SUMMARY
Rho GTPases have been implicated in diverse cellular functions and are potential therapeutic targets. By virtual screening, we have identified a Rho specific inhibitor, Rhosin. Rhosin contains two-aromatic rings tethered by a linker, and it binds to the surface area sandwiching Trp58 of RhoA with a submicromolar Kd and effectively inhibits GEF-catalyzed RhoA activation. In cells Rhosin specifically inhibited RhoA activity and RhoA-mediated cellular function without affecting Cdc42 or Rac1 signaling activities. By suppressing RhoA or RhoC activity Rhosin could inhibit mammary sphere formation by breast cancer cells, suppress invasion of mammary epithelial cells, and induce neurite outgrowth of PC12 cells in synergy with NGF. Thus, the rational designed RhoA subfamily specific small molecule inhibitor is useful for studying the physiological and pathologic roles of Rho GTPase.
Keywords: Rho GTPases, RhoA, signaling, small molecule, inhibitor, rational drug design, targeting
INTRODUCTION
Rho family GTPases are critical intracellular signaling molecules that regulate cytoskeleton organization, gene expression, cell cycle progression, cell motility, and other cellular processes (Etienne-Manneville et al., 2002; Ridley et al., 2001; Zohn et al., 1998). Abnormal Rho GTPase activities have been implicated in multiple human pathologies (Sahai et al., 2002; Boettner et al., 2002). RhoA and the closely related RhoC, in particular, are involved in cancer progression and metastasis. Upregulation of RhoA and/or RhoC activities have been reported in several human cancers (Bellizzi et al., 2008; Faried et al., 2007; Horiuchi et al., 2003). RhoA or RhoC deficiency suppresses the invasiveness of breast cancer cells (Pille et al., 2005; Chan et al., 2010), whereas elevated RhoA and/or RhoC activity can induce breast cancer cell migration, invasion and metastasis (Brantley-Sieders et al., 2008; Joshi et al., 2008). The RhoA/RhoC subfamily Rho GTPases have been suggested to serve as potential therapeutic targets.
RhoA subfamily GTPases cycle between the GTP-bound, active and the GDP-bound, inactive states in response to a variety of upstream signals, including growth factors, cytokines, adhesion molecules and cell intrinsic oncogenic cues. These incoming signals activate guanine nucleotide exchange factors (GEFs) that in turn activate Rho GTPases to elicit cellular responses. Within the Rho GTPase signaling module, several strategies of intervention have been proposed for effective suppression of Rho GTPase activities (Marchioni et al., 2009; vigil et al., 2010). Among them, targeting the GEF-Rho GTPase interactive surface essential for the guanine nucleotide exchange reaction has been successfully applied to the rational design of a lead Rac GTPse inhibitor, NSC23766, that specifically binds to the surface groove of Rac1 involved in GEF interaction and effectively inhibits Rac1 activity in diverse physiological and pathological systems (Gao et al., 2004). To date, however, in spite of extensive efforts devoted to the development of inhibitors targeting RhoA/RhoC (Genth et al., 2008; Naruniya et al., 2000; Evelyn et al., 2010), there has not been a reported success in identifying useful small molecules that work specifically on the RhoA subfamily Rho GTPases. Issues related to efficiency, specificity, and/or druggability have been a hindrance.
In the current work, we have employed a structure-based rational design strategy coupled with in silica and protein binding screening to identify a lead Rho specific inhibitor, Rhosin. Rhosin contains two aromatic chemical fragments tethered by a properly spaced linker, enabling it to bind to two adjacent shallow grooves on RhoA surface required for GEF interaction with submicromolar affinity. Rhosin specifically inhibits GEF activation of RhoA in vitro and in cells and can potently suppress breast cancer cell proliferation and invasion and induce neurite outgrowth in PC12 cells. It is useful for studying the physiological function of RhoA subfamily GTPases and for determining the therapeutic potential of Rho targeting in pathologic conditions.
RESULTS
Rational targeting the GEF - RhoA interactive surfaces
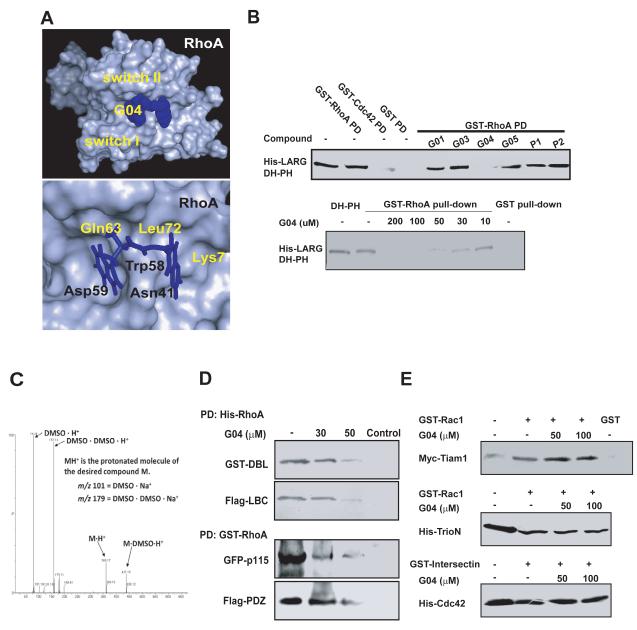
Based on a high resolution crystal structure of Rac1-Tiam1 complex, previously we have successfully identified a chemical compound, NSC23766, that specifically binds to the surface groove of Rac1 required for interaction with GEFs and effectively inhibits Rac1 activity in diverse physiological and pathological systems (Gao et al., 2004; Akbar et al., 2006). Extensive structural studies of Rho GTPase interaction with their activator GEFs (Rossman et al., 2005) led us to hypothesize that small molecules bound to the surface sites of RhoA GTPase involved in recognition by its GEFs could similarly inhibit RhoA activity and consequent downstream signaling. We used protein:protein interaction data from published x-ray crystal structures of the RhoA-LARG complex (PDB ID 1×86) (Kristelly et al., 2004) and virtual screening to search for small molecules that bind to a surface region of RhoA surrounding Trp58 that would predictably interfere with association with LARG (Figure 1A). Trp58 situates at the center of the LARG binding site of RhoA, as revealed in the LARG-RhoA co-crystal structure. Figure 1A shows a partial grid of the virtual screening targeting site, and depicts Trp58 at the position between two shallow pockets of RhoA surface involved in LARG recognition. From the docking of more than four million compounds from the ZINC library (International Zinc Association – Washington, DC), the top scoring (Krieger et al., 2004) 49 chemicals were tested for their ability to inhibit the interaction between RhoA and the DH-PH domain module of LARG in a complex formation assay. Purified LARG, which specifically binds to RhoA but not Cdc42 or Rac1 (Fukuhara et al., 2000), was incubated with RhoA in the presence of each individual compound. Among the chemicals tested, G04 was capable of suppressing LARG binding to RhoA (Figure 1B & Table S1). The inhibitory activity of G04 on RhoA/LARG interaction is dose-dependent with an effective concentration around 10 to 30 μM under the pulldown assay conditions (Figure 1B). Possible impurity and degradation of G04 and other compounds ware tested by a mass spec analysis, which showed no significant degradation product present (Fig 1C & S1 for representative MS data). G04 is specific to the interaction between RhoA and its GEFs including LARG, DBL, LBC, p115 RhoGEF or PDZ RhoGEF and does not interfere with the binding of Cdc42 or Rac1 to their respective GEFs (Figs. 1D & 1E), nor the interaction between RhoA and its effector/GAP/GDI ROCK, mDia, PKN, Rhoteckin, p190RhoGAP or RhoGDI (Figure S1). An examination of the structural analogs of G04 suggested that those compounds that contain the quinoxaline and indole/benzimidazole rings sharing a linker of sufficient length and flexibility retained the inhibitory activities (Table S2), whereas two analogs, A01 and A08, each containing only one aromatic head of G04, did not bind to RhoA (Figure S1). These results are consistent with the possibility that the tethered aromatic ring structures with proper linker length and flexibility are important for the effective binding to RhoA.
