Abstract
Despite our continued efforts to assert control over pathogens, more and more bacteria are saying “no” to drugs. It is becoming increasingly apparent that microbial environments, influenced by intracellular and extracellular metabolic processes, modulate antibiotic susceptibility in bacteria. A deeper understanding of these environmental processes may prove critical for the development of new antibacterial therapies.
The development of antibiotics to combat infectious diseases stands as one of the most significant advancements in medicine and public health. Since the discovery of penicillin in 1929, our mechanistic understanding of antibiotics has centered on the chemical inhibition of essential bacterial proteins. To date, this drug-target model has been a success. It has led to the isolation and subsequent chemical enumeration of additional antimicrobials that inhibit DNA, RNA, protein or cell wall biosynthesis. However, the efficacy of these antibiotics is continually undermined by the preferential selection of resistant mutants, an unfortunate result of antibiotic use. As a consequence, we are constantly breeding new strains of antibiotic-resistant bacteria. Additionally, even in the absence of genetic changes, antibiotic tolerance mechanisms allow subpopulations of bacteria to withstand repeated treatments of lethal antibiotics, leading, in some cases, to persistent and chronic infections.
These antibiotic-evasion tactics are particularly worrisome in light of the declining number of new antimicrobials in the developmental pipeline. The scarcity of new drugs reflects, in part, the limits of our drug-target model1. Novel therapies, based on a deeper understanding of bacterial physiology, are needed to counter these antibiotic-insensitive microbes. As we highlight below, there is a growing appreciation for the role that microbial environments, metabolic stresses within and metabolic influences between microbes, play in the defense strategies utilized by bacteria to deal with antibiotics.
Antibiotics induce metabolic stress in bacteria
While the direct targets of most antibiotics are well characterized, less is known about the events following drug-target binding which lead to cell death. Systems-level investigations of these events have implicated the role of intracellular metabolism in antibiotic-mediated cell death. A recent study by Dwyer et al. captured and analyzed the genome-wide transcriptional response of E. coli following exposure to lethal concentrations of a quinolone antibiotic, norfloxacin2. Detailed biochemical studies had previously shown that quinolones inactivate topoisomerases, DNA-bound proteins that control chromosome topology. Double-stranded DNA breaks, coupled with stalled DNA replication machinery at these topoisomerase sites, halt cell division and eventually lead to cell death3. However, reconstruction of stress responses using biological network analyses revealed that quinolones also induce cell death via the generation of reactive oxygen species2. Specifically, it was found that quinolones promote generation of superoxide, a byproduct of aerobic respiration, which attacks iron-sulfur clusters to release ferrous iron. Ferrous iron, in turn, reacts with hydrogen peroxide via Fenton chemistry to generate hydroxyl radicals, which contribute to the lethal effects of norfloxacin.
Building on these findings, Kohanski et al. discovered that different classes of bactericidal antibiotics (quinolones, beta-lactams, and aminoglycosides), regardless of their primary targets, utilize this oxidative damage cell death pathway4. Importantly, exposure to each antibiotic induces superoxide-mediated destabilization of iron-sulfur clusters, by stimulation of central metabolism, eventually leading to the generation of cytotoxic hydroxyl radicals (Fig. 1a). This common mechanism of killing was demonstrated in both Gram-negative and Gram-positive bacteria.
