Abstract
The human gut pathogen Clostridium difficile contains many conjugative transposons that have an array of accessory genes. In the current study, recently sequenced genomes were analyzed to identify new putative conjugative transposons. Eleven new elements in 5 C. difficile strains were identified and all had a similar structure to the previously described elements CTn1, CTn5 and CTn7 in C. difficile strain 630. Each element identified did however contain a new set of accessory genes compared with those previously reported; including those predicted to encode ABC transporters, a toxin/antitoxin system and multiple antibiotic resistance genes.
Keywords: Clostridium difficile, CTn, ICE, bioinformatics, conjugative transposon, host adaptation, integrative and conjugative element
Clostridium difficile is a Gram-positive, anaerobic bacterium that has emerged as a major nosocomial pathogen. It produces toxins during infection that affect gut epithelia leading to a range of diseases from mild, self-limiting diarrhea to the often fatal pseudomembranous colitis.1C. difficile disease usually follows antibiotic treatment which disrupts the protective gut microbiota. The organism has very low genome conservation with an estimated core genome of 947 to 1033 coding sequences (CDS) which encompasses approximately 23–26% of the total genome of the reference strain 630, calculated based on the genomic sequences of 15 strains.2 This is much lower than the predicted core genome of many other bacteria with low genome conservation such as Streptococcus pneumoniae with 46.5%3 or Campylobacter jejuni with 59.2%.4 In fact, the pan genome of C. difficile is estimated at approximately 9640 CDS,2 an order of magnitude greater than the number in the core genome and the mobile genetic elements in the C. difficile genome may contribute to this.5 For example, in strain 630, 11% of the genome is made up of various types of mobile genetic elements or horizontally acquired DNA.6 A large number of these elements are conjugative transposons, the conjugation and regulatory regions of which are related to the well-studied elements Tn9167 and Tn1549.6,8
In a recent publication, we reported the conjugative transfer of CTn1, CTn2, CTn4, CTn5 and CTn7 from strain 630 into CD375 all of which are conjugative transposons predicted from the genomic sequence of strain 630.6 We also searched the sequence of published genomes for the presence of these and other mobile genetic elements. Eighteen novel elements were described in nine different strains.5 All but one of these has a similar structure to the elements in C. difficile 630.5,9 The only completely novel element is a putative mobilizable transposon in strain QCD-63Q42, designated Tn6115, containing a predicted ABC-transporter system of unknown function and a predicted two component system. Another interesting finding was that eight of the elements contained putative sigma factors; it will be interesting to determine if these genes are expressed and whether their protein products affect the host transcriptome.5
Since the publication of our last study,5 several more C. difficile genomes have been published (Table 1) which we have analyzed using a similar methodology. Putative conjugative transposons were identified using the sequences of a library of recombinase genes5 as well as the sequences of the previously identified transposons CTn1, CTn2, CTn4, CTn5, CTn6, CTn7, Tn5397 and Tn6115 using BlastN.10 Comparisons were made using DoubleAct11 and visualized using the Artemis Comparison Tool.12 Regions containing novel CDS were annotated manually using SMART13 and PRODOM.14
Table 1. Bacterial strains for which the genomic sequences were used in this study and their putative conjugative transposons. All accessory functions are predictions based on bioinformatic analysis. The cell surface proteins described in several elements are homologs of the cell surface proteins previously described in CTn1 and CTn75.