Figure 1. Identification of G04 as an inhibitor of RhoA– LARG interaction.
(A) A simulated docking model of G04 on RhoA surface. (upper) Top view of the binding pocket of RhoA bound to G04. (lower) Top view of the predicted structural contacts of G04 in the binding pocket around Trp58 of RhoA. (B-upper) The inhibitory effect of a panel of compounds predicted by virtual screening on RhoA interaction with LARG DH-PH module was tested in a complex formation assay. (His)6-tagged LARG (1μg) was incubated with GST alone or GST-RhoA (1μg) immobilized on glutathione agarose beads in the presence or absence of the1mM indicated compounds. After an incubation at 4°C for 1 hour, the beads associated (His)6-LARG were detected by anti-His Western blotting. (B-lower) Dose-dependent specific inhibition of LARG binding to RhoA by G04. (His)6-tagged LARG (1μg) was incubated with GST alone or GST-fused RhoA on glutathione agarose beads in a binding buffer containing different concentrations of G04. The beads associated (His)6-LARG were detected by anti-His Western blotting. (C) G04 in DMSO solution was dissolved into methanol at 1:100 ratio. 50μl of the methanol solution is dissolved into 450μl of 1:1 water:acetonitrile with 0.1% formic acid. The prepared compound solution was processed by Thermo Scientific LTQ-FT™ hybrid mass spectrometer consisting of a linear ion trap used for tandem mass spectrometry and a Fourier transform ion cyclotron resonance mass spectrometer. (D) The inhibitory effects of G04 on the interaction between RhoA and multiple Rho GEFs. NIH3T3 cells stably over-expressing GST-DBL, Flag-LBC or transiently over-expressing GFP-p115 RhoGEF or Flag-PDZRhoGEF, were harvested and the cell lysates were subjected to the His-RhoA or GST-RhoA pull-down assay in the absence or presence of G04 at the indicated concentrations. (E) G04 does not affect Rac1 or Cdc42 binding to respective GEFs. Purified, immobilized GST-Rac1 or GST-intersectin were incubated with cell lysates expressing myc-Tiam1, (His)6-TrioN or (His)6-Cdc42 in the presence or absence of indicated concentrations of G04. The co-precipitates were subject to Western blotting to detect Rac1 binding to Tiam1 or triN, and cdc42 binding to intersectin.
The lead inhibitor G04 specifically binds to RhoA to inhibit GEF reaction of RhoA
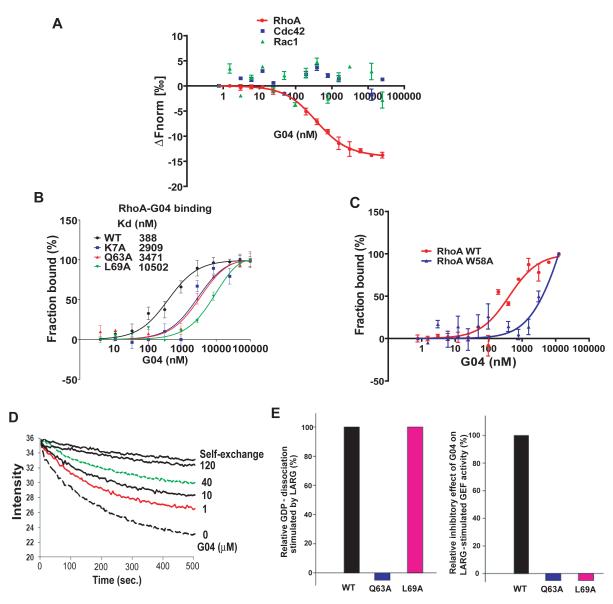
The unique Trp58 residue at the GEF recognition site of RhoA allowed us to use its intrinsic fluorescence to monitor the direct interaction of G04 with RhoA. Titration of increasing concentrations of G04 readily quenched the tryptophan fluorescence emission of RhoA dose-dependently, whereas an analog of G04, A03, was not effective (Figure S2), suggesting a binding of G04 to RhoA that affects Trp58 fluorescence. To more strictly quantify the direct binding interaction between RhoA and G04, a microscale thermophoresis analysis (Duhr et al., 2006) was carried out using purified RhoA protein. This assay shows that G04 specifically binds to WT RhoA with an affinity ~0.4 uM Kd (Kd = 354±48 nM, Figure 2A), whereas it does not detectably interact with Cdc42 or Rac1, nor the GEF, LARG (Figures 2A & S2). As positive controls, Cdc42 and Rac1 were found to interact with their inhibitors CASIN and NSC23766, respectively, in similar assays (data not shown). To further confirm the structural motif of RhoA involved in G04 binding, RhoA point mutants bearing Ala mutations around the predicted G04 binding site, i.e. K7A, Q63A and L69A, were examined for their binding affinity to G04 by thermophoresis analysis. G04 showed significantly reduced affinity towards L69A (Kd =10502 ± 2310 nM), K7A (Kd =2909 ± 1030 nM), and Q63A (Kd =3471 ± 912 nM, Figure 2B), indicating that these residues participate in the G04 binding. We also have analyzed the interaction between G04 and the RhoAW58A mutant by an affinity binding assay and found that mutation of Trp58 of RhoA to alanine partially inhibits G04 binding, yielding a Kd of 6.2 μM compared with G04 binding to WT RhoA with a Kd of ~0.4 μM (Fig. 2C). These data, together with the Trp fluorescence assay of G04 titration to WT RhoA protein (supplemental Fig S3), strongly suggest that G04 interaction with RhoA involves Trp58 and/or the surrounding region. To examine whether G04 could specifically inhibit GEF-catalyzed guanine nucleotide-exchange of RhoA, a GDP/GTP exchange assay was performed in the presence of increasing concentrations of G04. G04 was able to inhibit the nucleotide exchange of RhoA catalyzed by LARG dose-dependently (Figure 2D) but was inactive in affecting Cdc42 and Rac1 exchange catalyzed by their selective GEFs, intersectin and TrioN, respectively (data not shown). Consistent with the significantly reduced G04 binding activity, although L69A still retained the WT activity to respond to LARG in the GEF reaction, it has lost responsiveness to G04 inhibition at the GEF assay conditions (Figure 2E). Collectively, these results indicate that G04 binds to RhoA with micromolar affinity at the predicted docking pocket involved in GEF recognition.
