Abstract
Recent analyses suggest that transposable element-derived transcripts are processed to yield a variety of small RNA species that play critical functional roles in gene regulation and chromatin organization as well as genome stability and maintenance. Here we report a mass spectrometry analysis of an RNA-affinity complex isolation using a piRNA homologous sequence derived from Alu retrotransposal RNA. Our data point to potential roles for piALU RNAs in DNA repair, cell cycle and chromatin regulations.
Keywords: Alu, DNA repair, PIWI, SINE, TE (transposable element), cell cycle, centromere, chromatin modifiers, histone modifiers, kinetochore assembly, microtubule dynamics, piRNA, transcriptional factors
Introduction
Genome-wide analyses have shown that a large proportion of the genome is transcribed to generate non-coding RNAs (ncRNAs), the biological role and significance of which remains largely unknown.1 The genomes of humans and other complex eukaryotes have many transposon-derived sequences of various classes, commonly referred to as “repetitive elements” or simply “repeats.” Until recently, the most prevalent view was that these repeats represent non-functional “junk DNA.” However, this assumption is now being challenged.
A number of studies indicate that transcripts derived from transposable element (TE) sequences are processed through the cellular RNAi machinery.2 The dis-regulation of TE-RNA processing is associated with diseases such as cancer,3 macular degeneration,4 autoimmunity5,6 and human adult stem cell aging.7 It has been proposed that RNA processing is necessary to silence TE transcription, and is mediated by several pathways including Argonaute-associated short interfering RNAs “siRNAs” in Arabidopsis8 and Saccharomyces pombe,9 Dicer-independent short RNA piRNA machinery in Drosophila melanogaster10 and Dicer1 in mice.4,11 All of these pathways converge at the level of chromatin to drive the heterochromatinization of TEs. However, modern analyses of retrotransposal RNA processing pathways have expanded our understanding of the degree to which TE elements are intimately linked to essential cellular signal-response and developmental functions. The RNAi-mediated processing of TE transcripts (in cis- or in trans-) is most likely involved in a broader spectrum of genomic regulations than simply TE silencing.
In light of new investigations, transcriptional activity from a number of retrotransposon families has been proposed to exert a beneficial function, suggesting that ncRNAs from retrotransposons, in a similar manner to the nuclear function of lncRNAs, may facilitate epigenetic regulation by orienting chromatin modifying factors/complexes to specific genomic locations in a cell type-, developmental stage- and signal-specific manner.12-16 For example, mammalian LINE retrotransposons facilitate X chromosome inactivation17 and LINE RNA products are essential structural and functional components for the formation of neocentromeric chromatin.18 Also, SINEB2 and B1 elements define chromosomal boundaries19-21 and regulate both mRNA production22 and the heat shock response.22,23 Interestingly, Alu elements provide a novel class of enhancer signals by mediating interchromosomal interactions24-26 and also participate in chromatin remodeling27 and DNA damage repair.7,28,29 It is tempting to speculate that the assembly of protein complexes onto retrotransposal RNAs might be nucleotide-specific, implying that RNA-binding protein complexes may undergo conformational and/or functional changes dependent on the sequence composition, RNA length or frame of the processed intermediates, and could be sensitive to the presence of ssRNA, dsRNA or DNA-RNA hybrids.
Unlike the majority of long ncRNAs, which remain permanently in the nucleus, ncRNA from retrotransposons can be found in both nuclear and cytoplasmic compartments29 where their regulation and processing, although scarcely understood, likely involves intricate feed-back loops between these two cellular compartments. This was elegantly demonstrated for the production of type I interferons in autoimmune disease.6
