for the diagram of transcriptional reinforcement (E): red, transcriptional level of the pol II launched from the SSR; blue, transcriptional level of the pol II launched from dispersed Pr77; dotted-line, global pol II transcriptional level.
The L1Tc Pr77 internal promoter may guarantee the transcription of the L1Tc copies present in the antisense orientation with respect to the polycistronic clusters (Fig. 2B) as well as those that lie outside them (Fig. 2C). The existence of many SIDER antisense-inserted copies in the intergenic regions within the clusters of Leishmania genome supports this idea.29 In addition, only two RNA polymerase II-dependent promoters have been described in trypanosomatids: the SL RNA external promoter30 and the L1Tc Pr77 internal one.17 It has been suggested that, due to the high density of L1Tc and NARTc copies found within SSRs, the initiation of the polycistronic transcription may be a result of the firing from the Pr77 promoters (Fig. 2D).31 Thus, the Pr77 promoters may be partially domesticated. Finally, the dispersed copies in sense orientation could also be preventing a decay of the transcription level of distal regions, ensuring the correct expression of the last genes of each cluster (Fig. 2E).
The origin of the Pr77-dependent transcription has been determined by primer extension using extracted RNA from epimastigote cells and found to be located at the +1 nucleotide.17 Since the L1TcRz cleaves at the 5′-side of the mentioned +1 nucleotide of the element, the result of the primer extension previously referred above could be a consequence of the L1Tc HDV-like ribozyme activity. This finding may suggest, therefore, that L1TcRz is active in vivo.
In contrast to the hammerhead or hairpin ribozymes that cleave within their catalytic core, the particular cleavage characteristic of the HDV-like ribozymes allows to keep intact the catalytic core at the mRNA 3′-product, ensuring its complete preservation within the de novo synthesized copies given the single reverse transcription step of the TPRT. Moreover, the existence of L1TcRz enables the generation of L1Tc mRNAs from polymerase II polycistronic transcripts. The L1TcRz would preserve the individuality of each L1Tc/NARTc copy, guaranteeing the accurate definition of the 5′-end of the element and preventing the mobilization of host sequences located upstream of the parental copy.
Taking into account all these data, we propose that the L1Tc mRNAs lack in vivo SL sequences, since their 5′-end start at the nucleotide +1 of the element.17,18 Presumably the L1Tc mRNAs have an uncapped 5′-hydroxyl group. Many other cytoplasmic RNAs are naturally uncapped, like tRNAs, 5S rRNA or viral RNAs of pestivirus and narnavirus, among others. For example, tRNAs are transcribed as longer precursors, and their 5′- and 3′-ends are maturated by RNase P and RNase Z, respectively. The 5′-end and the 3′-end of tRNAs and 5S rRNA are usually constrained in a GC pair rich helix32-34 like the 5′-end of narnavirus RNA.35 Actually, the disruption of the GC pairs of the 5′-stem loop of narnavirus RNA makes it to be susceptible to exonuclease SKI1/XRN1 degradation, thus reducing its half-life.35 The in silico analysis of the folding of the 5′-UTR of the L1Tc and ingi RNAs resulting from the L1TcRz cleavage predicts that it adopts a 5′-stem loop rich in GC pairs (SL1 of Fig. 1B and ref. 36). Thus, the L1TcRz could be responsible of some kind of 5′-end RNA maturation to ensure the stability of the uncapped L1Tc mRNA in vivo.
As previously shown, the Pr77-derived transcripts lack SL and are abundant although poorly translatable.17 The confirmation of an in vivo 5′-hydroxyl end in the mature L1Tc mRNAs would point to a cap-independent translation in which other sequences, apart from Pr77, must be involved. The best known cap-independent translation system is the IRES (internal ribosome entry site). Some IRES have been described in many viral and other retrotransposon RNAs.37-40 An IRES consist of a complex RNA structure able to mediate a cap-independent ribosome recruitment. However, in some cases, this recruitment may require additional cellular factors, associated or not, to cap-dependent translation. We have found an attenuator sequence downstream of the L1TcRz in L1Tc (Fig. 1B), but not in NARTc, revealing that the downstream sequences of L1Tc and NARTc induce different refoldings.18 The L1Tc 5′-UTR RNA, but not the NARTc RNA, adopts a tRNA-like structure susceptible of being cleaved by RNase P. The RNase P is a natural ribozyme responsible for the maturation of the 5′-end of tRNAs by the catalysis of an endonucleolytic cleavage that removes the 5′-leader sequence of pre-tRNAs. The variety of tRNAs strongly indicates that the recognition of the substrates is mediated by three-dimensional tRNA folding. Interestingly, RNAs of some IRES are also recognized by RNase P, revealing the existence of a so-called tRNA-like structure within them.41-43 Although other tRNA-like structures unrelated to IRES have been described, the identification of one of them within the 5′-UTR RNA of the coding L1Tc element, and not in the 5′-RNA region of the non-coding NARTc element, is consistent with the hypothesis of the existence of an IRES in L1Tc. Thus, we propose a model in which after L1TcRz cleavage, the L1Tc mRNA elongation leads to a RNA refolding toward an uncapped hidden 5′-end and an IRES structure (Fig. 1A and B).
