Abstract
Transposable elements provide a convenient and flexible means to disrupt plant genes, so allowing their function to be assessed. By engineering transposons to carry reporter genes and regulatory signals, the expression of target genes can be monitored and to some extent manipulated. Two strategies for using transposons to assess gene function are outlined here: First, the PCR can be used to identify plants that carry insertions into specific genes from among pools of heavily mutagenized individuals (site-selected transposon mutagenesis). This method requires that high copy transposons be used and that a relatively large number of reactions be performed to identify insertions into genes of interest. Second, a large library of plants, each carrying a unique insertion, can be generated. Each insertion site then can be amplified and sequenced systematically. These two methods have been demonstrated in maize, Arabidopsis, and other plant species, and the relative merits of each are discussed in the context of plant genome research.
The rapid accumulation of sequence data from chromosomal DNA and expressed sequence tags means that a large number of plant genes have been discovered for which a function has yet to be established (1, 2). In Arabidopsis thaliana, for example, only 2% of the genes identified by genomic sequencing can be assigned a genetic function on the basis of mutation. Homology searches can be used to indicate likely functions in many of the remaining genes, but the phenotypic role of individual gene family members is usually obscure without assessing gene function by genetic analysis.
A number of strategies have been proposed to probe gene function. In yeast, targeted gene disruption is the primary tool for this purpose (3, 4) because of the high levels of homologous recombination in haploid and to some extent diploid yeast, which makes gene disruption an exquisitely precise and efficient process. Although there have been recent successes in gene replacement in Arabidopsis, this method is still laborious, involving the generation of hundreds or thousands of transgenic plants for every gene assayed (4). Gene silencing via sense or antisense suppression also has been a popular method over the last 10 years (5). However, this method requires that several independent transgenic lines be generated for every gene. Important to note, essential genes cannot be down-regulated in this way because suppression would lead to dominant lethal phenotypes that cannot be maintained. Chemical, radiation, and transposon mutagenesis offer the most versatile methods for assessing gene function. Transposon and T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] insertions offer the additional advantage of tagging the target gene molecularly and in many cases genetically via reporter and selectable marker genes carried by the insertion (6–10). These features can be combined with conventional mutagenesis to provide a comprehensive strategy for probing plant gene function.
Transposable elements mutagenize genes by insertion into coding and regulatory regions (11). This can lead to loss-of-function, and occasionally gain-of-function, phenotypes that reflect the function of the target gene in its normal chromosomal context. In many cases, loss-of-function mutations can be used to reveal essential genes, provided they lead to recessive lethality so that they can be maintained in heterozygous diploids (12, 13). Two examples of gene disruption systems in plants are discussed below, in maize and A. thaliana. Each has been demonstrated to provide an efficient and practical method for assessing gene function, and the relative merits are discussed. First, though, the scale of the problem is explored in a brief review of plant genes and plant genomes.
Plant Genes and Genomes.
The Arabidopsis genome comprises ≈120 Mb of DNA that encodes roughly 20,000 genes. Physical mapping and systematic sequencing have revealed that most genes reside in ≈105 Mb of “euchromatic” DNA, with ≈15 Mb of highly repeated sequences comprising centromeric satellite, nucleolar organizers, and other highly reiterated regions (1). Transposons are relatively rare and are found dispersed among genes. Transcription units occupy 2.5 kb of every 4.5 kb on average.
The maize genome is a segmental allotetraploid, so all unique genes have been duplicated at some time in the distant past (14). Many of these new copies have evolved unique functions over time, however, so maize behaves as a diploid organism in many respects. Small scale sequencing and mapping of repeats suggests that maize may have ≈5–10 times lower gene density than in Arabidopsis (15–17). Because the duplicated maize genome is 20 times the size of the Arabidopsis genome, these “back-of-the-envelope” calculations lead to the prediction that most genes in maize will be represented by an ortholog in Arabidopsis. This prediction has been borne out; it is generally accepted that any gene in maize can be used to identify at least one homolog in Arabidopsis.
Most of the additional DNA found in maize can be attributed to high copy repeated sequences that are interspersed between genes (18). Recently, these repeats have been found to be nested retrotransposons that comprise a small number of families of highly re-iterated transposons of conserved structure and therefore relatively recent origin (19–21). Similar observations have been made in other cereals including wheat (22–24). These transposons are heavily methylated, when compared with genes (25).