Recently, we have reported that the majority of the repairable DNA damage sites in self-renewing human adult stem cells is associated with the retrotransposal portion of the genome, in particular, Alu retrotransposons.7 Moreover, it has been shown that the upregulation of transcriptional activity from Alu can be triggered by genotoxic stress-induced damage30 as well as upon in-vivo and ex-vivo aging in human retinal pigment epithelium (RPE)31 and human adult stem cells,7 respectively. Intriguingly, evidence indicates that cellular cytotoxicity resulting from increased accumulation of Alu RNA is directly linked to Dicer-1 deficiency,4 suggesting that such cytotoxicity may not necessarily be due to an increase in transcriptional activity, but rather the absence of concomitant transcript processing. Our recently published data indicate that the accumulation of unprocessed Alu transcripts triggers chromatin deterioration, loss of DNA repair in pericentromeric areas eliciting the persistent DNA damage response and, ultimately, cellular senescence.7 Further, we have shown that human adult stem cells stably expressing an shRNA against Alu transcripts, sh-132Alu, override cellular senescence and reinstate their DNA repair capacity,7 suggesting that RNAi machinery is involved in these events. This published data also implies two equally possible mechanisms through which an shRNA against Alu might mediate the observed function: (1) through the post transcriptional gene silencing (PTGS) Dicer-dependent pathway via the cytoplasmic degradation of Alu RNA or (2) through facilitating transcriptional silencing by recruiting either the Dicer-dependent or Piwi-dependent arms of the RNAi pathway to act directly on the chromatin as shown in Figure 1A. Both of these pathways are plausible and either could depend on the assembly of single or multiple protein complexes capable of cross-talk with DNA-damage-sensing/repair and centromeric maintenance pathways. This hypothesis is plausible, considering that a recent study indicates that transcripts generated from telomere-repeat-encoded RNA (TERRA) interact with heterochromatin protein 1 (HP1), trimethlyated histone H3 lysine 9 (H3K9me3), core components of the Shelterin complex, as well as members of the DNA-damage-sensing pathway.32
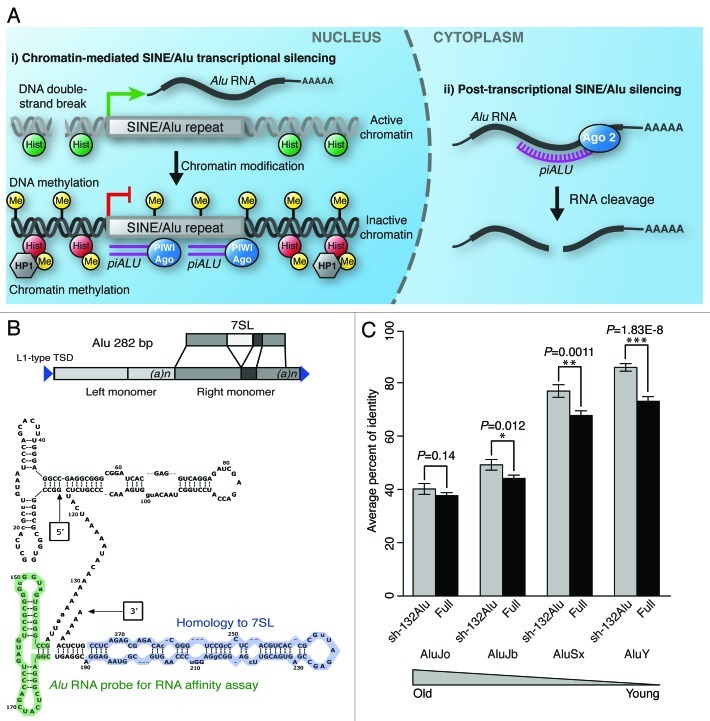
Figure 1. (A) Alternative models of sh-132Alu action7, (i) shRNA against Alu, which directs either nuclear PIWI or Dicer machinery to the genomic SINE/Alu repeat location, initiating transcriptional silencing via heterochromatinization involving both DNA methylation and histone modification. (ii) sh-132Alu acts through the PTGS Dicer-dependent Ago2 pathway, leading to the cytoplasmic degradation of unprocessed Alu RNA transcripts. (B) Top: Diagram representation of 7SL-conserved region within Alu full-length sequence. Bottom: Secondary structure of generic full-length Alu RNA. The segment highlighted in blue represents highly conserved 7SL-derived portion, while the segment highlighted in green represents region used for RNA affinity assay. (C) Comparison of sequence conservation for the region of Alu elements corresponding to the sh-132Alu sequence (grey bars: sh-132Alu) versus sequence conservation for the rest of the Alu sequence (black bars: Full). Average levels of sequence identity (y-axis) among dispersed Alu elements are shown for four Alu subfamilies of different relative ages (x-axis).
Similarly, we hypothesize that this region of Alu RNA targeted by sh-132Alu, which is functionally relevant to overriding the senescent phenotype of human adult stem cells,7 can mediate a broad array of downstream effects associated with retrotransposal transcription. Since the cytoplasmic role of Alu RNA in the assembly of the signal recognition particle (SRP) has been previously reported,33 we have focused our efforts specifically on the discovery of nuclear complexes assembled on Alu RNA. Using an unbiased RNA affinity assay coupled with mass spectrometry, we provide evidence for the composition of molecular complexes assembled on processed Alu RNA transcripts. Our data implicate a number of molecular pathways through which processed intermediates of Alu RNA may participate in a multitude of nuclear processes within human adult stem cells.
Results
Alu RNA and its homology to human PIWI-interacting RNA (piRNA)
Previously it was reported that Alu RNA participates in the cytoplasmic assembly of SRP in mammalian cells.33 The mammalian SRP is composed of a single RNA, the 7SL RNA and six proteins.34 SRP9 and SRP14 bind to the 5′ end of the RNA (Alu-domain), which functions in translational arrest.35,36 However, the functional sh-132Alu reported by Wang et al.7 has no complementarity to 7SL RNA (Fig. 1B). In addition, the region of Alu that corresponds to the shRNA is more highly conserved between Alu repeats than is the rest of the element (Fig. 1C). This conservation is consistent with the reported ability of the shRNA to effectively knock down transcription from multiple Alu repeats,7 including members of evolutionarily distinct young and old Alu subfamilies (Fig. 1C). All together, these data illuminate the functional relevance of the sh-132Alu sequence analyzed here, and indicate that this sequence may represent a naturally occurring processed Alu RNA species that functions in the nucleus.