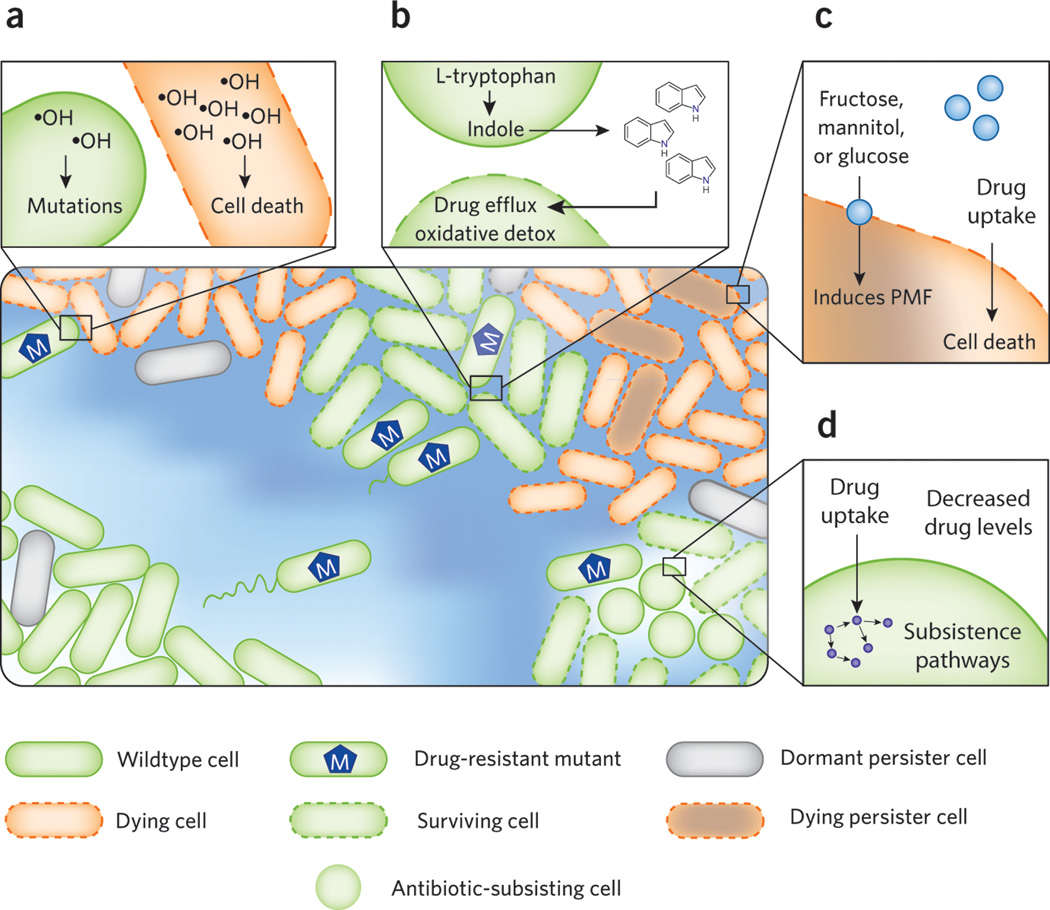
Figure 1. Microbial environments enable antibiotic resistance and tolerance.
A bacterial population subjected to a gradient of antibiotic concentration is shown in the main figure. (a) Antibiotics induce formation of deleterious hydroxyl radicals. Under sublethal antibiotic stress, these mutagenic hydroxyl radicals promote the emergence of antibiotic-resistant mutants. Lethal concentrations of antibiotic lead to high levels of hydroxyl radicals, which contribute to antibiotic-mediated cell death. (b) An antibiotic-resistant mutant produces indole by catabolizing tryptophan. Indole boosts the survival capacity of the population by inducing drug efflux pumps and oxidative stress detoxification pathways in the more vulnerable cells. (c) A subpopulation of dormant cells, persisters, is tolerant to antibiotics. Addition of specific metabolites, such as fructose, mannitol, or glucose, to the extracellular environment sensitizes persisters to aminoglycoside antibiotics via generation of proton motive force. (d) Some bacteria are capable of subsisting on antibiotics as a sole carbon source. They may allow a microbial community to evade treatments by reducing the local drug concentration, leading to formation of antibiotic-resistant mutants via mutagenic hydroxyl radicals, or by eliminating the antibiotic altogether.
The common oxidative damage cell death pathway provides an important framework for the development of new antibacterial therapies. For example, compounds that target bacterial systems that remediate hydroxyl radical damage or increase the production of hydroxyl radicals beyond the cell’s detoxification capacity could be powerful adjuvants for existing antibiotics. They may also form the basis for novel, and more effective, antibacterial compounds.
The above mechanism also provides insight into how bacteria may evade antibiotics through changes in metabolism. We now have a context for understanding how genetic mutations that seem unrelated to an antibiotic’s primary target protect a microbe against antibiotic attack. For example, bacteria with genetic mutations that alter pathways involved in superoxide response5, citric acid cycle6, or aerobic respiration7 result in reduced antibiotic susceptibility and this may be due to attenuation of hydroxyl radical production.
More work is needed to improve our understanding of how reactive oxygen species (ROS) are generated and how they modulate antibiotic lethality. For example, some Gram-positive bacteria can generate nitric oxide, another type of reactive oxygen species. Gusarov et al. showed that endogeneously produced nitric oxide in bacteria can be cytoprotective against a wide range of antibiotics, and acts as such by alleviating the oxidative stress induced by the drugs8. Nitric oxide generation, produced by the oxidation of l-arginine to l-citrulline, is thus another mechanism by which the intracellular metabolic environment can alter antibiotic susceptibility and may represent a useful pathway to target for antibacterial therapies.