Multicopy and Single-Copy Insertional Mutagenesis.
Insertional mutagenesis represents a powerful way to determine the function of plant genes identified in systematic genome and expressed sequence tag sequencing projects. Essentially, two types of approach can be considered: (i) the use of plants with high copy numbers of highly active transposons per genome and (ii) the use of plants with a single copy of a given transposon per genome. In each case, the object is to recover at least one informative insertion per gene. High-copy methods have the advantage that relatively small populations of plants need to be maintained for complete genome coverage. Single- copy methods require much larger populations, but they have a number of other advantages. First, single-copy transposons can be used to integrate reporter genes into the genome, allowing gene expression to be observed at each integration. Multicopy reporter gene insertions would have mixed patterns of expression. Second, single-copy transposons also can be used to integrate genetic and physical markers into the genome, allowing their use in map-based genomic strategies. Finally, single-copy transposon insertions can be sequenced directly thus reducing the number of manipulations required to isolate insertions in every gene. These aspects of single- and multicopy systems are explored below with respect to transposon systems in maize and Arabidopsis.
Site-Selected Transposon Mutagenesis in Maize.
Transposons have been used widely to generate populations of model organisms (libraries) that can be searched for those individuals with transposon insertions in any given sequence (26–32). Searches are performed by amplifying DNA from these populations by using specific primers from the transposon and from the gene. Positive pools are rescreened by sib selection to identify individuals that have the desired insertion. To have a 95% probability of finding an insertion into any gene in Drosophila, Arabidopsis, or C. elegans (which have similar genome sizes and gene densities) such a library needs to contain ≈100,000 insertions (32). Even so, most insertions are into noncoding regions and have no phenotypic effect. Secondary insertions or deletions must be generated at the locus by remobilizing the transposon to disrupt gene function (28–30).
Similar methods have been developed for isolating insertion alleles in maize (30). For example, we used the Robertsons’ Mutator system of transposons as insertional mutagens. These transposons exist in hundreds of copies per genome, subdivided into six classes that share 200-bp terminal inverted repeats but are otherwise unrelated. Transposition is facilitated by the MuDR autonomous transposon that encodes genes required for the transposition of the other Robertson’s Mutator transposon (Mu) elements (33, 34).
We used Mutator to isolate null alleles of the maize gene hcf106, which encodes a chloroplast protein involved in membrane biogenesis (35). The reference allele at the hcf106 locus has a Mu1 element inserted in the promoter region, which results in a form of epigenetic instability (36). We reasoned that deletions and insertions into the locus would likely be more stable than the original allele (33, 34). We used a PCR strategy to isolate derivatives of hcf106 by screening two-dimensional pools of seedlings by using primers from the Mu1 element and from the hcf106 gene (30).
Three new alleles were identified. The first allele had a 200-bp deletion that removed the first exon of the gene. The second and third alleles, however, had new insertions, of Mu1 and dMuDR, respectively, inserted at the same location in the first intron of the hcf106-mum1 and wild-type genes, respectively. The two derivative alleles were no longer subject to epigenetic instability (30).
This method can be used to isolate insertions in any DNA sequence in the maize genome, even those that might be lethal or those that might have no phenotypic consequence. Another advantage is that germinal excisions are very rare in Mutator lines (33, 34), so new alleles are typically stable in this sense. A major disadvantage is that many (perhaps most) insertions will fall in introns and other flanking regions. These insertions typically confer weak or undetectable phenotypes. In these cases, our method can be used to generate deletion and secondary insertion alleles in order to stabilize and enhance any mutant phenotype (30).
Our results show that new insertions into maize genes can be obtained from a very small sample size (2 insertions into 1,500 potential chromosomes screened). Pioneer Hi-Bred (Des Moines, IA) has developed a large collection of pooled Mu lines to enable PCR screening of a standard array of Mu families, each corresponding to a self-pollinated parent plant (R. Meeley and S. Briggs, personal communication). This centralized collection is a very useful resource, provided screening procedures are used that conserve DNA samples and seed stocks after multiple screens. However, our results show that a few thousand Mu seedlings can be screened efficiently to obtain Mu insertions into genes identified by sequence alone. By self-pollinating the plants after DNA has been collected, the DNA pools can be screened repeatedly with primers from different genes. It is thus feasible for individual laboratories to perform site-selected mutagenesis on a manageable scale (30).