Furthermore, we queried the sh-132Alu sequence analyzed here against PIWI-interacting RNA (piRNA) sequences previously reported by Girard et al.37 The exact sequence of the shRNA was identified to be a PIWI-interacting RNA, pir-52207 and pir-5746037 (NCBI accession #DQ585095), and is referred to herein as piALU. This suggests that Alu RNA is subjected to processing by the human PIWI complex, the molecular function of which is not fully understood. Nevertheless, the existence of this piALU, derived from processed Alu transcripts, indicates a possible nuclear function.
piALU RNA interaction with nuclear factors
We attempted to obtain a comprehensive list of nuclear proteins that interact, both directly and indirectly, with piALU. For this purpose, we utilized an unbiased RNA-affinity assay using synthetic biotinylated RNA oligos encompassing Alu nucleotides 140–185, shown in green in Figure 1B, with nuclear extracts from self-renewing human adult stem cells. This portion of Alu is conserved among different types of Alu retrotransposons: AluSx, AluJb, AluJo and AluY (Fig. 1C).
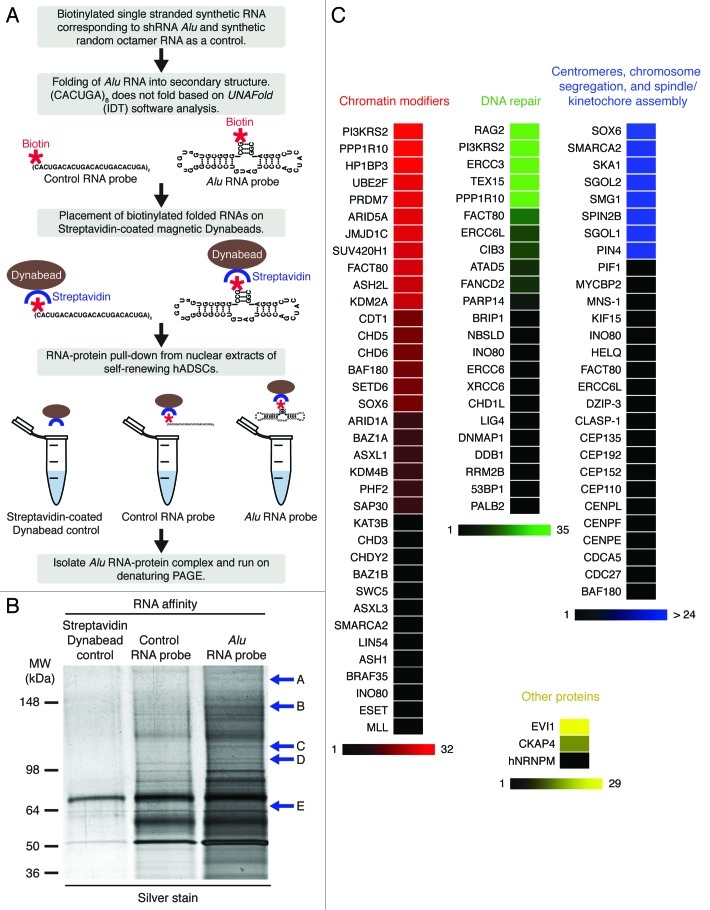
Our piALU RNA affinity assay was combined with an LC-MS/MS (liquid chromatography-electrospray tandem mass spectrometry) analysis of interacting proteins. To differentiate from non-sequence-specific interactions occurring with components of the nuclear RNA surveillance pathway protecting from transcriptional noise, we included a control hexanucleotide repeat control RNA “bait,” as described in Materials and Methods. The experimental procedure was previously reported by Deng et al.32 and is outlined in Figure 2A. A comparative analysis of RNA-protein binding by denaturing PAGE identified the presence of multiple piALU probe-specific bands (Fig. 2B) that were excised for LC-MS/MS identification. A list of the proteins identified is given in Table S1. A list of all identified peptides can be found in Table S2.