A radical source of antibiotic resistance
ROS can also catalyze the development of antibiotic resistance. Hydroxyl radicals, produced as a metabolic byproduct of antibiotic stress, can be mutagenic as they readily damage DNA (Fig. 1a)9. A recent study showed that treating E. coli with sublethal concentrations of antibiotic induces hydroxyl radical formation, which leads to mutations that enable multidrug resistance10. Of note, it was found that this radical-based mechanism can lead to mutant strains that are sensitive to the applied antibiotic but resistant to other antibiotics.
Interestingly, it was also shown that treating a bacterial population with low levels of ampicillin, an antibiotic that inhibits cell wall synthesis, can result in the development of mutants that are resistant to ampicillin as well as kanamycin, a ribosome inhibitor10. Multidrug resistance is usually attributed to mutations that increase drug efflux or the accumulation of mutations specific to the primary antibiotic targets. However, in this case, the strain resistant to ampicillin and kanamycin was found to contain a mutation in a redox sensing system, an important element in the common mechanism that influences the regulation of aerobic respiration. This finding indicates that mutations in the common oxidative cell death pathway can afford multidrug resistance, and suggests that deep sequencing studies are needed to identify a broader range of mutations involved in the emergence of antibiotic resistance. This work also suggests that one may help reduce and contain the spread of new antibiotic-resistant bacteria by developing compounds that target ROS-producing systems or error-prone DNA damage repair systems.
Crowd-sourcing resistance
Bacteria utilize a variety of cooperative schemes to survive in diverse conditions11, including antibiotic stress. In a recent study, we found that bacteria can coordinate the development of antibiotic resistance within a population by sharing indole, a secondary metabolite produced by catabolism of l-tryptophan12. Bacterial populations that are continuously challenged by increasing concentrations of an antibiotic result in the stratification of resistance in the population. The vast majority of isolates in the culture are less resistant than the population as a whole. In contrast, a small subpopulation of highly resistant cells exist, due to the accumulation of mutations in antibiotic-specific alleles, and they condition the extracellular environment by producing and sharing indole at a fitness cost to themselves. Indole protects the less resistant cells from antibiotic lethality, by activating drug pumps and pathways responsible for detoxification of reactive oxygen species, until they can accrue mutations to boost their innate resistance (Fig. 1b). Indole effectively serves as a capacitor for the evolution of antibiotic resistance.
Intriguingly, bis-indoles, novel compounds based on indole chemistry, have been shown to have antibacterial properties and similar modes of efflux13. Lethal indole-like compounds that compete with the uptake and activity of endogenous indole may thus form the basis for powerful antibacterials with reduced rates of resistance.
Bacteria generate a wealth of secondary metabolites which have unknown biological function, and further investigation may reveal a variety of biogenic molecules in the extracellular environment that modulate antibiotic sensitivity. A recent study by Bernier et al. found that gaseous ammonia influences multidrug resistance between physically distant bacterial colonies14. It was shown that an E. coli colony can generate gaseous ammonia, by catabolism of l-aspartate, which increases polyamine levels in physically separated colonies of either Gram-positive or Gram-negative bacteria. Increases in intracellular polyamines lead to alterations in membrane permeability to different antibiotics and enhance protection against oxidative stress. These findings indicate that ammonia production and polyamine synthesis may be important pathways to target as part of new therapies.
Together, these studies suggest that efforts to combat antibiotic resistance may be complicated by bacterial signaling mechanisms and cooperative survival strategies. Thus, a better understanding of how bacteria work together to survive stressful, unpredictable environments may prove critical for the design of effective clinical interventions.
Engineering reduced antibiotic tolerance
Persisters are dormant bacteria within a population that can withstand lethal concentrations of antibiotic in the absence of genetic changes. This tolerant subpopulation has been implicated as a driving force behind chronic and recurrent infections15–17. Diverse processes and conditions contribute to persister formation, and antibiotic tolerance is achieved, in part, by the induction of pathways that promote cellular quiescence18. It was recently shown that metabolic stimulation can sensitize bacterial persisters to antibiotic treatment. Specifically, Allison et al. found that addition of specific metabolites (such as fructose, mannitol or glucose) to the extracellular environment could potentiate the activity of aminoglycosides, antibiotics that target the ribosome, in Gram-positive and Gram-negative persisters19. These metabolites induce the generation of proton motive force, which is required for the uptake of aminoglycosides (Fig. 1c). This work showed that bacterial persisters, although dormant, are primed for metabolite uptake, central metabolism and respiration. Importantly, the effectiveness of this metabolite-based strategy was validated in a mouse urinary tract infection, demonstrating that the approach can be utilized in complex, in vivo environments.