Enhancer and Gene Trap Mutagenesis in Arabidopsis.
Gene traps and enhancer traps are reporter genes that are not normally expressed unless they are integrated near or within a chromosomal gene. Enhancer traps are equipped with a minimal promoter that can respond to nearby enhancers, and gene traps are equipped with a splice acceptor so that integration within introns leads to readthrough transcription and splicing (37–39). In each case, the expression of the reporter gene closely mimics that of the chromosomal gene. Large collections of enhancer trap and gene trap lines, each carrying a unique reporter gene insertion somewhere in the genome, have been established in Drosophila and in the mouse. They are being used extensively for both developmental biology and genomic research (40, 41).
We have devised a system for gene trap and enhancer trap transposon mutagenesis in Arabidopsis (12, 42). We are using Dissociation transposons (Ds) from maize that we have engineered to carry a uidA [β-glucuronidase (GUS)] reporter gene and an NPTII kanamycin resistance gene. In our DsE (enhancer trap) construct, the reporter gene is preceded by the −46 region of the CaMV 35S promoter, which has been shown to have no detectable transcriptional activity unless it is in the vicinity of an enhancer. In our DsG (gene trap) construct, the reporter gene is preceded by a triple splice acceptor and by a short intron so that insertion into chromosomal introns leads to reporter gene expression via alternate splicing in each reading frame (12, 42). Additional splice donor sites in the end of Ds means that reporter gene expression also can result when the DsG element is inserted into an exon (43). The elements are mobilized by crosses to transgenic plants carrying Activator transposase gene (Ac) transposase driven by the 35S promoter from CaMV. This results in high frequencies of transposition early in development (44, 45). Transposed elements are selected by using a positive marker within the Ds element (the NPTII gene) and a negative marker adjacent to it (the iaaH gene). Positive–negative selection on naphthalene acetamide and kanamycin results in seedlings that have retained the Ds element but lost the donor site from whence it came (42, 46). Because selection depends on recombination between the transposed element and the donor site, the transposed element is almost always unlinked to the donor locus. We have found that 90% of gene trap lines carry a single transposed element at essentially random locations in the genome (42). The other lines have multiple elements (5%) or insertions that disrupt the negative marker gene on the T-DNA (5%). Each line is referred to as an ET (enhancer trap) or GT (gene trap) line depending on the transposon.
The bacterial gene uidA (gusA) that encodes GUS has been the reporter of choice for plant studies and is used in our system (42, 47). Although there is very little background in plant tissues, GUS is inhibited by oxidative catalysts that prevent diffusion of the indigo reaction products before dimerization, requiring a compromise between sensitivity and precision. Nonetheless, the huge variety of very specific patterns that we have observed indicates that GUS is an efficient and reliable marker gene. Recently, Haseloff and colleagues have shown that modified forms of the green fluorescent protein can be equally sensitive in Arabidopsis (48). Green fluorescent protein is much more challenging to use in large scale screens that involve nontransparent tissues, but the extraordinary versatility of this marker in following reporter gene expression makes it very appealing.