Figure 2. (A) Step-by-step schematic representation of unbiased RNA affinity assay. (B) RNA affinity assay precipitants separated on a denaturing NuPAGE Novex 4–12% Bis-Tris gel, visualized by silver staining. Excised bands are labeled A-E. (C) Color-coded peptide representation of members in major functional categories. Proteins with the highest peptide representation within each group are depicted with the brightest color, fading to black for those with a representation of only 1. An extensive list of peptides and their spectra shown in Table S2, and all spectral data has been uploaded to the Tranche database.
piALU RNA interacts with major chromatin modifiers, DNA repair complexes, centromeric chromatin/kinetochore assembly and transcriptional factors
Our mass spectrometry analysis of unbiased piALU RNA-affinity precipitates revealed the presence of major protein categories such as chromatin modifiers, DNA repair complexes, centromeric chromatin/kinetochore assembly and “others” (Fig. 2C). Although multiple peptides were identified for each protein with variable confidence scores (low, medium and high), shown in Table S2, only proteins with high confidence scores were included in Figure 2C. For example, the MS spectra for MLL protein can be viewed as Figure S1 and others can be found in data online. All LC-MS/MS data has been uploaded to the Tranche database, and can be accessed as described in Materials and Methods.
The peptide representation of each component is depicted as a color gradient fading from true hues to black for singly represented peptides. It is important to note that the number of identified peptides does not necessarily indicate the abundance of the identified protein, as it is determined by the number of available tryptic cleavage sites. A larger protein with more tryptic sites has a greater chance of more peptide identifications than a shorter protein with fewer tryptic sites, even if their abundance is the same. Similar to protein interactions with telomeric RNA repeats (TERRA),32 piALU binding proteins include the DNA damage response proteins DDB1, PARP and hnRNPM, suggesting that piALU RNA may be integral to DNA repair complexes.
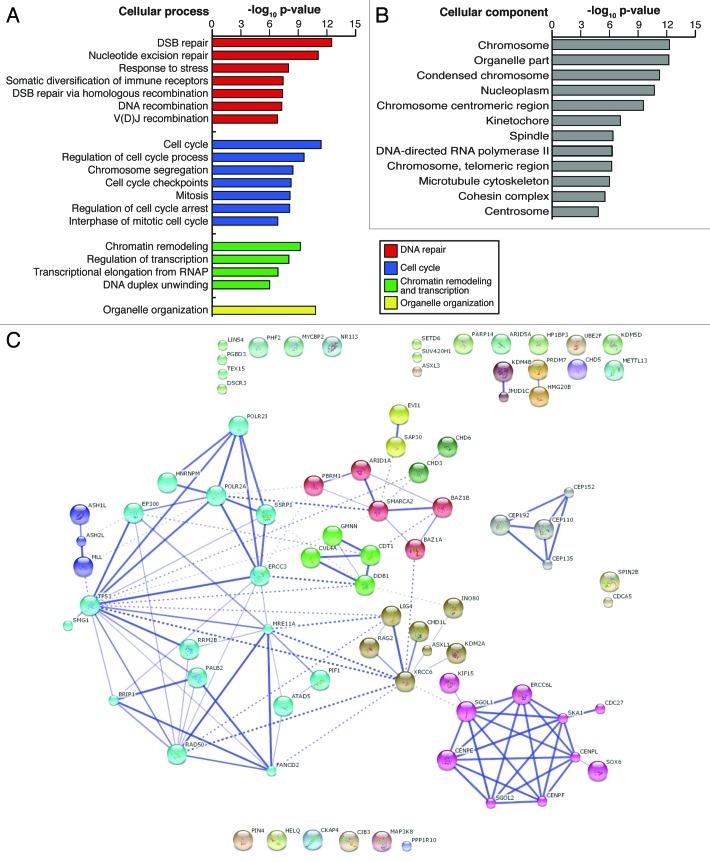
To assess the relevance of piALU RNA to biological processes, we applied a Gene Ontology (GO)38,39 analysis to the list of interacting proteins. All together, 19 cellular processes were significantly (3.12 × 10−13 ≤ p ≤ 1.5 × 10−3) enriched among piALU-interacting proteins including: DNA repair, organelle organization, cell cycle, chromatin remodeling and transcription (Fig. 3A). These broader categories encompass nucleotide excision repair, double-stranded break (DSB) repair via homologous recombination, DNA recombination, V(D)J recombination, response to stress, chromosome segregation, cell cycle check-points, regulation of cell cycle checkpoint arrest, interphase of mitotic cycle and mitosis (Fig. 3B). Complexes related to DNA duplex unwinding, regulation of transcription and transcriptional elongation from RNA polymerase II promoters were also significantly enriched.
Figure 3. (A) Confidence levels suggesting the involvement of isolated complex proteins in various cellular processes, represented as a log10 (p value), p values calculated by STRING v9.0 software. (B) Confidence levels for the presence of isolated complex proteins in various cellular components, represented as a log10 (p value), as calculated using STRING v9.0 software. (C) Interaction web of isolated proteins produced in STRING 9.0 software. Thicker lines represent higher confidence (experimentally derived) interactions while dotted lines represent inferred (lower confidence) interactions.