| Strain | Ribotype | Accession number | Isolation details | Source | Family of element | Element | Accessory genes, predicted properties |
|---|---|---|---|---|---|---|---|
| BI1 |
027 |
FN668941 |
USA, 1988 |
Human |
Tn916 |
CTn1-like |
Identical to R20291 CTn1-like element,5 contains ABC transporter of unknown function and a cell surface protein |
| BI9 |
001 |
FN668944 |
USA, 2001 |
Human |
Tn916 |
CTn1-like |
ABC transporter of unknown function and a cell surface protein |
| |
|
|
|
|
Tn1549 |
CTn5-like |
ABC transporter of unknown function, nearly identical to the CTn5-like element in QCD-63Q425 |
| |
|
|
|
|
Tn916 |
CTn7-like |
ABC transporter of unknown function, part of conjugation module is not present due to a gap between contigs |
| 2007855 |
027 |
FN665654 |
USA, 2007 |
Bovine |
Tn916 |
CTn1-like |
Identical to R20291 CTn1-like element,5 contains ABC transporter of unknown function and a predicted cell surface protein |
| |
|
|
|
|
Tn916 |
CTn1-like |
Erythromycin resistance gene and toxin-antitoxin system, 99% identical to M68 CTn1-like element |
| |
|
|
|
|
Tn1549 |
CTn5-like |
ABC transporter of unknown function, insertion of a putative mobilizable transposon containing aminoglycoside resistance gene |
| M68 |
017 |
FN668375 |
Ireland, 2006 |
Human, associated with an outbreak |
Tn916 |
Tn916-like |
Tetracycline resistance and β-lactamase, cell surface protein |
| |
|
|
|
|
Tn916 |
CTn1-like |
Erythromycin resistance and toxin-antitoxin system, 99% identical to 2007855 CTn1-like element |
| |
|
|
|
|
Tn1549 |
CTn5-like |
ABC transporter of unknown function, identical to CF5 CTn5-like element |
| CF5 | 017 | FN665652 | Belgium, 1995 | Human, asymptomatic patient | Tn1549 | CTn5-like | ABC transporter of unknown function, identical to M68 CTn5-like element |
Strains BI1 and 2007855 both contain an element with 100% nucleotide sequence identity to the CTn1-like element described in R20291 (ribotype 027). The target sites for both elements are identical and contained within an intergenic region.
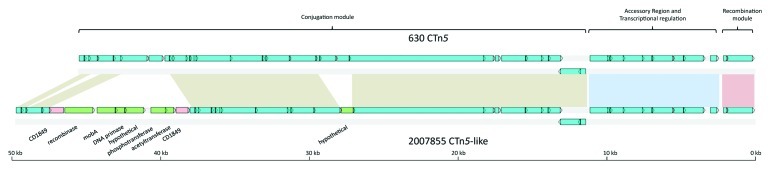
Strain 2007855 also contains a CTn5-like element present between homologs of CD3369 and CD3393; this intergenic region is the target site of the CTn5-like elements, Tn6110 and Tn6111 in strains QCD-66C26 and QCD-32G58, respectively, and interestingly, is also the target site of CTn7 in strain 630.5 The element in strain 2007588 shares 95% nucleotide sequence identity with CTn5, but contains in addition a putative mobilizable transposon inserted within the homolog of CD1849 (Fig. 1) This element contains six ORFs encoding a predicted serine recombinase, a mobilization A protein, a truncated DNA primase and a hypothetical protein. Furthermore, the element also contains an aminoglycoside phosphotransferase and an aminoglycoside acetyltransferase, which may be responsible for resistance to aminoglycosides.15 Although aminoglycosides are currently not used in the treatment of C. difficile, some strains have been shown to be susceptible to antibiotics in this class such as spectinomycin.16 It is possible that these resistance genes may provide such strains with an opportunity to colonize the gut following treatment with these antibiotics, once the protective immunity of the normal microbiota is disrupted.
Figure 1. Schematic comparison of CTn5 and the homologous element present in strain 2007855. Blue ORFs have homologs present in CTn5, green ORFs are not present in CTn5, pink ORFs are interrupted by insertion of new sequence compared with CTn5. Regions of homology are indicated by colored boxes: the red box shows the recombination module, the blue box shows the accessory module, the brown boxes show the conjugation module.