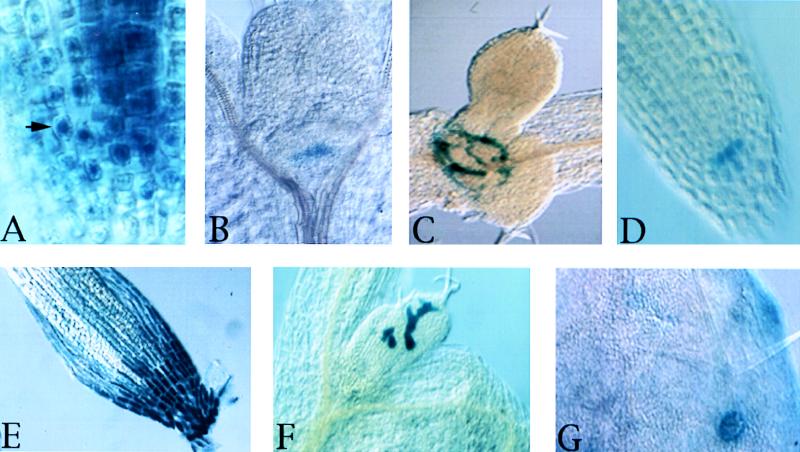
Screening of nearly 2,000 lines has revealed that 32% of gene trap insertions exhibit reporter gene expression in seedlings, with an additional 10% exhibiting reporter gene expression specific to floral and reproductive tissues (P. Springer, Q. Gu, D. Bush C. Yordan, and R.A.M., unpublished results). Only half of all gene trap insertions in genes will be in the appropriate orientation to result in reporter gene expression, so ≈80% of all gene trap insertions lie within genes (allowing 5% artifactual insertions into selectable marker genes). Similarly, 45% of enhancer trap insertions give rise to reporter gene expression in seedlings, but up to 80% give rise to reporter expression somewhere in the plant. These numbers seem high, but in fact they are not inconsistent with gene density estimates derived from genome sequencing (one 2.5-kb gene every 4.5 kb, or 55%), with only a slight bias for insertions into genes (a bias that has been observed for transposon insertions in many organisms). We have recovered a very wide range of expression patterns, including genes expressed in specific cell-types, genes expressed in cells undergoing cell division or cell death, and genes expressed in prepatterns that predict rather than reflect morphogenesis in the plant. A few examples are shown in Fig. 1.
Figure 1.
Examples of reporter gene expression (blue) in various gene trap and enhancer trap lines illustrating cell type-specific expression, expression in prepatterns, and subcellular gene trap localization. Seedlings were grown in continuous light for 7 days, stained for GUS activity, cleared in 70% ethanol, and mounted for differential interference contrast microscopy in 25% glycerol (12, 42). (A) Nuclear staining (arrow) in roots from heterozygous prolifera/+ plants. (B) Meristem staining in the shoot. (C) GUS expression at the basal boundaries of leaf primordia. (D) GUS expression in columella initials in the root. (E) GUS expression in root cap cells. (F) GUS expression in immature trichomes. (G) GUS expression in trichome accessory cells.
Systematic Insertion Site Sequencing: Multicopy vs. Single- Copy Libraries.
As described above, libraries of plants that have one or more transposon insertions per plant can be readily screened for insertions into genes of interest by site-selected mutagenesis. This approach involves pooling the plants and doing hundreds of PCRs for each given gene. However, if most of the genes in a given genome are to be targeted in this way, it is considerably more efficient to obtain insertion site sequences systematically instead. Each sequence only requires a handful of PCRs to obtain in this way (see below). The database of sequences can then be screened by computer for insertions into genes of interest.
In principle, both multicopy transposon libraries and single- copy libraries can be used to develop insertion site sequence databases of this sort. The relative merits of each method are explored below, taking the Mutator system in maize and the gene trap system in Arabidopsis as examples. Of course, the principles apply equally to other multicopy and single-copy approaches in each species.
Mutator insertion sites in maize can be sequenced in a systematic way. Individual PCR products from multiple elements are obtained by one of a number of anchored PCR approaches and are displayed by gel electrophoresis. Individual bands are excised, and the products are purified for sequencing. The idea is to build up a catalog of insertions into recognizable genes so that affected families can be examined for the phenotypic consequences of a given insertion. However, a number of problems might be anticipated. First, somatic transpositions that are not transmitted germinally also will lead to PCR products. These will not be represented in the progeny of selected families. Second, each plant carries several hundred insertions and typically a handful of visible mutations. Sorting through the mutations to determine which phenotype is caused by which insertion takes several generations and multiple PCR reactions. Finally, many insertions are into introns and noncoding regions that are difficult to recognize in the absence of systematic whole-genome sequencing. Amplification of cDNA by rapid amplification of cDNA end and PCR might be preferable in this case.
The multicopy approach may be necessary in large genomes like maize when plant growth space is limiting because a small library of plants can harbor a large number of elements. However, in Arabidopsis, single-copy approaches have a number of distinct advantages as illustrated below.
A Gene Trap Tag Database.