Several nuclear proteins were associated with multiple GO terms (n = 12) designated to cellular components including the centrosome, nucleoplasm, kinetochore, spindle, microtubule cytoskeleton, centromeric and telomeric regions of chromosomes, as well as the cohesin and DNA-directed RNA polymerase complexes (Fig. 3B).
These data suggest a possible role for piALU RNA in the regulation of chromatin organization of centromeric/pericentromeric regions, DNA repair processes, and the transcriptional regulation of genes. All of these cellular processes have been proven to be indispensable to the regulation of cell cycle progression.
Protein-protein interaction network of the piALU RNA-affinity complex
An essential prerequisite for understanding the cellular function(s) of an isolated protein complex(es) is the ability to correctly uncover and annotate all functional interactions among LC-MS/MS identified proteins. To build a protein-protein interaction network among piALU RNA-affinity isolated proteins, we used the online database Search Tool for the Retrieval of Interacting Genes (STRING) version 9.0. This tool provides uniquely comprehensive coverage and access to both experimental and predicted interaction information.40 The STRING model of our LC-MS/MS data of nuclear piALU RNA-protein interactions is shown in Figure 3C. Only high confidence level (experimental) evidence of interactions/associations in curated databases in conjunction with the Markov clustering algorithm (MCL) were used to build the network and assign proteins into families (stronger associations are represented by thicker lines in Figs. 3C and 4).41 Strikingly, nine out of the 64 piALU RNA interacting partners are involved in chromosome segregation and include SGOL1, SGOL2, CENPE, CENPF and CDCA5, with four more (PBRM1, CENPL, CDC27 and MAP3K8) directly linked to mitosis, while the addition of PIN4 and KIF15 depicts a complex protein network regulating spindle assembly. This observation suggests that piALU transcripts might form a molecular connection between centromere cohesion and microtubule dynamics at the kinetochore. We recently demonstrated that the accumulation of unprocessed Alu RNA in adult stem cells blocks the recruitment of the cohesin complex to centromeric areas, triggering cellular senescence.7 Another group recently reported the loss of outer kinetochore proteins CENP-E and CENP-F upon knockdown of Sgo.42 As CENP-E and CENP-F are members of the piALU interactome, it seems that Alu RNA, in its processed form, may be an RNA adaptor in spindle microtubule dynamics. This suggested involvement of piALU in chromosome segregation is intriguing in light of the high levels of cellular senescence and aneuploidy, due to a spindle checkpoint defect, seen in BubR1 deficient mice.43 Further investigation into the regulation of chromosome segregation/spindle checkpoints by piALU may elucidate a novel role for TE-RNAs in cellular aging.
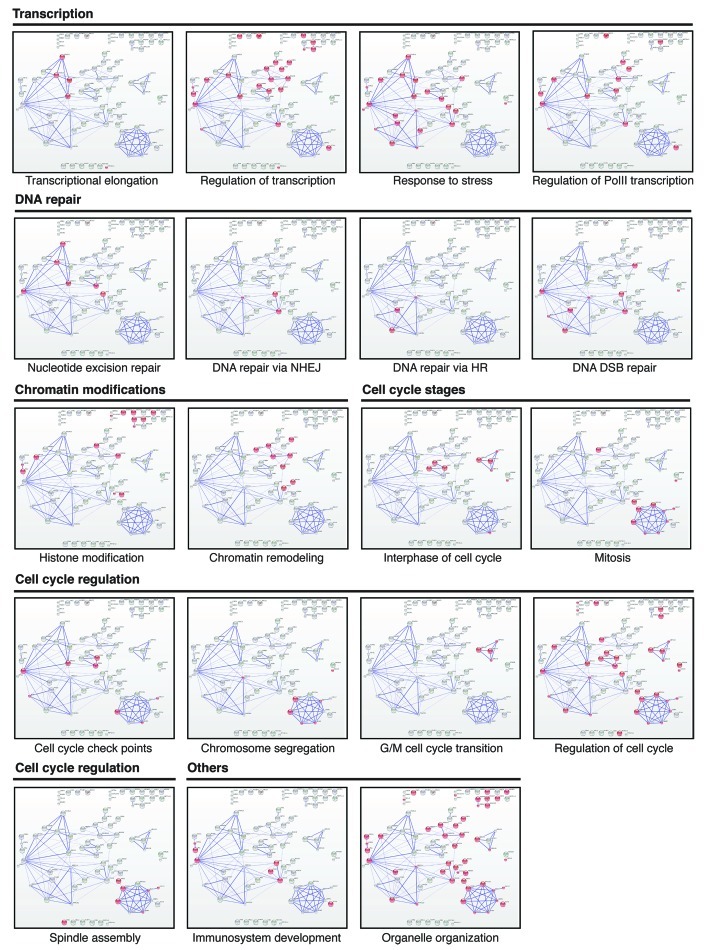
Figure 4 (See opposite page). Representation of protein complex using Gene Ontology (GO) terms in String 9.0. For each process, involved proteins are highlighted in red. Each specific cellular process is grouped into one of the following five categories: transcription, DNA repair, chromatin modifications, cell cycle regulation and others.