Figure 2. Biochemical characterizations of G04 interaction with RhoA.
(A-C) Microscale thermophoresis analysis of G04 to RhoA. Purified RhoA, Cdc42, Rac1 or RhoA mutants were first labeled with Alexa-647 fluorescence dye. G04 was titrated between 4nM to 100,000 nM or 0.76 nM to 250,000 nM to a constant amount of labeled RhoA, RhoA mutants, Cdc42 or Rac1 proteins (100 nM). The reaction was performed in 50 mM Hepes, 50 mM NaCl, 0.01% Tween20 and 2 mM MgCl2. The samples were incubated in room temperature for 1 hour before the measurements. Data were normalized to either ΔFnorm [‰] (10*(Fnorm(bound) - Fnorm (unbound))) or Fraction bound (ΔFnorm [‰]/amplitude).The average Kd values ± SD were calculated from three replicates. All the data are representative of three independent experiments. (D) G04 was effective in inhibiting RhoA GDP/GTP exchange stimulated by LARG in a dose-dependent manner. 50 nM RhoA loaded with BODIPY FL-GDP was incubated at 25°C in an exchange buffer containing 100 mM NaCl, 5 mM MgCl2, 50 mM Tris·HCl (pH 7.6), and 0.5 mM GTP in the absence (top line) or presence of 30 nM LARG. Increasing concentrations of G04 were included in the exchange buffer as indicated. (E) Examination of the binding residues of G04 on RhoA through the GDP/GTP exchange assays of RhoA mutants. (E-left) The GDP/GTP exchange activities of WT RhoA and RhoA mutants K7A, Q63A and L69A were examined in the presence of LARG. The relative GDP dissociation of each mutant at 10 min was normalized to that of WT RhoA without LARG in a parallel reaction. (E-right) The inhibitory effects of G04 (10 μM) on the GDP/GTP exchange activities of WT RhoA and the respective RhoA mutants were examined. The GEF reaction conditions were similar to that in (D). The relative inhibitory effects by G04 were normalized to that of WT RhoA.
The lead inhibitor G04 specifically inhibits RhoA subfamily Rho GTPases in cells
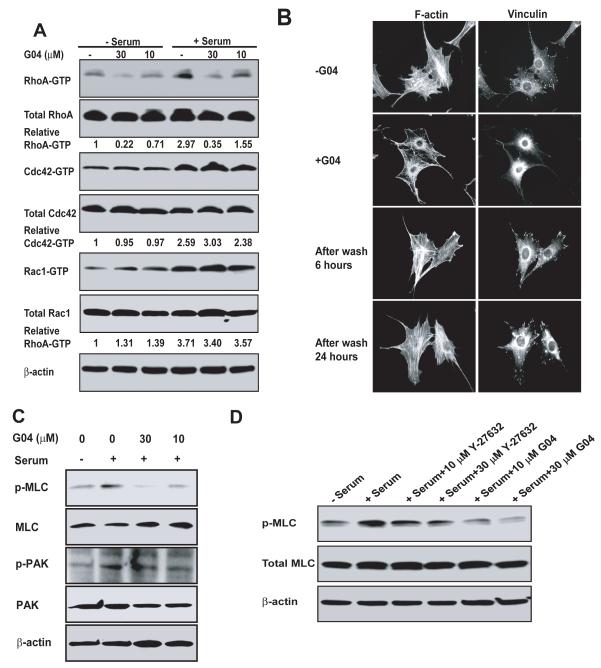
To examine whether G04 is effective in specifically suppressing RhoA activity in cells, NIH 3T3 cells grown in serum-free media were treated with G04 in different concentrations, followed by stimulation with or without 10% calf serum. As shown in Figure 3A, G04 strongly inhibited RhoA-GTP formation in both serum-free and serum stimulation conditions with an EC50 ~10-30μM under the assay conditions, but did not affect the activities of endogenous Cdc42 and Rac1 in the same cells. To evaluate the ability of G04 to inhibit RhoA-mediated cell functions, we next examined actin cytoskeleton structures of cells stimulated by serum or LPA in the absence or presence of G04. Figure 3B & Figure S3 show that in the presence of 30 μM G04, both stress fiber and focal complex formation of the cells were significantly reduced, whereas the lamellipodia and filapodia stimulated by PDGF and Bradykinin, respectively, were not affected. To examine whether G04 may act reversibly, we washed the G04 treated cells and then analyzed the effects on actin polymerization and focal adhesion complex at 6-hour and 24-hour time points. The results showed that removal of G04 by the wash could reverse the inhibitory effects of G04 (Fig. 3B). In addition, G04 significantly inhibited integrin-mediated cell adhesion to fibronectin (Figure S3). Under these assay conditions, G04 did not appear to be cytotoxic as it did not affect the survival status of the cells at the concentrations that show efficacy in disrupting actin stress fiber and focal adhesions (Figure S3). Given the implicated role of Rho activity in actin cytoskeleton organization and adhesion (Hall, 1998), these results indicate that G04 is active in specifically inhibiting RhoA-mediated cellular events.
Figure 3. Cellular validation of G04 as a specific inhibitor of RhoA activity.
(A) NIH3T3 cells were treated with G04 at the indicated concentrations for 24 hours in serum-free media. Cells were subsequently stimulated with or without 10% calf serum for 15 min and were subjected to GST-Rhotekin or GST-PAK1 effector domain pull-down assays, and the activities of RhoA, Cdc42 and Rac1 were examined. Blotting of the respective total cell lysates was carried out in parallel. Relative amounts of GTP-bound form of the GTPases were quantified by densitometry measurements and normalized to those of the unstimulated cells. (B) Effect of G04 on cell stress fiber and focal complex assembly. NIH3T3 cells were treated with or without 30 μM G04 in serum-free media for 24 hours, and subsequently were stimulated with 10% calf serum for 15 minutes. Medium containing G04 was removed and cells were washed for 3 times. The cells were then fixed 6 and 24 hours after the wash, respectively, and stained with Rhodamine-phalloidin for F-actin and anti-vinculin for focal adhesion complexes. Images shown are representative of more than 100 cells examined. (C) G04 treatment affects signaling downstream of RhoA, but not that of Rac1/Cdc42. Western blots are of p-PAK and p-MLC and relevant controls of NIH 3T3 cells treated with G04 at indicated concentrations in serum-free media and subsequently stimulated by 10% calf serum for 10 min. (D) A comparison of G04 and ROCK inhibitor, Y-27632. The relevant control NIH3T3 cells or cells treated with G04 or Y-27632 of indicated concentrations were stimulated with 10% calf serum for 10 minutes prior to blotting for p-MLC.