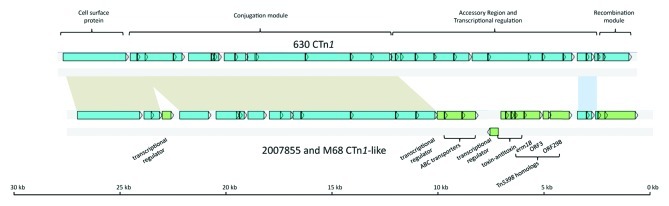
Strain 2007855 also contains a second CTn1-like element that has 99% homology to a CTn1-like element in strain M68 (Table 1). Two CDS at the 3′ end of the elements (Fig. 2) are predicted to function as tyrosine integrases and excisionases based on amino acid homology. Within the accessory module of both elements is a region showing nucleotide sequence homology (97%) to part of Tn5398, an erythromycin resistance-conferring element present in strain 630.17 This region contains an ermB gene with 98% amino acid identity to ermB1 of Tn5398; homologs of ORF3 and ORF298 of Tn5398 are also present, (Fig. 2). Also, a homolog of the omega–epsilon–zeta operon is present, encoding a toxin-antitoxin (TA) system required for plasmid maintenance in Gram positive bacteria.18 A similar system has also been shown to stabilize a transposon in Vibrio cholerae when it excises from the chromosome.19 However, in 2007855 and M68, the zeta toxin gene has acquired a mutation resulting in an early stop codon at R52, halfway into the conserved toxin domain. The epsilon antitoxin and omega regulatory gene are intact and could have alternative roles in regulatory pathways or may act as anti-addiction systems against incoming DNA containing similar TA systems.20 The fact that the two zeta toxin genes in these strains contain the same mutation suggests that this occurred in the gene before its dissemination to these two strains. In addition, the accessory regions of both elements in 2007855 and M68 contain genes encoding putative ABC-transporters of unknown function and putative transcriptional regulators of the GntR (IPR000524) and Xre (IPR001387) families.
Figure 2. Schematic comparison of CTn1 and the homologous elements present in strain 2007855 and M68. Blue ORFs have homologs present in CTn1, green ORFs are not present in CTn1. Regions of homology are indicated by colored boxes: the blue box shows the accessory module, the brown boxes show the conjugation module.
Strains M68 and CF5 contain identical elements that have 99% nucleotide sequence identity with CTn5, excluding the region between CD1865 to CD1868 (in 630) encoding putative conjugative transfer proteins, which has been replaced in both M68 and CF5 by two genes encoding hypothetical proteins. It is possible that the loss of the two conjugative transfer proteins will prevent these elements from conjugating to a new host although this needs to be determined experimentally.
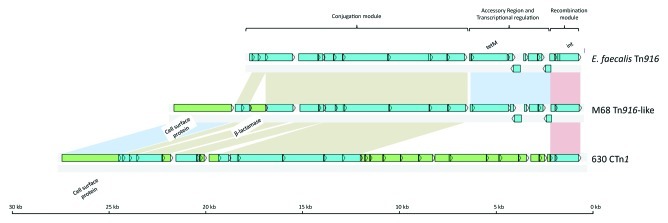
Strain M68, contains an element with 98% nucleotide sequence identity with the Tn916 core (Fig. 3) but with two insertions compared with the prototype Tn916. The first insertion between homologs of ORF21 and ORF22 is predicted to encode a metallo β-lactamase-domain. Proteins containing this domain are sometimes involved in genetic competence and antibiotic resistance (IPR001279). The second insertion replaces ORF24, and is likely to encode a surface-located protein. It is predicted to have the “stalk-like” structure of collagen binding protein domain CnaB of Staphylococcus aureus21 but importantly, lacks identifiable ligand-binding domains. Furthermore, it contains a predicted LPXTG domain, a signal for sortase-mediated cell wall anchoring, commonly found in surface proteins of Gram positive bacteria.22 ORFs predicted to encode similar proteins have been found on CTn1 and CTn7 of 630 (Fig. S1).5,6 It is interesting that all the strains studied here and in our previous communication, except CF5, contain a CTn1-like element with a gene encoding a homolog of this surface protein. Furthermore, E. faecium Tn5386, a Tn916-like element, is also predicted to encode a similar cell surface protein, however, this element contains an intimin/invasion domain.23 Insertional mutagenesis, has shown that this gene is not required for conjugative transfer of CTn1 and CTn7.5 The fact that a surface protein is so highly conserved in CTn1-like elements and yet is not required for transfer is intriguing, and suggests that it could play a role in the host-pathogen interaction.
Figure 3. Schematic comparison of Tn916, the homologous element present in strain M68 and CTn1. Blue ORFs have homologs present in Tn916, green ORFs are not present in Tn916. Regions of homology are indicated by colored boxes: the red box shows the recombination module, the blue box shows the accessory module, the brown boxes show the conjugation module.
Strain BI9 contains a CTn1-like element exhibiting 99% nucleotide sequence identity with the Tn916-like element previously described in strain QCD-63Q42;5 the predicted ORFs of the elements are identical, including the ABC transporter genes in the accessory region, and the element is present in a homologous target site.