The gene trap collection we are developing in Arabidopsis represents a library of individual insertions carried by individual plants whose progeny are maintained as a separate seed packet and is thus an example of a single-copy insertion library. During selection of the transpositions, DNA is prepared from each individual plant and then subjected to amplification by using TAIL (thermal assymetric interlaced) PCR (13, 49). This PCR procedure uses seminested primers from within the transposon and arbitrary degenerate primers to amplify genomic sequences flanking each insertion. By using a combination of different primers, it is possible to amplify ≈95% of all insertion sites by following a hierarchical tiered procedure by using a minimal number of PCRs. These products are sequenced directly without further resolution on gels. The seed from each plant is stored and catalogued, and the sequence is entered into a gene trap tag database. By comparing the sequence to known genomic and transcribed sequences from Arabidopsis, insertions into genes, and their relative orientation, can be identified readily.
The high frequency of insertions that give rise to reporter gene expression means that preselecting lines on the basis of their staining patterns, even if all tissues were screened systematically, only enriches for insertions into genes by ≈25%. Furthermore, mapped insertions in genes and nongenic regions are extremely powerful tools even without the bonus of having the expression pattern determined via the enhancer and gene trap. This is because, unlike in maize, the complete Arabidopsis genome sequence is going to be determined and publicly available within the next few years. Thus, even very short insertion site sequences can be used to precisely map each insertion in the genome with nucleotide precision. The context of each insertion thus can be determined by using standard sequence analysis of the surrounding region.
So far, we have sequenced a few hundred insertions selected in this way. The results are very encouraging concerning the range of sequences that are obtained. Approximately 15% match Arabidopsis expressed sequence tags, and 5–10% match genomic sequence currently in the database. These frequencies are in line with expectations on the basis of how much of the genome is represented in public databases. An additional 25% of the sequences match a protein sequence when they are translated, with matches at various levels of significance. We have recovered a few instances of two independent insertions into the same gene. Although this is not yet evidence for insertional bias, these biases are expected, so some regions of the genome may need to be mutagenized in a more directed fashion.
Eventually, the gene trap database can be used to infer the function and in many cases the pattern of gene expression associated with most of the genes in the Arabidopsis genome. A major challenge in the future will be in systematically screening these lines for phenotypes associated with gene disruption. One advantage of a sequence-based database is that gene redundancy can be addressed directly. That is, combinations of insertions in different members of a multigene family can be made by crossing individual lines together. This approach can be further refined by combining insertions in genes that are expressed in similar tissues and might therefore be expected to have overlapping roles. For example, an enhancer trap insertion in the AGL8 (agamous-like) MADS box transcription factor gene is expressed early during inflorescence development and late during development of the fruit, in agreement with RNA expression studies (50) Homozygous fruitfull-1 mutant plants have a null mutation in the AGL8 gene and only have a fruit phenotype (Q. Gu, C. Ferrandiz, M. Yanofsky, and R.A.M., unpublished work). However, when combined with mutations in related MADS box genes expressed in the early inflorescence, a redundant role for this gene in meristem identity is uncovered (C. Ferrandiz, Q. Gu, R.A.M., and M. Yanofsky, unpublished observations).
Lethal Insertions.
Transmission studies using deficiencies indicate that at least 1 gene every 100 kb is essential to the haploid Arabidopsis gametophyte and/or the early diploid embryo (51). This means that Arabidopsis has 1,000–2,000 essential genes, compared with 2,000 in haploid yeast (3, 4). Essential genes are required for cell growth and division, so almost all are expected to have a function elsewhere in the diploid plant body (12). However, adult functions of essential genes cannot be assessed if homozygous mutants do not survive to maturity. Gene traps and enhancer traps can give some indication of these adult functions because of reporter gene expression in viable heterozygotes. Somatic mosaics and secondary screens then can be performed to confirm these roles. The ability to identify essential genes with roles in later development has proven to be one of the most important applications of enhancer traps in Drosophila developmental biology (40, 52).
