Abstract
Progress in agricultural and environmental technologies is hampered by a slower rate of gene discovery in plants than animals. The vast pool of genes in plants, however, will be an important resource for insertion of genes, via biotechnological procedures, into an array of plants, generating unique germ plasms not achievable by conventional breeding. It just became clear that genomes of grasses have evolved in a manner analogous to Lego blocks. Large chromosome segments have been reshuffled and stuffer pieces added between genes. Although some genomes have become very large, the genome with the fewest stuffer pieces, the rice genome, is the Rosetta Stone of all the bigger grass genomes. This means that sequencing the rice genome as anchor genome of the grasses will provide instantaneous access to the same genes in the same relative physical position in other grasses (e.g., corn and wheat), without the need to sequence each of these genomes independently. (i) The sequencing of the entire genome of rice as anchor genome for the grasses will accelerate plant gene discovery in many important crops (e.g., corn, wheat, and rice) by several orders of magnitudes and reduce research and development costs for government and industry at a faster pace. (ii) Costs for sequencing entire genomes have come down significantly. Because of its size, rice is only 12% of the human or the corn genome, and technology improvements by the human genome project are completely transferable, translating in another 50% reduction of the costs. (iii) The physical mapping of the rice genome by a group of Japanese researchers provides a jump start for sequencing the genome and forming an international consortium. Otherwise, other countries would do it alone and own proprietary positions.
Plant Genomics Is Key to Food Supply, Human Health, and a Sustainable Environment
To understand and treat human diseases, the health sciences can focus resources on the determination of the entire DNA sequence of a single animal genome, the human genome. To engineer plants as a source for food, fiber, renewable energy, and bioremediation, the genomes of many plant species have to be analyzed. Therefore, it may appear that, whereas the human genome project is timely, comparable plant genome projects are too costly with current technology. This premise has completely changed with the recent realization that the synteny of genomes of related species (e.g., grasses) provides the opportunity to also focus on anchor genomes in the agricultural sciences (1).
Moreover, the recently developed gene transfer methods allow genes to be moved with ease from one plant species to another, not achievable by classical breeding programs. Examples include the recent successful isolation of resistance genes, and the transfer of the N-gene for viral resistance across genetically incompatible borders from tobacco to tomato, thereby conferring viral resistance in the latter (2). This success establishes that this strategy offers a natural, effective, and environmentally safe procedure for protection of crops against disease without chemicals. Sequencing the plant genome of one of the most important crops will provide the necessary encyclopedia of genes that will form the backbone or framework for gene discovery, not only for the genome sequenced, but for the genomes of other plants by a procedure called “gene cloning by position.” This pool of genes will form a national resource for insertion of genes, via biotechnological procedures, into crop plants lacking them, thereby adding or subtracting desirable or undesirable properties as in the case of the N-gene for viral resistance.
There is also a clear urgency to accelerate the process of plant gene discovery. Food, essential to feed the world, also is becoming a critical factor to our health. For instance, crop production depends heavily on environmentally and health-threatening levels of chemicals, which could be reduced or replaced by moving genes between germ plasms of crops and wild species by using biotechnology.
From One Genome to Many Genomes
Cereals are the staple diet of the world population. They also serve as animal feed, thereby contributing indirectly to our meat supply as well. For many years we have assumed that each genome of the most important cereals would differ from each other and that corn, wheat, and rice would have to be analyzed separately. However, it appears that these genomes differ by the amount and nature of repetitive DNA sequences; many of them are species-specific retrotransposons (3, 4). When we analyzed a 250-kb region on the short arm of maize chromosome 4 by a set of overlapping cosmid clones, gap closure by a chromosome walking step was prevented because of the high density of repetitive DNA sequences in the center of this region. However, these sequences did not hybridize to sorghum DNA. In contrast, if gene sequences or low-copy sequences of maize were used as probes on sorghum DNA cross-hybridization occurred (Fig. 1). To detect, then, unique sequences in maize cosmids, sorghum DNA cloned in bacterial artificial chromosomes (BACs) and orthologous to the maize 4S chromosomal region was used to probe cosmid DNA (V.L., R. Wing, and J.M., unpublished work) after separation of various restriction digests (Fig. 2). This method has revealed new maize probes for chromosome walking and has allowed gap closure by linking two sorghum BAC clones (Fig. 3). Moreover, the distance of two unique sequences that are more than 25 kb apart in maize are only 5 kb apart in sorghum (Fig. 4). This example illustrates how a closely related genome that is smaller in size can provide DNA probes to detect unique or low-copy sequences in the larger genome and shrink the large physical distances between genetic markers to smaller DNA fragments (Fig. 5).