The Pr77/L1TcRz Sequence is Widely Distributed among Different Trypanosomatid Retrotransposons: An Indicator of Pr77/L1TcRz’s Activity in Self-Mobilization and Host Genetic Regulation
The Pr77/L1TcRz sequence of 77 nt in length is also found in other trypanosomatid’s non-LTR retrotransposons like ingi, RIME, DIREs24 and SIDERs29,44 with a high level of identity. Consequently, it has been called the 77–79 bp signature. However, there are no data reporting that the catalytic RNA and the internal promoter functions of the 77–79 bp signature are maintained across various members of the ingi/L1Tc clade of retrotransposons. Ingi is a supposedly active LINE element that resides within the T. brucei (Tbingi)45 and T. vivax (Tvingi)24 genomes like the L1Tc element does in both the T. cruzi and T. congolense (L1Tco24). RIME46 is a truncated version of ingi (like NARTc of L1Tc) and also resides within T. brucei (TbRIME) and T. vivax (TvRIME) genomes.24 To date, there is not data reporting the existence of NARTc homologous elements in the T. congolense genome. DIREs (degenerated ingi/L1Tc-related elements) are long (approx. 5kb) interspersed elements with high nucleotide divergence among their copies.36 The presence of abundant stop codons and misreading mutations indicate that DIREs have not coding capacity. SIDERs (short interspersed degenerated retroposons) are short non-coding degenerated elements of approximately 600 bp in length that reside, together with DIREs, in the trypanosomatid’s genomes.44,47 DIREs and SIDERs are thought to be mobilized in trans by the ingi/L1Tc machinery. However, ingi/L1Tc homologs in the genomes of various Leishmania spp, where SIDERs are especially abundant, have not been described. Consequently, it is believed that LmSIDERs and LmDIREs are not active elements.36
Extensive phylogenetic analyses of the ingi/L1Tc-related elements’ lineage of trypanosomatids have been preformed.23,25,26 The transposition mechanism of the non-LTR elements involves two single and asymmetric cleavages at the target insertion site that lead to the generation of a direct repetition (of 11–12 nt in L1Tc) flanking each element, called target site duplication (TSD) (Fig. S1B).2 The conservation degree of the TSDs provides a hint of the elapsed time from the transposition, since ancient copies would have accumulated mutations at their TSDs, while recent ones would not. Based on that, it was established that TvSIDER1c, TcoSIDER1, TbSIDER1, LmSIDER and LmDIRE have not been recently mobilized, suggesting they are not active elements.26
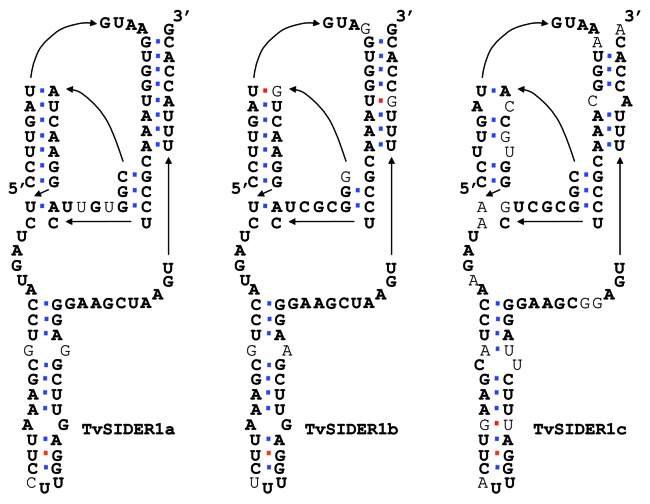
Since the HDV-like ribozyme activity has low sequence requirements but needs a consistent folding, we have analyzed the predicted folding of the 77 bp signature of the ingi/L1Tc clade members as an indicator of the element activity (Fig. S2). The results indicate that the TvSIDER1c consensus sequence has severe deficiencies for a proper HDV-like ribozyme folding as it has two mispairings in the P1 helix and a complete extinction of the pseudoknot P1.1 (Fig. 3). However, the TvSIDER1a and TvSIDER1b consensus sequence has an HDV-like ribozyme folding, although the latter bears a mispairing in the P3 pseudoknot (Fig. 3). Interestingly, all the foldings compatible with a functional HDV-like ribozyme correlate with the deduced active elements by the analysis of the TSDs.26
Figure 3. Folding predicted for the Pr77-transcribed RNAs corresponding to the consensus sequence of different families of the Trypanosoma vivax SIDER1(a-c) described by Bringaud, F. et al., 2011.26 The nucleotides in bold are conserved in the three consensus sequences. Watson-Crick basepairs are in blue and wobble basepairs are in red. The TvSIDER1c consensus presents severe misfolding mutations like two mismatches at P1 helix, one mismatch at P2 and complete disappearance of the P1.1 pseudoknot. The TvSIDER1b presents one mismatch at P3 pseudoknot. Folding of TvSIDER1a has not divergences with an expected active folding.