To further define the cellular specificity of G04, we next examined its effect on the closely related RhoB and RhoC proteins of the RhoA subfamily and downstream signaling. Consistent with its mode of action, G04 inhibited RhoB and RhoC activities in cells similarly to that of RhoA (Figure S3), since these RhoA subfamily members share identical surface residues required for G04 binding. G04 efficiently inhibited serum-induced phosphor-MLC formation, an event mediated by the RhoA effector ROCK, without affecting p-PAK level that is regulated by Rac/Cdc42 (Figure 3C). We compared the effect of G04 with the ROCK inhibitor, Y27632, and found that G04 and Y27632 had a similar effect in inhibiting MLC-phosphorylation (Fig. 3D).
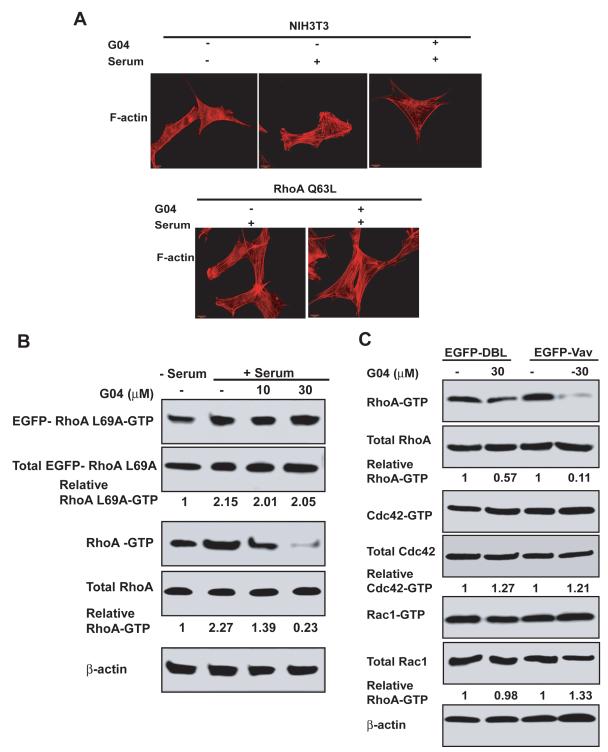
To examine whether G04 works specifically through the RhoA-mediated pathway, we performed mutant rescue experiments using constitutively active RhoA or ROCKII mutant and a G04 binding defective RhoA mutant, RhoAL69A. G04 treatment did not alter actin stress fiber and focal adhesion complex formation induced by a constitutively active ROCKII or RhoA mutant, nor the constitutively active RhoA mutant-induced downstream signaling to p-MLC and p-FAK (Figure 4A & Figure S4). Furthermore, the RhoA mutant, RhoAL69A, defective in G04 binding but not GEF activation (Fig. 2B & 2E), was resistant to G04 in cells by maintaining unaffected RhoAL69A-GTP level, whereas the endogenous WT RhoA-GTP was responsive to G04 inhibition (Fig. 4B). Together, the RhoAL69A, RhoAQ63L, and active ROCKII rescue experiments suggest that G04 can specifically suppress RhoA activity and RhoA-mediated signaling function. Finally, DBL and Vav are two GEFs that can activate a broad spectrum of Rho GTPases including RhoA, Cdc42 and Rac1. We found that G04 could readily inhibited RhoA-GTP formation, but not that of Cdc42 or Rac1, in serum-starved DBL- or Vav-expressing cells, further supporting the specificity of G04 towards RhoA (Fig. 4C). Based on these in vitro and cellular properties, we termed G04 a Rho activity specific inhibitor, or Rhosin.
Figure 4. Cellular validation of G04 specificity by ectopic expression of RhoA mutants or RhoGEFs.
(A) WT NIH 3T3 cells and RhoAQ63L expressing cells were grown on tissue culture dish. After an 24-hour starvation in the presence of 30μM G04, cells were stimulated with 10% calf serum and were co-stained with Rhodamine-phalloidin for F-actin. (B) NIHT3 cells were transduced retrovirus expressing EGFP-RhoAL69A mutant. Cells were then treated with G04 at indicated concentrations in serum-free medium. Cells were subsequently stimulated with 10% calf serum for 15 min and were subjected to GST-Rhotekin effector domain pull-down assay. The RhoA-GTP and EGFP-RhoAL69A-GTP levels were examined by RhoA and EGFP antibodies, respectively. Blotting of the respective total cell lysates was carried out in parallel. Relative amounts of the GTP-bound form of the GTPases were quantified by densitometry measurements and normalized to those of the unstimulated cells. (C) NIH3T3 cells stably expressing DBL or Vav were treated with G04 at the indicated concentrations for 24 hours in serum-free medium. Cells were subsequently stimulated with or without 10% calf serum for 15 min and subjected to GST-Rhotekin or GST-PAK1 effector domain pull-down assay, and the activities of RhoA, Cdc42 and Rac1 were determined. Blotting of the respective total cell lysates was carried out in parallel. Relative amounts of the GTP-bound form of the GTPases were quantified by densitometry measurements and normalized to those of the unstimulated cells.
Rhosin effectively reverses the functions of breast cancer cells and neuronal cells regulated by RhoA
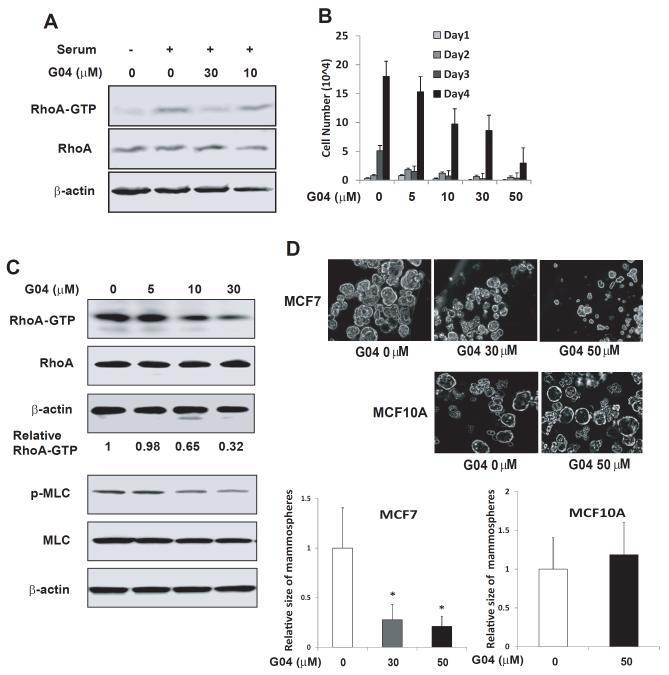
Elevated RhoA and RhoC activities have been associated with cancer cell hyperproliferative and invasive behaviors (Sahai et al., 2002; van Golen et al., 2000; Clark et al., 2000). Next, we investigated the effect of Rhosin on the mammosphere formation of breast cancer MCF7 cells, a property associated with their tumorigenic potential. Rhosin showed a dose-dependent inhibition of endogenous RhoA activity (Figure 5A) and cell growth of MCF7 cells (Figure 5B). To assess the cellular effect of Rhosin, we performed cell apoptosis assay and cell cycle analysis. As shown in Figure S5, Rhosin could significantly induce cell apoptosis but did not affect cell cycle progression. These results suggest that a partial inhibition of RhoA activity may not affect cytokinesis as that shown by a complete removal of RhoA gene by gene targeting (Melendez et al., 2011). Moreover, Rhosin dose-dependently reduced RhoA and p-MLC1 activities of MCF7 cell-derived mammospheres with an EC50 ~30-50μM, and caused decreased size and reduced number of mammospheres in MCF7 cells (Figure 5C & 5D). Interestingly, Rhosin has no effect on the non-tumorigenic epithelial MCF10A cells derived mammosphere growth (Figure 5D). These results indicate that Rhosin is effective in targeting RhoA mediated breast cancer cell proliferation.