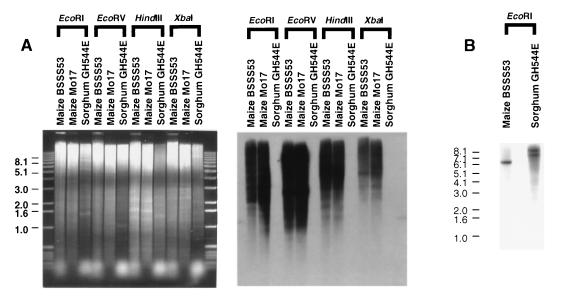
Figure 1.
Maize and sorghum genomes exhibit differential cross-hybridization to low- and high-copy DNA sequences. (A) Genomic Southern analysis on DNA isolated from the maize inbred lines BSSS53 and Mo17 and the sorghum hybrid GH544E. Total genomic DNA was hybridized at moderate stringency with the maize highly repetitive element V.9C11-6.2 (V.L., R. Wing, and J.M., unpublished work). There is no detectable cross-hybridization of V.9C11-6.2 to sorghum DNA. (B) Genomic Southern analysis on DNA isolated from the maize inbred BSSS53 and the sorghum hybrid GH544E. DNA was hybridized at high stringency to the maize restriction fragment length polymorphism (RFLP) marker php20725, which maps to a single location in maize and in sorghum.
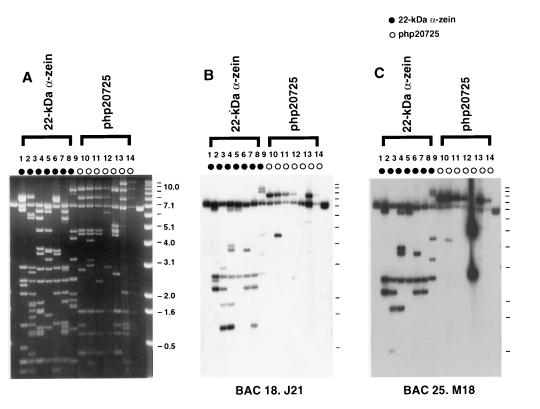
Figure 2.
The differential hybridization between maize and sorghum can be used as a tool in the identification of single- and low-copy sequences. In maize, the small arm of chromosome 4 contains a large cluster of 20–22 pseudogenes and genes encoding α-zeins, the major group of storage proteins in kernels. Most of the intergenic region in the cluster consists of highly repetitive DNA, many of them retrotransposons (V.L., R. Wing, and J.M., unpublished work). BACs containing sorghum DNA homologous to the 22-kDa cluster were hybridized to 14 cosmid clones from the cluster region isolated from a maize BSSS53 library. (A) Agarose gel electrophoresis of the maize cosmids digested with EcoRI. (B and C) Southern blots from the same gel, hybridized to two different sorghum BACs, M18 and J21, respectively. Fragments cross-hybridizing to sorghum BACs contained single- or low-copy sequences, including zeins and the RFLP marker php20725.