By contrast, the existence of a conserved ribozyme activity in a degenerated retrotransposon might mean that both the insertion and the catalytic activity would have been positively selected along the evolution as they provide some kind of favorable genetic regulation for the host. This may well be the case of SIDER copies inserted within the 3′-UTR of many genes of Leishmania major and L. infantum.48,49 In fact, it has been shown that SIDER2 copies within the 3′-UTRs of two genes in L. major (SIDER3810 and SIDER1270) and L. infantum (SIDER4000 and SIDER1222) cause a downregulation of the mRNAs via an endonucleolytic cleavage of such RNAs without prior deadenylation.49 In the L. major SIDER2 copies two in vivo endonucleolytic cleavages were detected within the 77 bp signature (so called “signature II” in LmSIDER2).49 The existence of an active ribozyme within this signature could explain the generation of these endonucleolytic cleavages that lead to the RNA degradation (Fig. 2F). The experimental evidences needed to confirm this hypothesis are currently being pursued.
Supplementary Material
Aknowledgments
This work was supported by the Plan Nacional I+D+I (Ministerio de Ciencia e Innovación—MICINN)—Spain [grant numbers BFU2007/65095/BMC, BFU2007-64999, BFU2010-1670; and the Instituto de Salud Carlos III (ISCIII)—Redes Temáticas de Investigacio´n Cooperativa en Salud (RETIC)—Spain [grant numbers RD06/0021/0014, RD06/0021/0008] and FEDER.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/19233
References
- 1.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 3.McLean C, Bucheton A, Finnegan DJ. The 5′ untranslated region of the I factor, a long interspersed nuclear element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol Cell Biol. 1993;13:1042–50. doi: 10.1128/mcb.13.2.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizrokhi LJ, Georgieva SG, Ilyin YV. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–91. doi: 10.1016/S0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- 5.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–29. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 2010;30:3142–50. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruminski DJ, Webb CH, Riccitelli NJ, Luptak A. Processing of insect retrotransposons by self-cleaving ribozymes. Nature Precedings 2010; Available from Nature Precedings <http://hdl.handle.net/10101/npre.12010.14333.10101>
- 8.Thomas MC, Macias F, Alonso C, Lo´pez MC. The biology and evolution of transposable elements in parasites. Trends Parasitol. 2010;26:350–62. doi: 10.1016/j.pt.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Marti´n F, Maraño´n C, Olivares M, Alonso C, Lo´pez MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J Mol Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 10.Olivares M, Alonso C, Lo´pez MC. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J Biol Chem. 1997;272:25224–8. doi: 10.1074/jbc.272.40.25224. [DOI] [PubMed] [Google Scholar]
- 11.Garci´a-Pe´rez JL, González CI, Thomas MC, Olivares M, Lo´pez MC. Characterization of reverse transcriptase activity of the L1Tc retroelement from Trypanosoma cruzi. Cell Mol Life Sci. 2003;60:2692–701. doi: 10.1007/s00018-003-3342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olivares M, Garci´a-Pe´rez JL, Thomas MC, Heras SR, Lo´pez MC. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J Biol Chem. 2002;277:28025–30. doi: 10.1074/jbc.M202896200. [DOI] [PubMed] [Google Scholar]