Figure 5. Rhosin inhibits RhoA activity and suppresses proliferation of breast cancer cells.
(A) Rhosin inhibits RhoA activity in MCF7 cells. MCF7 cells were treated with G04 or Analog3 of indicated concentrations for 24 hours in a serum-free media. Cells were subsequently stimulated with 10% fetal bovine serum for 15 min and were subjected to Rhotekin effector domain pull-down assays, and the activities of RhoA were determined. Blotting of the respective total-cell lysates was carried out in parallel. Results shown are representative of three independent experiments. (B) Rhosin inhibits MCF7 cell growth. MCF7 cells were plated at 1.5 ×104 /24 well in the presence of G04. Cell numbers were determined at the indicated times. (C) Rhosin inhibits RhoA and its downstream signaling activities in MCF7-derived mammospheres. MCF-7 cells were dissociated to single cells with trypsin and cultured for 10 days at the density of 2×104/mL in suspension. The spheres were collected and subjected to GST-Rhotekin effector domain pull-down assays, and the activities of RhoA were examined. Blotting of the respective total cell lysates was carried out in parallel. Relative amounts of GTP-bound form of RhoA were quantified by densitometry measurements and normalized to those of the untreated cells. Lower panel: Western blots are p-MLC of MCF7-derived spheres treated with G04 at indicated concentrations. (D) Rhosin inhibits MCF-7 cell-derived mammosphere formation. MCF7 and MCF10A cells were dissociated to single cells with trypsin and cultured in the media with G04 at the indicated concentrations for 10-14 days at the density of 2×104/mL in suspension. Photos were taken after 10-14 day’s culture. Images shown are representative of five to ten fields containing a total of at least 100 spheres which were chosen randomly. The average size of mammary sphere were measured and calculated relative to the control.
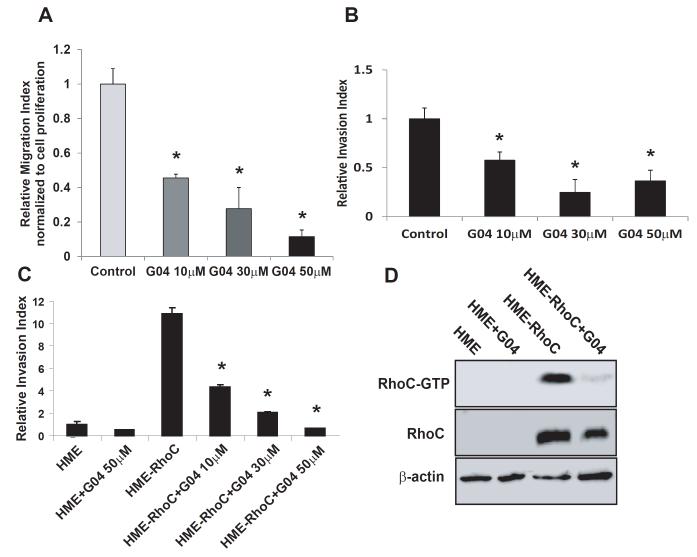
Both RhoA and its closely related subfamily member, RhoC, have been implicated in promoting breast cancer cell migration, invasion and metastasis (Pille et al., 2005; Chan et al., 2010; Brantely-Sieders et al., 2008). We next examined the effect of Rhosin on breast cancer cell migration and invasion properties. The results showed that Rhosin potently inhibited MCF7 cell migration and invasion activities, dose-dependently (Figs. 6A & 6B). In human mammary epithelial cells that were endowed with a highly invasive phenotype by RhoC expression (van Golen et al., 2000), Rhosin readily inhibited RhoC-GTP formation and the resulting invasive activity (Figs. 6C & 6D). These results indicate that Rhosin can suppress RhoA and RhoC mediated breast cancer cell migration and invasion.
Figure 6. Rhosin suppresses the migration and invasion of breast cancer cells.
(A) Rhosin inhibits MCF7 cell migration. MCF7 cells were subjected migration assays in the presence of G04. The percentages of the cells that migrated towards a 10% FBS gradient for 16 hours in a transwell migration chamber were calculated by dividing the number of migrated cells into the number of input equivalents plated on the transwell. (B) Rhosin inhibits MCF7 cells invasion. The invasive activities were assayed in a Matrigel-coated transwell. The MCF7 cells that succeeded in invasion into the Matrigel were stained with 5% Giemsa solution for visualization and quantification 16 hours after plating in the presence of G04 of the indicated concentrations. (C-D) Rhosin supresses HME-RhoC cells invasion activity via inhibiting the RhoC activity. Human mammary epithelial (HME) cells and RhoC expressing HME cells were grown in Ham’s F-12 medium supplemented with 5% FBS, insulin, hydrocortisone, epidermal growth factor, and cholera toxin. (C) The invasive activities were assayed in a Matrigel-coated transwell. The cells that succeeded in invasion into the Matrigel were stained with 5% Giemsa solution for visualization and quantification 16 hours after plating in the presence of G04 of the indicated concentrations. (D) The cells were treated with 30 μM for 24 hours in serum-free media. Cells were subsequently stimulated with 10% fetal bovine serum for 15 min and were subjected to a pull-down assay, and the activity of RhoC was determined. Blotting of the respective total-cell lysates was carried out in parallel. Results shown are representative of three independent experiments.