Lethal insertions in Arabidopsis can be lethal either to the diploid embryo or to the haploid gametophyte. They are identified by opening self-pollinated siliques from heterozygous transposants under a dissection scope and looking for white (rather than green) seed or unfertilized ovules, respectively (12). Only a proportion of lethal mutations recovered in our screen are caused by the transposon (unpublished results). These exhibit segregation distortion for the kanamycin resistance gene contained within the Ds element (12). Lethal mutations caused by the transposon can be confirmed readily by transposon-mediated reversion. Each transposant with reduced seed set is test-crossed to plants carrying Ac. F1 plants are then examined for the presence of occasional branches with full seed set (12). This mosaicism reflects somatic excision of the Ds element and is a very strong indication that the lethal phenotype is caused by Ds insertion (12). The prolifera gene, and other genes involved in cell division cycle control, are good illustrations of essential genes that have roles throughout development (12). PROLIFERA encodes a homolog of the CDC47 (MCM7) DNA replication licensing factor from yeast and mammalian cells. It was identified as a lethal mutation caused by the insertion of DsG into the last intron of the gene. In viable heterozygotes, the resulting reporter gene fusion is located in the nucleus as expected for a DNA replication factor (Fig. 1A) and is expressed widely in dividing cells (12).
Remobilization: Mosaics and Local Mutagenesis.
Unlike T-DNA and other types of insertional mutagen, transposons can be remobilized by re-introduction of the relevant transposase. The resulting reversion of the mutation can be used to confirm that it is caused by the transposon. Remobilization of transposons also can be used to generate phenotypic mosaics. That is, homozygous mutants that carry Ac will have somatic sectors that have lost the Ds transposon and so have restored gene function. This can be a very important analytical tool in determining the site of action of a given gene, in combination with its expression pattern.
Even a large collection of arbitrarily selected insertions is unlikely to represent disruption of every gene in the Arabidopsis genome. In cases in which a transposon has integrated near, but not within, a desired gene, it can then be used as a “launch pad” for local mutagenesis. This is because Ac/Ds transposons, in common with most eukaryotic invert-repeat transposons, have a pronounced preference to integrate within a few centimorgans of their starting location after transposition (53, 54).
We demonstrated this technique inducing new DsG transpositions around the prolifera gene on the short arm of chromosome 4 (C. Yordan, J. Montagu, D. Bush, P. Springer, and R.A.M., unpublished work and ref. 46). Excision frequencies are so high (44, 45) that local transpositions can be recovered by selecting for the transposon (kanamycin) and against Ac (napthalene acetamide). Approximately one-quarter of the selected plants have one or more transposed elements, and the remainder retain the parental element. Pooling strategies similar to those described for maize (30) allow the recovery of transposons into nearby regions by virtue of PCR-mediated site-selection. This remobilization strategy allows launch pads to be used to disrupt genes that have been mapped genetically to any given region of the genome.
Using an Insertion Library in Positional Cloning.
Once a catalog of 1–2,000 insertions has been mapped, it is possible to use them to map any new mutation at a very fine scale (40). Typically, most new mutations are mapped to a 10- to 20-cM interval of the genome by using molecular markers, soon after they are first isolated. Once our program is complete, such a 10- to 20-cM interval will have approximately 20 transposons mapped within it. Each of these insertions carries a dominant genetic marker, the kanamycin resistance gene. The new mutation can be crossed to these insertion lines, and kanamycin-resistant seedlings carrying the mutation can be selected in the F2. These progeny will have undergone recombination between the mutation and the Ds element. This procedure will allow the new mutation to be mapped to an interval between two transposons whose physical positions in the genome are known. The recombination breakpoints will allow rapid positional cloning of the new mutation, and the transposons themselves can be used to tag the mutation via a short range transposition.
Transposable elements are powerful mutagens for functional genomics in plants. Multicopy transposons can be used effectively in plants with large genomes to heavily mutagenize and recover insertions into genes of interest. Single-copy transposons, on the other hand, allow the use of enhancer and gene trap reporter genes to monitor patterns of gene expression as well as gene disruption. Furthermore, libraries of single-copy insertions can be sequenced systematically and screened for mutant phenotypes. These libraries represent the most economical method for systematic function search in plant genomes. In the case of A. thaliana, the comparison of gene trap tag sequences with genomic and expressed sequence tag databases will allow the location of every insertion to be determined with nucleotide precision, allowing their use as tools in positional cloning as well as in wide scale gene disruption. By examining reporter gene expression patterns in viable heterozygotes, the role of essential genes can be assessed in later development even if insertions are homozygous or haploid lethal. By combining insertions in homologous genes, the function of redundant genes can be assessed also.