- 13.Heras SR, Lo´pez MC, Garci´a-Pe´rez JL, Martin SL, Thomas MC. The L1Tc C-terminal domain from Trypanosoma cruzi non-long terminal repeat retrotransposon codes for a protein that bears two C2H2 zinc finger motifs and is endowed with nucleic acid chaperone activity. Mol Cell Biol. 2005;25:9209–20. doi: 10.1128/MCB.25.21.9209-9220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heras SR, Thomas MC, Macias F, Patarroyo ME, Alonso C, Lo´pez MC. Nucleic-acid-binding properties of the C2-L1Tc nucleic acid chaperone encoded by L1Tc retrotransposon. Biochem J. 2009;424:479–90. doi: 10.1042/BJ20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heras SR, Thomas MC, Garci´a-Canadas M, de Felipe P, Garci´a-Pe´rez JL, Ryan MD, et al. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell Mol Life Sci. 2006;63:1449–60. doi: 10.1007/s00018-006-6038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bringaud F, Garci´a-Pe´rez JL, Heras SR, Ghedin E, El-Sayed NM, Andersson B, et al. Identification of non-autonomous non-LTR retrotransposons in the genome of Trypanosoma cruzi. Mol Biochem Parasitol. 2002;124:73–8. doi: 10.1016/S0166-6851(02)00167-6. [DOI] [PubMed] [Google Scholar]
- 17.Heras SR, Lo´pez MC, Olivares M, Thomas MC. The L1Tc non-LTR retrotransposon of Trypanosoma cruzi contains an internal RNA-pol II-dependent promoter that strongly activates gene transcription and generates unspliced transcripts. Nucleic Acids Res. 2007;35:2199–214. doi: 10.1093/nar/gkl1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sánchez-Luque FJ, Lo´pez MC, Macias F, Alonso C, Thomas MC. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5′-end of the L1Tc retrotransposon from Trypanosoma cruzi. Nucleic Acids Res. 2011;39:8065–77. doi: 10.1093/nar/gkr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res. 2008;41:1027–35. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 20.Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur J Biochem. 1997;247:741–53. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 21.Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–92. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 22.Chadalavada DM, Gratton EA, Bevilacqua PC. The human HDV-like CPEB3 ribozyme is intrinsically fast-reacting. Biochemistry. 2010;49:5321–30. doi: 10.1021/bi100434c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bringaud F, Bartholomeu DC, Blandin G, Delcher A, Baltz T, El-Sayed NM, et al. The Trypanosoma cruzi L1Tc and NARTc non-LTR retrotransposons show relative site specificity for insertion. Mol Biol Evol. 2006;23:411–20. doi: 10.1093/molbev/msj046. [DOI] [PubMed] [Google Scholar]
- 24.Bringaud F, Berriman M, Hertz-Fowler C. Trypanosomatid genomes contain several subfamilies of ingi-related retroposons. Eukaryot Cell. 2009;8:1532–42. doi: 10.1128/EC.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bringaud F, Biteau N, Zuiderwijk E, Berriman M, El-Sayed NM, Ghedin E, et al. The ingi and RIME non-LTR retrotransposons are not randomly distributed in the genome of Trypanosoma brucei. Mol Biol Evol. 2004;21:520–8. doi: 10.1093/molbev/msh045. [DOI] [PubMed] [Google Scholar]
- 26.Bringaud F, Berriman M, Hertz-Fowler C. TSIDER1, a short and non-autonomous Salivarian trypanosome-specific retroposon related to the ingi6 subclade. Mol Biochem Parasitol. 2011;179:30–6. doi: 10.1016/j.molbiopara.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bringaud F. Les trypanosomatides sous les feux du se´quençage. Med Sci (Paris) 2005;21:1027–8. doi: 10.1051/medsci/200521121027. [DOI] [PubMed] [Google Scholar]
- 28.Marti´nez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–17. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith M, Bringaud F, Papadopoulou B. Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genomics. 2009;10:240. doi: 10.1186/1471-2164-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunes LR, Carvalho MR, Shakarian AM, Buck GA. The transcription promoter of the spliced leader gene from Trypanosoma cruzi. Gene. 1997;188:157–68. doi: 10.1016/S0378-1119(96)00726-3. [DOI] [PubMed] [Google Scholar]