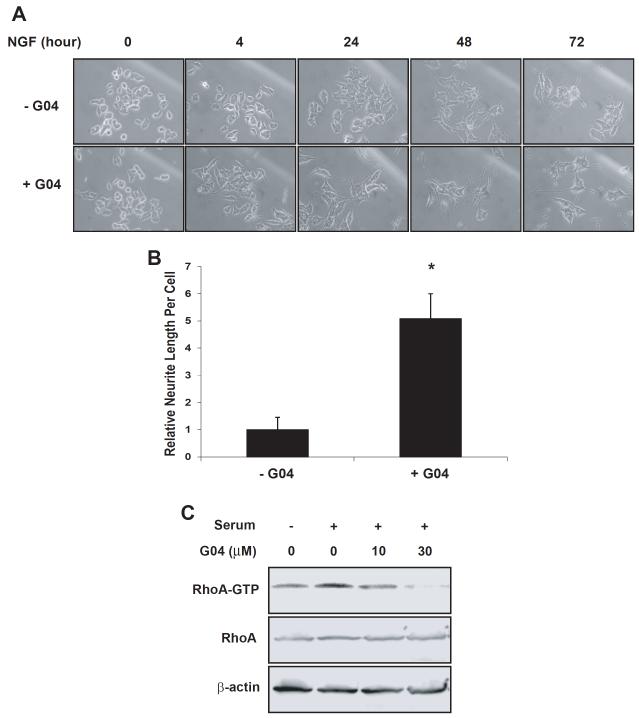
Inactivation of RhoA promotes the neurite outgrowth of neuronal cells (Sebok et al., 1999; Kozma et al., 1997). Next, we tested the effect of Rhosin on the neuritogenesis of a well characterized neuronal cell line, PC-12. In the presence of 50 mg/ML NGF and 30μM Rhosin, neurite initiation and branching were significantly increased compared with that following NGF treatment alone (Figs. 7A & 7B). A Rho GTPase activity pull-down assay confirmed that the RhoA-GTP level of PC12 cells was responsive to Rhosin (Figure 7C). These observations indicate that Rhosin can induce neurite outgrowth and branching of PC12 cells similar to that of previously reported C3 toxin treatment (Sebok et al., 1999).
Figure 7. Rhosin induces neurite outgrowth of PC12 cells in synergy with NGF.
(A-B) PC12 cells were treated with 30 μM G04 and 50 ng/mL NGF for 72 hours. Photos were taken at indicated time points (A) when relative neurite lengths per cell were measured (B). At least 100 cells were chosen randomly for the measurement. Total neurite output was determined by dividing the combined length of all neurites by the number of neurite-bearing cells. (C) PC12 cells were treated with G04 of indicated concentrations for 24 hours in serum-free media. Cells were subsequently stimulated with 5% fetal calf serum and 10% horse serum for 15 min and were subjected to effector domain pull-down assays, and the activity of RhoA was examined by GST-Rhoteckin pulldown. Relative amounts of RhoA-GTP were quantified by densitometry measurements and normalized to those of unstimulated cells.
DISCUSSION
Rho family GTPases belong to the Ras-like small GTPases superfamily and are intracellular signal transducers involved in diverse cell signal transduction processes from cell adhesion molecules, growth factor receptors, G-protein coupled receptors, and cytokines. It has become increasingly appreciated that Rho family members, RhoA and RhoC in particular, are often upregulated or hyperactivated in human diseases including cancer and inflammation, and these Rho GTPases can serve as useful targets in reversing pathologic conditions such as cancer cell proliferation and invasion (Faried et al., 2007; Horiuchi et al., 2003). Activation and signal transduction through Rho GTPase regulated pathways require cascades of protein-protein interactions, including enzymatic reactions of kinases and GTPases. However, Rho GTPases, like Ras, are considered undruggable by conventional means. This is in part because the small GTPases themselves are globular structures with limited surface structural areas suitable for high affinity binding by small molecules (Kristelly et al., 2004). Thus, extensive efforts have previously been devoted to the development of inhibitors of the upstream regulators (e.g. farnesyl or gerynyl transferases (Sebti, et al., 2003)), Rho GTPases themselves (e.g. by bacteria toxins (Genth et al., 2008) or small molecules (Gao et al., 2004; Onesto et al., 2008)) and downstream effectors (e.g. ROCK and SRF (Narumiya et al., 2000; Evelyn et al., 2010), of the GTPase signaling pathways, but none have achieved clinical utility.
Activation of Rho GTPases by the Dbl family GEFs is known to be one major mechanism leading to the activated state of multiple Rho family members including RhoA, RhoC, Cdc42 and Rac1. Extensive structural and biochemical work has provided detailed information on how this class of GEFs recognizes a Rho GTPase substrate and catalyzes the GDP/GTP exchange. The studies have presented a possibility that small molecules designed for specifically targeting the GEF - Rho GTPase interaction could be obtained to affect the Rho GTPase activities, not unlike previously identified brefeldin A that can bind to and inhibit an ARF small GTPase GEF (Louis et al., 2003). Applying this rationale, we previously identified a lead Rac activation-specific inhibitor, NSC23766, that displays a micromolar binding affinity to Rac1 at the GEF binding site and is active for inhibiting GEF activation of Rac1(Gao, et al., 2004). However, effort in enhancing the efficacy of this lead has proven difficult mostly due to the low affinity nature of the available targeting site on Rac1.
Here, we have identified a lead small molecule inhibitor that specifically targets RhoA and closely related RhoB and RhoC by structure-based rational design. This inhibitor, Rhosin, contains two aromatic chemical fragments that are tethered by a properly spaced linker, with each likely bound to a distinct, shallow groove of RhoA. In vitro it is able to specifically inhibit the RhoA-GEF interaction and RhoA activation by GEFs. In cells, Rhosin blocks RhoA activation and RhoA-mediated MLC phosphorylation, actin stress fiber formation and focal adhesion assembly without affecting the activity or signaling events of endogenous Cdc42 or Rac1. Moreover, this compound showed no cytotoxicity or non-specific effect on the constitutively active RhoA or ROCK mutant induced actomyosin changes. Finally, by suppressing RhoA or RhoC activity, Rhosin was active in decreasing mammosphere formation and inhibiting invasion of breast cancer cells, and promoting neurite outgrowth from neuronal cells. Thus, Rhosin constitutes a Rho-specific small molecule inhibitor that is useful in the study of the physiological role of Rho and for tackling Rho-mediated pathologies.
Cdc42 or Rac1 shares high homologies with RhoA in the region between switches I & II involved in Rhosin binding and GEF interaction. An analysis of the amino acid sequences and the 3D structure of the RhoA binding pocket indicates that Cdc42 contains a Phe56 instead of Trp58 in the critical site of Rhosin binding, whereas Rac1, although containing mostly conserved amino acid residues, has a minor structural conformation difference from RhoA (Worthylake et al., 2000; Kristelly et al., 2004). Likely due to similar reasons, the previously characterized Rac inhibitor, NSC23766, shows high specificity towards Rac1 (Gao et al., 2004). The detailed mode of action by Rhosin in interaction with RhoA awaits further structural illustrations.