Acknowledgments
I acknowledge the past and present members of his laboratory who have contributed to this work, particularly P. Springer, Q. Gu, C. Yordan, J. Montagu, J. Simorowski, R. Benton, and L. Das. Fig. 1 was assembled by P. Springer. The development of the gene trap tag database is a collaboration with W. Richard McCombie and members of his laboratory. The Cold Spring Harbor Laboratory gene trap system was developed by R. M. and V. Sundaresan. This work was supported by National Science Foundation Grants MCB-9408042 and IBN-9723671 and United States Department of Agriculture Grant 95–37300-1578 to R.M and W.R.M. The development of the gene trap system would not have been possible without the generous support of Westvaco, Inc., David L. Luke, III, and the Robertson Research Fund. I apologize to those whose work was not cited in this review because of space requirements.
ABBREVIATIONS
- GUS
β-glucuronidase
- Ac
Activator transposase gene
- Ds
transposon Dissociation
- Mu
Robertson’s Mutator transposon
References
- 1.Goodman H M, Ecker J R, Dean C. Proc Natl Acad Sci USA. 1995;92:10831–10835. doi: 10.1073/pnas.92.24.10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooke R, Raynal M, Laudie M, Grellet F, Delseny M, Morris P C, Guerrie T D, Giraudat J, Quigley F, Clabault G, et al. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- 3.Oliver S G. Nature (London) 1996;379:597–600. doi: 10.1038/379597a0. [DOI] [PubMed] [Google Scholar]
- 4.Kempin, S. A., Liljegren, S. J., Block, L. M., Rounsley, S. D., Yanofsky, M. F. & Lam, E. Nature (London) 389, 802–803. [DOI] [PubMed]
- 5.Baulcombe D C. Plant Mol Biol. 1996;32:79–88. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 6.Fedoroff N V, Smith D L. Plant J. 1993;3:273–289. doi: 10.1111/j.1365-313x.1993.tb00178.x. [DOI] [PubMed] [Google Scholar]
- 7.Feldmann K A. Plant J. 1991;1:71–82. [Google Scholar]
- 8.Topping J F, Agyeman F, Henricot B, Lindsey K. Plant J. 1994;5:895–903. doi: 10.1046/j.1365-313x.1994.5060895.x. [DOI] [PubMed] [Google Scholar]
- 9.Sundaresan V. Trends Plant Sci. 1996;1:184–191. [Google Scholar]
- 10.Wilson K, Long D, Swinburne J, Coupland G. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClintock B. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Springer P, McCombie W, Sundaresan V, Martienssen R A. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- 13.Tsugeki R, Fedoroff N. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- 14.Helentjaris T. Proc Natl Acad Sci USA. 1993;90:8308–8309. doi: 10.1073/pnas.90.18.8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Hsia A P, Zhang L, Nikolau B J, Schnable P. Plant Cell. 1996;7:2151–2161. doi: 10.1105/tpc.7.12.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, SanMiguel P, de Oliveira A C, Woo S S, Zhang H, Wing R A, Bennetzen J L. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avramova Z, Tikhonov A, SanMiguel P, Jin Y K, Liu C, Woo S S, Wing R A, Bennetzen J L. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- 18.Springer P S, Edwards K J, Bennetzen J L. Proc Natl Acad Sci USA. 1994;91:863–867. doi: 10.1073/pnas.91.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd N S, Schwarz-Sommer Z, Wienand U, Sommer H, Deumling B, Peterson P A, Saedler H. Mol Gen Genet. 1982;188:266–271. [Google Scholar]
- 20.White S E, Habera L F, Wessler S R. Proc Natl Acad Sci USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SanMiquel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, et al. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 22.Flavell R B, O’Dell M, Hutchinson J. Cold Spring Harb Symp Quant Biol. 1981;45:501–508. doi: 10.1101/sqb.1981.045.01.066. [DOI] [PubMed] [Google Scholar]
- 23.Harris N. Ph.D. dissertation. Cambridge, U.K.: Cambridge Univ.; 1987. [Google Scholar]
- 24.Abbo S, Dunford R P, Foote T N, Reader S M, Flavell R B, Moore G. Chromosome Res. 1995;3:5–15. doi: 10.1007/BF00711156. [DOI] [PubMed] [Google Scholar]