Data shown in Figure 4 also indicate that 34 out of the 64 analyzed piALU RNA interacting partners are somehow involved in the regulation of the cell cycle. The presence of centrosomal proteins CEP192, CEP110, CEP152 and CEP135 points to the intriguing possibility that Alu RNA intermediates play a role in this cellular organelle and together with CUL4A, GMNN and CTD1 (Fig. 4), may participate in regulating both interphase and the G2/M phase transition.
Excitingly, piALU RNA also pulled down components of the nucleotide excision repair (ERCC3, DDB1, MRE11A), non-homologous end joining (LIG4, XRCC6) and homologous recombination (PALB2, RAD50) DNA repair pathways as well as the BAZ1B protein, suggesting a putative role for piALU RNA in double-stranded break repair. The assembly of the DNA damage repair complexes described herein may be a critical event in adult stem cell aging.4,7 It is tempting to speculate that a lack of proper assembly/function of these complexes together or separately with chromatin remodeling components (CHD6, SMARCA2, FACT80, BAZ1A, ARID1A, INO80, ASXL1 and KDM2A) may explain the gradual loss of DNA damage repair capacity with aging, triggering the persistent DNA damage response and cellular senescence.44,45
Discussion
A recent genome-wide ChIP-RNA sequencing project revealed that over a third of all known large conserved ncRNAs can be pulled down with just four different chromatin modifying proteins,46 suggesting that most long ncRNAs associate with chromatin modifiers. The piALU RNA interactome described herein also includes a variety of chromatin modifiers, possibly adding another level of complexity to ncRNA/chromatin modifier protein complex formation with a preference for a shorter, processed form of Alu RNA. The full length sequence of Alu RNA can form four distinct stem-loop structures, each of which may act as individual nucleating centers for the formation of distinct RNA-protein complexes. Only one of the four stem-loops was used here as a synthetic RNA “bait,” since it encompasses the previously reported human PIWI-interacting RNA37 and is capable of nucleating protein complex(es) that are directly or indirectly responsible for reinstating the self renewal of senescent adult stem cells.7
Indeed, in accord with our previously published experimental data, the observed protein interactions support the relevance of these nuclear piALU RNA-protein complex(es) to cell cycle control, DNA repair, chromatin maintenance and the function of centromeres/pericentromeres in human adult stem cells. This data are currently lacking functional assessment, but nevertheless may hold the key to understanding how retrotransposal transcription/processing regulates human adult stem cell aging and senescence ex vivo.
Since no nuclear interactions with this portion of Alu RNA have been published, we cannot yet assess the technical or biological limitations of our reported data. It is possible that additional protein interactions were too unstable to withstand the stringent conditions used to purify Alu RNA-interacting components. In addition, some cellular proteins with unusual physical properties can be difficult to detect by LC-MS/MS analysis. Finally, some interactions could be cell type- or culture condition-dependent. Despite these drawbacks, our approach is suitable to probe nuclear Alu RNA-protein interactions in different cell types, culture conditions as well as upon various treatments that induce DNA-damage and/or cell cycle arrest.
The data reported in this manuscript point to a potentially interesting involvement of Alu retrotransposon-derived short RNA species in the regulation of cell cycle control, RNA-pol II transcription and the maintenance of chromatin architecture. It is also possible that DNA repair processes are intimately linked to piALU pathways. Further investigation will allow us to elucidate the functional significance of these intriguing observations.
Materials and Methods
Human ADSC isolation and expansion
Human adipose-derived stem cells (hADSCs) were isolated from human subcutaneous white adipose tissue collected during liposuction procedures. The lipoaspirate was suspended in Hank’s Buffered Salt Solution (HBSS), 3.5% Bovine Serum Albumin (BSA), 1% Collagenase, type II (Sigma) in 1:3 w/v ratio and shaken at 37°C for 50 min. The cells were filtered through a 70 μm mesh cell strainer (BD Falcon, 352350), treated with Red Blood Cell Lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.3) and expanded ex-vivo in DMEM/F12 complete medium (DMEM/F12, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2.5 μg/ml amphotericin B; Invitrogen) in 10% CO2 at 37°C and passaged at 80% confluency, changing medium every 72–96 h.
Nuclear chromatin extraction
Nuclear and cytoplasmic proteins were isolated from self-renewing hADSCs using the ProteoJET™ Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas #K0311), according to the manufacturer’s protocol.