We have performed multiples assays in this study to test the target binding affinity (i.e. Kd), the IC50 for the inhibition of GEF-RhoA binding, and the EC50 of cellular activity, of G04. In the assay conditions, G04 displays a Kd of ~0.4 uM to RhoA, and a weaker efficacy in inhibiting the GEF binding to RhoA reaction or cellular RhoA-GTP formation (at over 10 uM). This is not unlike most kinase inhibitors that have a high binding affinity to targets but may require higher concentrations to be effective to inhibit a kinase-substrate interaction or a kinase reaction in cells. Although the lead inhibitors of RhoA identified in the current studies remain a distance from pharmaceutical applications, our studies put forward a demonstrated example that the GEF reaction of small GTPase activation is a valid site for rationalized targeting. The discovery of Rhosin, a lead inhibitor targeting RhoA that contains two aromatic chemical fragments tethered by a properly spaced linker, with each likely bound to a distinct, shallow groove of RhoA, adds to the previously characterized Rac GTPase-specific small molecule inhibitors that shared the principle of targeting the GEF recognition site of a Rho GTPase (Kristelly et al., 2004). It further raises the possibility that design and search for double headed, low affinity binding structures tethered by a properly sized linker may be applicable to small GTPases or other difficult to target biological molecules.
SIGNIFICANCE
Small molecule chemicals, as one of the major structural classes of drugs, are broadly used in targeting oncoproteins and their signaling pathways (Verdine et al., 2007). However, only proteins containing deep hydrophobic pocket are considered druggable (Hopkins et al., 2002; Russ et al., 2005), which significantly limits the scope of small molecule drug development in targeted therapy. RhoA has a globular structure with limited surface pocket areas, which is a hinder for high affinity binding by small molecule compounds. The discovery of Rhosin, a lead inhibitor targeting RhoA that contains two aromatic chemical fragments tethered by a properly spaced linker, with each likely bound to a distinct, shallow groove of RhoA, adds to the previously characterized Rac GTPase-specific small molecule inhibitors that shared the principle of targeting the GEF recognition site of a Rho GTPase. It further raises the possibility that design and search for low affinity binding structures tethered by a properly sized linker may be applicable to small GTPases or other difficult to target biological molecules. Comparing with other inhibitors for RhoA or RhoA signaling pathway, Rhosin has higher affinity and specificity without detectable toxicity in normal cells. Our functional studies in breast cancer cells and neuronal cells further suggest its potential usage in cancer and neurologic disorders.
EXPERIMENTAL PROCEDURES
Virtual Screening
The atomic interactions between RhoA and LARG were obtained from PDB ID 1×86 (21). The concave surface to either side RhoA Trp58 meets these criteria and was chosen as the binding site for virtual screening (see Supplemental Materials). The in silico compound library was derived from the ZINC purchasable (International Zinc Association – America, Washington, DC) compound set. Single molecules were used to create conformers using OMEGA at default settings (OpenEye Scientific Software). The conformer library was screened across the W58 binding site using FRED, rigid ligand and receptor, default scoring (OpenEye Scientific Software), in parallel mode on an SGI Altix 4700 computer. The top 1000 hits were submitted for flexible-ligand, rigid-receptor docking and energy minimization using Molegro, default scoring (Molegro ApS). The top 100 compounds were selected and the top 49 were obtained for testing. Among the 49 compounds, Y1-Y17 were obtained from ChemBridge (San Diego, CA); G1-G16 were obtained from InterBioScreen Ltd.(Russia); O1-O12 were obtained from Enamine (Ukraine); P1-P5 were obtained from ChemDiv (San Diego, CA). All the analogs of G04 (A01-A16) were obtained from P&G Compound library (Cincinnati, OH).
In Vitro Complex Formation Assay
About 1 μg of (His)6-tagged LARG DH-PH was incubated with 1 μg of EDTA-treated (1mM) GST-fused RhoA or Cdc42 or GST alone in binding buffer (20 mM Tris·HCl (pH 7.6), 100 mM NaCl, 1% BSA, 1% Triton X-100, 1 mM MgCl2), and 15 μl of suspended glutathione-agarose beads. Compound G04 or other chemicals were added in the incubation buffer at the indicated concentrations. After incubation at 4°C for 1 hour under constant agitation, the glutathione beads were washed twice with the binding buffer. The amount of (His)6-tagged protein co-precipitated with the GST-fusion-bound beads was detected by anti-His Western blotting. Similarly, cell lysates containing myc-Tiam1, (His)6-Cdc42, (His)6- TrioN or (His)6- RhoA were mixed with purified GST-Rac1, GST-Cdc42, GST-mDia, GST-PKN, GST-Rhotekin, GST-ROCKII, GST-p190GAP or GST alone, and the pairwise association between the proteins was assessed in the presence of the indicated amount of G04 in the respective assays.
Microscale Thermophoretic Analysis
A NanoTemper Monolith Instrument (NT.015) was used for measuring thermophoresis (see Supplemental Materials). In this instrument an infra red-Laser (IR-Laser) beam couples into the path of light (i.e. fluorescence excitation and emission) with a dichroic mirror and is focused into the sample fluid through the same optical element used for fluorescence imaging. The IR laser is absorbed by the aqueous solution in the capillary and locally heats the sample with a 1/e2 diameter of 25 μm. Up to 24 mW of laser power where used to heat the sample, without damaging the bio-molecules (Wienken et al., 2010). Thermophoresis of the protein in presence of varying concentrations of compound was analyzed for 30 seconds. Measurements were performed at room temperature and standard deviation was calculated from three independent experiments. Data were normalized to either ΔFnorm [‰] (10*(Fnorm(bound) - Fnorm (unbound))) or Fraction bound (ΔFnorm [‰]/amplitude).
Guanine Nucleotide Exchange Assay
A 200 μl solution of Tris-HCl (20 mM, pH 7.6), NaCl (100 mM) and MgCl2 (1 mM) containing purified RhoA(50 nM), Cdc42 (2 μM) or Rac1(2 μM) protein was incubated with 25 nM of BODIPY FL-GDP (Invitrogen) at 25°C until the monitored fluorescence signal was constant, and then 10 μM GDP was added. Self-exchange was monitored by reading the change in fluorescence intensity. The GEF-catalyzed exchange was performed by adding purified LARG (10 nM), intersectin (10 nM) or TrioN (50 nM) to the BODIPY- FL-GDP-loaded RhoA, Cdc42 or Rac1 in the presence or absence of G04 (1-120 μM). Fluorescence intensity was measured using a Cary Eclipse fluorescence spectrophotometer (Gao et al., 2004).
Endogenous Rho GTPase Activity Assay
NIH3T3 cells, MCF7 cells, or HME cells were grown in log phase in a 10-cm dish or a six-well dish, and were starved in serum-free medium in the presence or absence of G04 at indicated concentrations for 24 hours and were subsequently stimulated with 10% calf serum or fetal bovine serum for 15 min. Cells were lysed in a buffer containing 20 mM Tris-HCL, pH 7.6, 100 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 2 mM NaF and protease inhibitors (2 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin). Lysates were clarified, the protein concentrations were normalized, and the GTP-bound RhoA, Rac1 or Cdc42 in the lysates were measured by respective anti-RhoA, Rac1 and Cdc42 Western blotting of the effector domain pull-downs.
Statistical Analysis
All experimental data were analyzed and compared for statistically significant differences by two-tailed Student’s t test. Data are presented as the averaged values ± standard deviations (SD) where applicable. For Microscale Thermophoresis assay, non-linear regression was used to fit curves to the mean and standard deviations (N=3) calculated with GraphPad Prism™ software. For Western blot quantification, one representative sample of three or more experiments is shown.