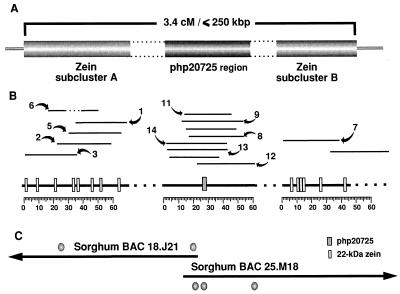
Figure 3.
Schematic representation of the 22-kDa α-zein cluster region in maize and utilization of syntenic sorghum BACs in gap closure and single-copy marker identification. (A) Using a BSSS53/Mo17×Mo17 backcross, the 22-kDa zein genes clustered in chromosome 4S have been mapped as far apart as 3.4 cM (5). However, using long-range restriction analysis, we have estimated that the cluster has a maximum size of only 250 kb. The 22-kDa zein genes in the cluster are further grouped in two subclusters, approximately 60 kb apart. The RFLP marker php20725 is located in the intermediate region, between the 22-kDa zein subclusters. (B) To characterize the organization of this gene cluster, we constructed a physical map based on cosmid overlaps. The high density of the nearly identical 22-kDa zein sequences in the cluster provided useful tags for ordering contigs, but two gaps remain in regions with high density of highly repetitive DNA. Cosmid numbers correspond to those in Fig. 2. (C) Incorporation of the sorghum BACs described in Fig. 2 into the physical map of the zein cluster in maize (V.L., R. Wing, and J.M., unpublished work). Filled circles on BACs indicate sequences found to cross-hybridize in both species, corresponding to single- and low-copy elements.
Figure 4.
The use of the smaller sorghum genome for gap closure and contig linking in maize can reduce significantly distances in walking strategies. In sorghum, the distance from the RFLP marker php20725 to the nearest zein-homologue sequences, called kafirins in sorghum, is only 5 kb, at least six times shorter than its counterpart in maize. Most of the difference in distance can be accounted for the presence of large sections of high-copy DNA that includes retrotransposon-like sequences.
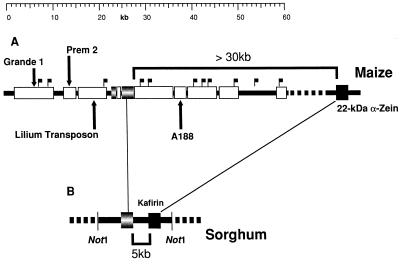
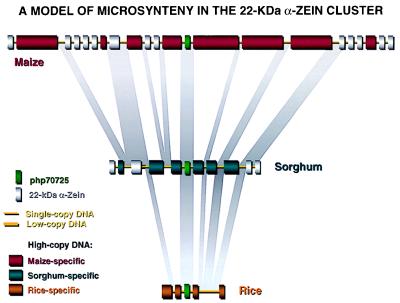
Figure 5.
Model of microsynteny between maize, sorghum, and rice in the 22-kDa α-zein cluster. In particular, maize and sorghum are highly related species. Most RFLP markers, genes and other known single- or low-copy sequences from both species cross-hybridize in highly stringent conditions and have a conserved genetic order, despite a 4-fold difference in genome size (6). However, most of the high-copy DNA does not cross-hybridize at the same conditions. Low- or single-copy sequences constitute a small fraction of the total genome in maize. They are immersed in large sections of high-copy DNA. The differential divergence between highly repetitive and low-copy DNA in maize and sorghum allows us to identify gene-like sequences in either direction among large segments of repetitive DNA in the cluster. Furthermore, the 22-kDa zein and kafirin multigene family is absent in rice. Therefore, the rice genome, in particular, should be useful to identify the nonzein, low-, or single-copy sequences in maize.
Which Genome First?
Although maize may be one of the preferential targets for entire genome sequencing in the United States, current cost of about $1.4 billion is still very high for such a project. As an interim strategy, one could develop a high inventory of maize cDNA sequences and compare those to the genome of a related cereal species with one of the smallest genomes. In addition, one could focus on the microsynteny of gene-rich regions, in particular, on gene clusters in a few cereal species regardless of their genome size. As shown in Fig. 5, such a comparative analysis has the advantage that progress will be rapid and cost at realistic levels. Moreover, we will learn which genes are expressed at which time, distribution of genes within chromosomes, and which genes escaped cDNA cloning techniques or have been deleted. Moreover, a set of orthologous probes will be obtained that can be used across cereals and noncrop grasses to enrich our knowledge about their synteny and the nature of chromosomal rearrangement that might have occurred during their evolution.