RNA secondary structure folding
5′ biotinylated single stranded synthetic RNA probes corresponding to the 140–185 nucleotide region of full-length Alu, piALU: (5′-/Bio/-GCCGGGCGUGAUGGUGGGCGCCUGUAGUCCCAGCUACUCGGGAGGC-3′) as well as a synthetic random octamer repeat 5′-/Bio/-(CACUGA)8 as a linear control, were purchased from Integrated DNA Technologies (IDT). These oligos were purified via RNase-Free HPLC. Eight micrograms of each oligo were heated in a PCR tube with 15 μl of 2X Folding Buffer (40 mM HEPES pH 7.9, 0.4 mM EDTA, 2 M NaCl) and 9 μl DEPC water with 50 U/ml SUPERase-In RNase inhibitor (Ambion, #AM2694). The RNA oligonucleotides were heated to 78 °C to induce a linear conformation, then the reaction temperature was reduced to and held at 54 °C to accommodate and retain desired secondary structure formation (mimicking the secondary stem-loop structure of the full length RNA), based on calculations by UNAFold software (IDT). The control repeat RNA 5′-/Bio/-(CACUGA)8 is not capable of forming a secondary structure.
Protein pulldown (unbiased RNA affinity assay)
One hundred microliters of streptavidin-coated Dynabeads (Dynabeads M-270 Streptavidin, #65306) per reaction were washed three times in 1× D150 buffer32 (20 mM HEPES pH 7.9, 20% glycerol, 0.2 mM EDTA, 150 mM NaCl, 0.05% NP-40, 1 mM PMSF, 10 μM β-mercaptoethanol) before a final high-salt wash with 1× Folding Buffer (containing 1 M NaCl). Thirty μl of folded RNA solution was added to each reaction tube and incubated at RT for 15 min, then washed twice with 100 μl of low-salt Folding Buffer (containing 150 mM NaCl). Bead-bound RNA probes were incubated for 30min at RT with nuclear extracts from self-renewing hADSCs, along with 1 μl of 0.01M PMSF and 3 μl SUPERase-In. After incubation, the Dynabeads were washed five times in 1× D150 buffer, leaving only directly and indirectly interacting partners attached to the beads. As a negative control, nuclear extracts were also incubated with Dynabeads alone to control for any interaction between nuclear molecules and the beads themselves.
Denaturing PAGE analysis of affinity purified protein complexes
XT Sample buffer 12 (BioRad, 161-0791) and XT Reducing Agent (BioRad, 161-0792) were added to 10 μg of protein. After denaturation for 5 min at 100°C, protein samples were loaded into a precast 4–12% NuPAGE Novex 4–12% Bis-Tris gel (Invitrogen, NP0321BOX) and run out at 150 V for two hours. Gels were stained with ProteoSilver Plus Silver Stain Kit (Sigma, Prot-SIL1). A SeeBlue Plus2 Pre-Stained Standard (Invitrogen, LC5925) was used for visualization and approximation of molecular weights of protein samples. Bands unique to the piALU probe lane were excised and the analyzed by LC-MS/MS for protein content.
Sample preparation for MS analysis
The gel samples were first rinsed with acetonitrile, and then reduced using DTT followed by alkylation using iodoacetamide. We rinsed gel samples with three alternating washes of 50 mM ammonium bicarbonate and acetonitrile, then cooled and resuspended each gel slice in trypsin (5.5 μg/mL in 50mM ammonium bicarbonate/10% acetonitrile) and incubated at 37°C for 24 h for digestion of proteins. We extracted peptides with one rinse of 50 mM ammonium bicarbonate/10% acetonitrile followed by one rinse of 50% acetonitrile/0.1% formic acid, and prepared samples for mass spectrometry by lyophilization and rehydration in 20 μL 5% acetonitrile/0.2% formic acid.
Identification of protein partners by high resolution LC-MS/MS
High resolution LC-MS/MS analysis was performed on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) as previously described.47 Briefly, we loaded excised and digested samples into 96-well plates for mass spectrometry analysis on an LTQ-Orbitrap XL (Thermo Fisher Scientific) instrument as previously described.47 We injected 10 μL of each re-constituted sample using a Thermo Scientific EASY-nLC Autosampler. Reverse phase chromatographic separations were performed using Hypersil GOLD™ C18™ 3 μm media packed into a fused silica 75 μm inner diameter, 20 cm long column running at 250 nL/min. A gradient was produced using 5–40% acetonitrile, 0.2% formic acid over 150 min. The LTQ-Orbitrap was run in a top 8 configuration at 60 K resolution for a full scan, with monoisotopic precursor selection enabled and +1 and unassigned charge state rejected. The analysis on the LTQ-Orbitrap instrument was performed with CID fragmentation.