Supplementary Material
HIGHLIGHTS.
A lead inhibitor, Rhosin, targeting the GEF-interactive site of RhoA was identified by virtual screen.
Rhosin binds to RhoA and inhibits RhoA-RhoGEF interaction,
Rhosin exhibits dose-dependent, reversible inhibitory activity towards RhoA.
Rhosin is useful in studying the role of RhoA subfamily GTPases and Rho-mediated cell functions.
Footnotes
Author contributions X.S. performed research, contributed vital new reagents or analytical tools, analyzed data, and wrote the paper; F.M., N.S., C.R.E., M.J.-W and S.D. analyzed data; W.S. and M.W. performed research and analyzed data; Y. Z. designed research, contributed vital new reagents or analytical tools, analyzed data, and wrote the paper.
Competing financial interests The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akbar H, Cancelas J, Williams DA, Zheng J, Zheng Y. Rational design and applications of a Rac GTPase-specific small molecule inhibitor. Methods Enzymol. 2006;406:554–565. doi: 10.1016/S0076-6879(06)06043-5. [DOI] [PubMed] [Google Scholar]
- 2.Bellizzi A, Mangia A, Chiriatti A, Petroni S, Quaranta M, Schittulli F, Malfettone A, Cardone RA, Paradiso A, Reshkin SJ. RhoA protein expression in primary breast cancers and matched lymphocytes is associated with progression of the disease. Int. J. Mol. Med. 2008;22:25–31. [PubMed] [Google Scholar]
- 3.Boettner B, Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- 4.Brantley-Sieders DM, Zhuang G, Hicks D, Fang WB, Hwang Y, Cates JM, Coffman K, Jackson D, Bruckheimer E, Muraoka-Cook RS, et al. The receptor tyrosine kinase EphA2 promotes mammary adenocarcinoma tumorigenesis and metastatic progression in mice by amplifying ErbB2 signaling. J. Clin. Invest. 2008;118:64–78. doi: 10.1172/JCI33154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, Wu J, Nakayama KI, Kang HY, Huang HY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat. Cell Biol. 2010;12:457–267. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 7.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duhr S, Braun D. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19678–19682. doi: 10.1073/pnas.0603873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 10.Evelyn CR, Bell JL, Ryu JG, Wade SM, Kocab A, Harzdorf NL, Showalter H.D. Hollis, Neubig RR, Larsen SD. Design, synthesis and prostate cancer cell-based studies of analogs of the Rho/MKL1 transcriptional pathway inhibitor, CCG-1423. Med. Chem. Lett. 2010;20:665–672. doi: 10.1016/j.bmcl.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faried A, Faried LS, Usman N, Kato H, Kuwano H. Clinical and prognostic significance of RhoA and RhoC gene expression in esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2007;14:3593–3601. doi: 10.1245/s10434-007-9562-x. [DOI] [PubMed] [Google Scholar]
- 12.Fukuhara S, Chikumi H, Gutkind JS. Leukemia-associated Rho guanine nucleotide exchange factor (LARG) links heterotrimeric G proteins of the G(12) family to Rho. FEBS Lett. 2000;485:183–188. doi: 10.1016/s0014-5793(00)02224-9. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genth H, Dreger SC, Huelsenbeck J, Just I. Clostridium difficile toxins: more than mere inhibitors of Rho proteins. Int. J. Biochem. Cell Biol. 2008;40:592–597. doi: 10.1016/j.biocel.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;9:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 17.Horiuchi A, Imai T, Wang C, Ohira S, Feng Y, Nikaido T, Konishi I. Up-regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab Invest. 2003;83:861–870. doi: 10.1097/01.lab.0000073128.16098.31. [DOI] [PubMed] [Google Scholar]
- 18.Joshi B, Strugnell SS, Goetz JG, Kojic LD, Cox ME, Griffith OL, Chan SK, Jones SJ, Leung SP, Masoudi H, et al. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008;68:8210–8220. doi: 10.1158/0008-5472.CAN-08-0343. [DOI] [PubMed] [Google Scholar]
- 19.Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieger E, Darden T, Nabuurs S, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- 21.Krieger E, Nielsen JE, Spronk CA, Kollman PA. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J. Biol. Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 23.Marchioni F, Zheng Y. Targeting rho GTPases by peptidic structures. Curr. Pharm. Des. 2009;15:2481–2487. doi: 10.2174/138161209788682334. [DOI] [PubMed] [Google Scholar]
- 24.Melendez J, Stengel K, Zhou X, Chauhan BK, Debidda M, Andreassen P, Lang RA, Zheng Y. RhoA GTPase is dispensable for actomyosin regulation but is essential for mitosis in primary mouse embryonic fibroblasts. J. Biol. Chem. 2011;286:15132–15137. doi: 10.1074/jbc.C111.229336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narumiya S, Ishizaki T, Uehata M. Use and properties of ROCK-specific inhibitor Y-27632. Methods Enzymol. 2000;325:273–284. doi: 10.1016/s0076-6879(00)25449-9. [DOI] [PubMed] [Google Scholar]
- 26.Nassar N, Cancelas J, Zheng J, Williams DA, Zheng Y. Structure-function based design of small molecule inhibitors targeting Rho family GTPases. Curr. Top. Med. Chem. 2006;6:1109–1116. doi: 10.2174/156802606777812095. [DOI] [PubMed] [Google Scholar]
- 27.Onesto C, Shutes A, Picard V, Schweighoffer F, Der CJ. Characterization of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. Methods Enzymol. 2008;439:111–129. doi: 10.1016/S0076-6879(07)00409-0. [DOI] [PubMed] [Google Scholar]
- 28.Pille JY, Denoyelle C, Vare J, Bertrand JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP, Malvy C, Soria C, Li H. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol. Ther. 2005;11:267–274. doi: 10.1016/j.ymthe.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 29.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 30.Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–477. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- 31.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 32.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell. Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 33.Russ AP, Lampel S. The druggable genome: an update. Drug Discov. Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 34.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nature Reviews Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 35.Sebok A, Nusser N, Debreceni B, Guo Z, Santos MF, Szeberenyi J, Tigyi G. Different roles for RhoA during neurite initiation, elongation, and regeneration in PC12 cells. J. Neurochem. 1999;73:949–960. doi: 10.1046/j.1471-4159.1999.0730949.x. [DOI] [PubMed] [Google Scholar]
- 36.Sebti SM, Der CJ. Opinion: Searching for the elusive targets of farnesyltransferase inhibitors. Nat. Rev. Cancer. 2003;3:945–951. doi: 10.1038/nrc1234. [DOI] [PubMed] [Google Scholar]
- 37.van Golen KL, Wu ZF, Qiao XT, Bao LW, Merajver SD. RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res. 2000;60:5832–5838. [PubMed] [Google Scholar]
- 38.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 39.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 40.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;19:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 42.Zohn IM, Campbell SL, Khosravi-Far R, Rossman KL, Der CL. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]
- 43.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408:682–8. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.