- 25.Bennetzen J L, Schrick K, Springer P S, Brown W E, SanMiguel P. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 26.Ballinger D G, Benzer S. Proc Natl Acad Sci USA. 1989;86:9402–9406. doi: 10.1073/pnas.86.23.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwaal R R, Broeks A, van Meurs J, Groenen J T M, Plasterk R H A. Proc Natl Acad Sci USA. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgkin J, Plasterk R H, Waterston R H. Science. 1995;270:410–414. doi: 10.1126/science.270.5235.410. [DOI] [PubMed] [Google Scholar]
- 29.Plasterk R H. Methods Cell Biol. 1995;48:59–80. doi: 10.1016/s0091-679x(08)61383-7. [DOI] [PubMed] [Google Scholar]
- 30.Das L, Martienssen R. Plant Cell. 1995;7:287–294. doi: 10.1105/tpc.7.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKinney E C, Ali N, Traut A, Feldmann K A, Belostotsky D A, McDowell J M, Meagher R B. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser K, Goodwin S F. Proc Natl Acad Sci USA. 1990;87:1686–1690. doi: 10.1073/pnas.87.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandler V L, Hardeman K. Adv Genet. 1992;30:77–122. doi: 10.1016/s0065-2660(08)60319-3. [DOI] [PubMed] [Google Scholar]
- 34.Bennetzen J L. Curr Top Microbiol Immunobiol. 1996;204:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- 35.Settles A M, Yonetani A, Baron A, Bush D, Cline K, Martienssen R A. Science. 1998;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 36.Martienssen R A, Barkan A, Taylor W C, Freeling M. Genes Dev. 1990;4:331–343. doi: 10.1101/gad.4.3.331. [DOI] [PubMed] [Google Scholar]
- 37.O’Kane C, Gehring W J. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson C, Bellen H J, Gehring W J. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 39.Skarnes W C. Curr Opin Biotechnol. 1993;4:684–689. doi: 10.1016/0958-1669(93)90050-7. [DOI] [PubMed] [Google Scholar]
- 40.Spradling A C, Stern D M, Kiss I, Roote J, Laverty J, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans M J, Carlton M B L, Russ A P. Trends Genet. 1997;13:370–375. doi: 10.1016/s0168-9525(97)01240-7. [DOI] [PubMed] [Google Scholar]
- 42.Sundaresan V, Springer P, Volpe T, Haward S, Dean C, Jones J, Ma H, Martienssen R A. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 43.Nussaume L, Harrison K, Klimyuk V, Martienssen R, Sundaresan V, Jones J D. Mol Gen Genet. 1995;249:91–101. doi: 10.1007/BF00290240. [DOI] [PubMed] [Google Scholar]
- 44.Swinburne J, Balcells L, Scofield S R, Jones J D G, Coupland G. Plant Cell. 1992;4:583–595. doi: 10.1105/tpc.4.5.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long D, Swinburne J, Martin M, Wilson K, Sundberg E, Lee K, Coupland G. Mol Gen Genet. 1995;241:627–636. doi: 10.1007/BF00279905. [DOI] [PubMed] [Google Scholar]
- 46.Martienssen R, Springer P. In: Insertional Mutagenesis: A Practical Approach. Coupland G, editor. Oxford: Academic; 1998. , in press. [Google Scholar]
- 47.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haseloff J, Amos B. Trends Genet. 1995;11:328–329. doi: 10.1016/0168-9525(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y G, Mitsukawa N, Oosumi T, Whittier R. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 50.Mandel M A, Yanofsky M F. Plant Cell. 1995;7:1763–1771. doi: 10.1105/tpc.7.11.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vizir I Y, Anderson M L, Wilson Z A, Mulligan B J. Genetics. 1994;137:1111–1119. doi: 10.1093/genetics/137.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mlodzik M, Hiromi Y, Weber U, Goodman C S, Rubin G M. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- 53.Bancroft I, Dean C. Genetics. 1993;134:1221–1229. doi: 10.1093/genetics/134.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James D W, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner H K. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]