- 31.Ghedin E, Bringaud F, Peterson J, Myler P, Berriman M, Ivens A, et al. Gene synteny and evolution of genome architecture in trypanosomatids. Mol Biochem Parasitol. 2004;134:183–91. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Tan TH, Pach R, Crausaz A, Ivens A, Schneider A. tRNAs in Trypanosoma brucei: genomic organization, expression, and mitochondrial import. Mol Cell Biol. 2002;22:3707–17. doi: 10.1128/MCB.22.11.3707-3716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenardo MJ, Dorfman DM, Reddy LV, Donelson JE. Characterization of the Trypanosoma brucei 5S ribosomal RNA gene and transcript: the 5S rRNA is a spliced-leader-independent species. Gene. 1985;35:131–41. doi: 10.1016/0378-1119(85)90165-9. [DOI] [PubMed] [Google Scholar]
- 34.Hern´ndez-Rivas R, Marti´nez-Calvillo S, Romero M, Hern´ndez R. Trypanosoma cruzi 5S rRNA genes: molecular cloning, structure and chromosomal organization. FEMS Microbiol Lett. 1992;71:63–7. doi: 10.1111/j.1574-6968.1992.tb05235.x. [DOI] [PubMed] [Google Scholar]
- 35.Esteban R, Vega L, Fujimura T. 20S RNA narnavirus defies the antiviral activity of SKI1/XRN1 in Saccharomyces cerevisiae. J Biol Chem. 2008;283:25812–20. doi: 10.1074/jbc.M804400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bringaud F, Ghedin E, Blandin G, Bartholomeu DC, Caler E, Levin MJ, et al. Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements. Mol Biochem Parasitol. 2006;145:158–70. doi: 10.1016/j.molbiopara.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–76. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim Biophys Acta. 2009;1789:542–57. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Li PW, Li J, Timmerman SL, Krushel LA, Martin SL. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition. Nucleic Acids Res. 2006;34:853–64. doi: 10.1093/nar/gkj490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo´pez-Lastra M, Ulrici S, Gabus C, Darlix JL. Identification of an internal ribosome entry segment in the 5′ region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors. J Virol. 1999;73:8393–402. doi: 10.1128/jvi.73.10.8393-8402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadal A, Martell M, Lytle JR, Lyons AJ, Robertson HD, Cabot B, et al. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J Biol Chem. 2002;277:30606–13. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- 42.Lyons AJ, Robertson HD. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. J Biol Chem. 2003;278:26844–50. doi: 10.1074/jbc.M304052200. [DOI] [PubMed] [Google Scholar]
- 43.Serrano P, Gomez J, Marti´nez-Salas E. Characterization of a cyanobacterial RNase P ribozyme recognition motif in the IRES of foot-and-mouth disease virus reveals a unique structural element. RNA. 2007;13:849–59. doi: 10.1261/rna.506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bringaud F, Müller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimmel BE, ole-MoiYoi OK, Young JR. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol Cell Biol. 1987;7:1465–75. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasan G, Turner MJ, Cordingley JS. Complete nucleotide sequence of an unusual mobile element from trypanosoma brucei. Cell. 1984;37:333–41. doi: 10.1016/0092-8674(84)90329-5. [DOI] [PubMed] [Google Scholar]
- 47.Requena JM, Folgueira C, Lo´pez MC, Thomas MC. The SIDER2 elements, interspersed repeated sequences that populate the Leishmania genomes, constitute subfamilies showing chromosomal proximity relationship. BMC Genomics. 2008;9:263. doi: 10.1186/1471-2164-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller M, Padmanabhan PK, Papadopoulou B. Selective inactivation of SIDER2 retroposon-mediated mRNA decay contributes to stage- and species-specific gene expression in Leishmania. Mol Microbiol. 2010;77:471–91. doi: 10.1111/j.1365-2958.2010.07226.x. [DOI] [PubMed] [Google Scholar]
- 49.Müller M, Padmanabhan PK, Rochette A, Mukherjee D, Smith M, Dumas C, et al. Rapid decay of unstable Leishmania mRNAs bearing a conserved retroposon signature 3′-UTR motif is initiated by a site-specific endonucleolytic cleavage without prior deadenylation. Nucleic Acids Res. 2010;38:5867–83. doi: 10.1093/nar/gkq349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.