Current DNA sequencing techniques (Fig. 6), through computing and automation, could be raised to a production of 40–50 million bp annually by a few highly equipped production sites. With a current rate of about 50 cents per base, it would cost about $3 billion to sequence wheat (the largest crop in the world in metric tons). To get it done in 12 years would require 25 highly equipped sites, each producing 20 million bp annually. In contrast, rice would cost only $200 million, less than the human genome budget for a single year, and would require less than two sites over the same length of time. The rice genome is about three times the size of the Arabidopsis thaliana, which currently is being sequenced by an international consortium from the United States, Japan, and the European Union. Although the consortium was only recently formed, it appears that the majority of the Arabidopsis genome will be sequenced within 6 years. Similarly, if an international consortium could be formed for rice now, its sequence analysis also could be accelerated, and it should be possible to have the sequence of both a dicotelydoneous and a monocodyledoneous plant available in less than 10 years.
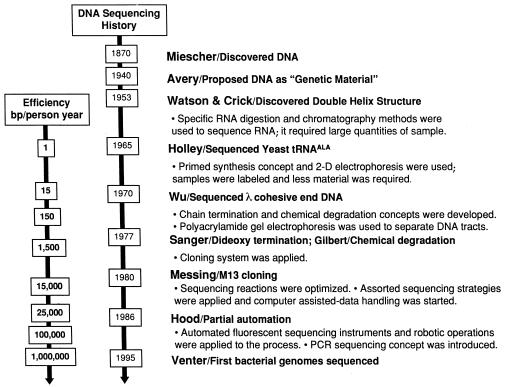
Figure 6.
History of DNA sequencing is related to the combination of new technologies. Applications of new technologies to DNA sequencing have played a major role in the increase of the amount of DNA sequence generated per person per year at a greater cost efficiency. The introduction of chain terminators to DNA polymerase-based sequencing and separation of DNA tracts by PAGE were based on new nucleic acid chemistries. However, without the integration of the DNA sequencing chemistry and DNA cloning, the sequencing of whole genomes would not be possible today. Two other technologies greatly accelerated the speed of DNA sequencing, partial automation with laser detection systems and the progress in computational hardware and software.
Such a coordinative effort would allow us to rapidly disseminate valuable increments of information on plant sequences that will aid academia and industry to further develop and commercialize agricultural and environmental (green) technologies.
The popularization of advances in plant gene discovery can be a natural extension of such genome efforts and help the public to understand improvements in food safety and quality, and in environmental technologies.
Acknowledgments
We thank Peter Quail of the Plant Gene Expression Center for his critical discussion on this subject and Ellson Chen of Applied Biosystems, Inc. for a copy of the history of DNA sequencing depicted in Fig. 6. The microsynteny work was supported by a grant from the U.S. Department of Energy DE-FG05-95ER20194 to J.M.
ABBREVIATIONS
- RFLP
restriction fragment length polymorphism
- BAC
bacterial artificial chromosome
References
- 1.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 2.Whitham S, McCormick S, Baker B. Proc Natl Acad Sci USA. 1996;93:8776–8781. doi: 10.1073/pnas.93.16.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu W, Das O P, Messing J. Mol Gen Genet. 1995;248:471–480. doi: 10.1007/BF02191647. [DOI] [PubMed] [Google Scholar]
- 4.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zkharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri S, Messing J. Mol Gen Genet. 1995;246:707–715. doi: 10.1007/BF00290716. [DOI] [PubMed] [Google Scholar]
- 6.Whitkus R, Doebley J, Lee M. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]