BI9 also contains a CTn5-like element that has 99% nucleotide sequence identity with the CTn5-like element described in QCD-63Q42.5 In both elements, homologs of 5 genes (CD1864 – CD1868 from CTn5 in C. difficile 630) are absent, however a predicted restriction and modification system is present in its place. Restriction modification systems, when present on mobile genetic elements, have been shown to function as addiction systems, helping to maintain the element within the cell.24 In common with strain QCD-63Q42,5 the element in BI9 has integrated in tandem with CTn7 in the homolog of the target site of CTn7 in 630. Most of the sequence of CTn7 appears to be present in this homologous element though a gap in the genomic sequence of strain BI9 makes a full analysis of the element impossible at this time.
In conclusion, we show here that putative conjugative transposons are present in all strains of C. difficile that have been sequenced so far. Although the accessory regions of the conjugative transposons vary, there are two types of core elements i.e., Tn916-like and Tn1549-like. The pan genome of C. difficile contains a limited repertoire of conjugative transposons but each has the ability to acquire and potentially disseminate a range of different accessory genes. We suggest therefore that it is these conjugative transposons, which give C. difficile the ability to sample the C. difficile pan genome. Furthermore, as some of these conjugative transposons are able to transfer to and from other genera it is possible that they provide C. difficile (and other organisms) with the ability to sample at least part of the metagenome of the intestinal tract.
Supplementary Material
Acknowledgments
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 223585 and the Medical Research Council (grant no. G0601176). The authors are grateful to the Wellcome Trust Sanger Institute for releasing the genomic sequences through GenBank.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/19297
References
- 1.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–36. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 2.Scaria J, Ponnala L, Janvilisri T, Yan W, Mueller LA, Chang YF. Analysis of ultra low genome conservation in Clostridium difficile. PLoS One. 2010;5:e15147. doi: 10.1371/journal.pone.0015147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, et al. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol. 2007;189:8186–95. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champion OL, Gaunt MW, Gundogdu O, Elmi A, Witney AA, Hinds J, et al. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc Natl Acad Sci U S A. 2005;102:16043–8. doi: 10.1073/pnas.0503252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One. 2011;6:e23014. doi: 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–86. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 7.Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–8. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Garnier F, Taourit S, Glaser P, Courvalin P, Galimand M. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology. 2000;146:1481–9. doi: 10.1099/00221287-146-6-1481. [DOI] [PubMed] [Google Scholar]
- 9.Mullany P, Wilks M, Tabaqchali S. Transfer of macrolide-lincosamide-streptogramin B (MLS) resistance in Clostridium difficile is linked to a gene homologous with toxin A and is mediated by a conjugative transposon, Tn5398. J Antimicrob Chemother. 1995;35:305–15. doi: 10.1093/jac/35.2.305. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 11.Underwood A, Green J. DoubleAct v2 Available at http://www.hpa-bioinfotools.org.uk/pise/double_act.html
- 12.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–6. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–71. doi: 10.1016/S0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 15.Young PG, Walanj R, Lakshmi V, Byrnes LJ, Metcalf P, Baker EN, et al. The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2″-phosphotransferase-IIa [APH(2″)-IIa] provide insights into substrate selectivity in the APH(2″) subfamily. J Bacteriol. 2009;191:4133–43. doi: 10.1128/JB.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, et al. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods. 2010;80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Farrow KA, Lyras D, Rood JI. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology. 2001;147:2717–28. doi: 10.1099/00221287-147-10-2717. [DOI] [PubMed] [Google Scholar]
- 18.Zielenkiewicz U, Ceglowski P. The toxin-antitoxin system of the streptococcal plasmid pSM19035. J Bacteriol. 2005;187:6094–105. doi: 10.1128/JB.187.17.6094-6105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rich RL, Demeler B, Ashby K, Deivanayagam CC, Petrich JW, Patti JM, et al. Domain structure of the Staphylococcus aureus collagen adhesin. Biochemistry. 1998;37:15423–33. doi: 10.1021/bi981773r. [DOI] [PubMed] [Google Scholar]
- 22.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice LB, Carias LL, Marshall SH, Hutton-Thomas R, Rudin S. Characterization of Tn5386, a Tn916-related mobile element. Plasmid. 2007;58:61–7. doi: 10.1016/j.plasmid.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–56. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.