LC-MS/MS data analysis and protein identification
LC-MS/MS data analysis, protein identification and peaklist generation were performed using Proteome Discoverer (Thermo Fisher Scientific) algorithm incorporating the SEQUEST® search engine and Percolator™48 as previously described.47 MS/MS data were searched using 10 ppm mass accuracy on precursor m/z and a 0.5 Da window on fragment ions. Fully enzymatic tryptic searches with up to three missed cleavage sites were allowed. Oxidized methionines were searched as a variable modification and alkylated cysteines were searched as a fixed modification. Human databases were downloaded from NCBI and supplemented with common contaminants. We filtered peptides for each charge state to a false discovery rate (FDR) of 1%, and then grouped peptides into proteins using Occam’s razor logic.
Protein interaction network analysis
All highly represented members of the RNA-protein complex that were isolated were analyzed using STRING 9.0 software.40 The obtained interaction network was subsequently analyzed with BINGO2 plug-in to determine statistical enrichments for 14 Gene Ontology (GO) categories.38,39
LC-MS/MS data deposition in Tranche database
The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash:
tgT9dlthcEjHrjgXb7syYw37XxF8qlIbhlffHXYyxbs2IrcUAT2axC3qlPM+yyyAEIGMthap579qtgdh9JSvXjkMqqQAAAAAAAAB+A==
More information about this upload:
Network: ProteomeCommons.org Tranche
Title: piALU_Interacting Proteins Complete
Description: Supplementary spectral database for publication.
Uploaded: Dec 7 2011, 07:42:08.457 p.m.49
Supplementary Material
Acknowledgments
We thank Michelle Ohlson for help with manuscript preparation. For critical reading of the manuscript we are grateful to Drs. B. Kennedy and Regina Brunauer. This work was supported by the Buck Institute Trust Fund to V.V.L., The Alfred P. Sloan Research Fellowship in Computational and Evolutionary Molecular Biology (BR-4839 to J.W. and I.K.J.) and Georgia Tech Integrative BioSystems Institute pilot program grant (to J.W. and I.K.J.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/19032
References
- 1.Mattick JS. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–16. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–92. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–30. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhoj VG, Chen ZJ. Linking retroelements to autoimmunity. Cell. 2008;134:569–71. doi: 10.1016/j.cell.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–98. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, et al. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011;10:3016–30. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djupedal I, Kos-Braun IC, Mosher RA, Soderholm N, Simmer F, Hardcastle TJ, et al. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. Embo J. 2009;28:3832–44. doi: 10.1038/emboj.2009.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonelig M. Developmental functions of piRNAs and transposable elements: A Drosophila point-of-view. RNA Biol. 2011;8 doi: 10.4161/rna.8.5.16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 13.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 17:629–34. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 17:620–8. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–69. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 18.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–51. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 20.Roman AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc Natl Acad Sci U S A. 2008;105:1632–37. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roman AC, Gonzalez-Rico FJ, Molto E, Hernando H, Neto A, Vicente-Garcia C, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–32. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–6. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 24.Shankar R, Grover D, Brahmachari SK, Mukerji M. Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol Biol. 2004;4:37. doi: 10.1186/1471-2148-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, et al. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–68. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, et al. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci U S A. 2009;106:11564–9. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–8. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 28.Marti´nez-Lo´pez W, Di Tomaso MV. Chromatin remodelling and chromosome damage distribution. Hum Exp Toxicol. 2006;25:539–45. doi: 10.1191/0960327106het650oa. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2011;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat Genet. 2003;35:219–20. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 471:325–30. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–13. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He XP, Bataille N, Fried HM. Nuclear export of signal recognition particle RNA is a facilitated process that involves the Alu sequence domain. J Cell Sci. 1994;107:903–12. doi: 10.1242/jcs.107.4.903. [DOI] [PubMed] [Google Scholar]
- 34.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–61. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegel V, Walter P. Functional dissection of the signal recognition particle. Trends Biochem Sci. 1988;13:314–6. doi: 10.1016/0968-0004(88)90127-2. [DOI] [PubMed] [Google Scholar]
- 36.Siegel V, Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature. 1986;320:81–4. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- 37.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 38.The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–5. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–68. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res . 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–78. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 44.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–9. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodier F, Munoz DP, Teachenor R, Chu V, Le O, Bhaumik D, et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez MF, Kuppusamy R, Sarracino DA, Prakash A, Athanas M, Krastins B, et al. Mass spectrometric discovery and selective reaction monitoring (SRM) of putative protein biomarker candidates in first trimester Trisomy 21 maternal serum. J Proteome Res. 2011;10:133–42. doi: 10.1021/pr100153j. [DOI] [PubMed] [Google Scholar]
- 48.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–5. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 49.The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash: tgT9dlthcEjHrjgXb7syYw37XxF8qlIbhlffHXYyxbs2IrcUAT2axC3qlPM+yyyAEIGMthap579qtgdh9JSvXjkMqqQAAAAAAAAB+A== The hash may be used to prove exactly what files were published as part of this manuscript's data set and the hash may also be used to check that the data has not changed since publication. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.