Provoking diverse energetic pathways via extracellular metabolite dosing may enable the use of other antibiotics in eradicating persisters. Intriguingly, a recent chemical screening effort to find compounds to awaken persisters identified a novel molecule that sensitizes persisters to antibiotics by selectively inducing their growth20. The specific mechanism by which this compound induces the reversion of persisters to antibiotic-sensitive cells is currently unknown. Further investigation of the pathways activated by this compound as well as more extensive studies into the basic physiology of bacterial persisters may provide critical insights that could be harnessed to engineer extracellular environments to reduce or eliminate the occurrence of antibiotic tolerance.
Natural environments and ecological niches
Our understanding of how microbial environments contribute to antibiotic resistance and tolerance remains incomplete. Diverse species of bacteria co-exist in nature, subject to a variety of physical constraints. Deconstructing natural bacterial communities may reveal insights into how diverse bacteria modulate their extracellular environments to enable antibiotic resistance and tolerance. As an example, Dantas et al. recently isolated bacteria, from natural soil samples, that can subsist on antibiotics as a sole carbon source21. Surprisingly, subsistence pathways exist for diverse classes antibiotics of natural and synthetic origin. It is intriguing to consider that these isolates may serve to alleviate antibiotic stress for a bacterial community, by devouring the antibiotics and thereby removing them from the environment (Fig. 1d).
While antibiotic catabolism has not yet been identified in human pathogens, it may be an unrecognized mechanism that promotes the antibiotic resistance and tolerance found in recurrent polymicrobial infections22,23. Identification of additional subsistence pathways and the conditions that lead to their activation will be required to ascertain their pertinence to the development of problematic infections.
Natural niches are composed of complex microenvironments that can be utilized by bacteria to enhance their survival capacity. Simulation of these natural niches may reveal the temporal dynamics by which antibiotic resistance occurs. In a recent study, Zhang et al. found that microenvironments accelerate the emergence and fixation of antibiotic-resistant mutants24. A microfluidic device, consisting of a simplified network of interconnected microenvironments, was developed to mimic metabolic gradients that bacteria naturally encounter. Resistant mutants, that emerge as rapidly as 10 hours following antibiotic treatment, preferentially move to regions with large gradients of antibiotic stress. These regions confer a growth advantage to the resistant strains that subsequently outgrow their antibiotic-sensitive progenitors to dominate the bacterial population. This phenomenon, which is not observable with traditional laboratory techniques, illustrates the value of developing new methodologies that mimic natural bacterial environments.
Summary
The increasing prevalence of bacteria that are insensitive to our current antibiotics amplifies the need for new antimicrobial therapies. Our traditional approach to antibacterial development based on the inhibition of essential processes appears to have reached the point of diminishing returns. The discovery that diverse antibiotics stimulate a common oxidative cell death pathway represents a fundamental shift in our understanding of bactericidal antibiotic modes of action. A number of studies, as discussed above, also provide the first hints of how intracellular and extracellular metabolism enable antibiotic resistance and tolerance (see Table 1).
Table 1.
Chemicals in microbial environments that affect antibiotic resistance or tolerance.
| Biomolecules | Source | Activity |
|---|---|---|
|
Reactive oxygen species |
||
| superoxide | aerobic respiration |
leaches ferrous iron from iron-sulfur clusters |
| hydroxyl radicals | Fenton reaction | low levels are mutagenic and high levels are lethal, causing damage to DNA, lipids, and proteins |
| nitric oxide | L-arginine | reduces hydroxyl radical concentrations by suppressing the Fenton reaction and activating catalase, an antioxidant enzyme |
| Biogenic molecules | ||
| indole | L-tryptophan | induces drug efflux pumps and oxidative stress protection pathways |
| gaseous ammonia | L-aspartate | modification of membrane permeability and increased resistance to oxidative stress |
| Carbon sources | ||
| mannitol, glucose, pyruvate, fructose |
supplied | stimulates generation of proton motive force in persisters for uptake of aminoglycosides, antibiotics which target ribosomes |
We have, nonetheless, just begun to understand the repertoire of antibiotic-evasion tactics used by bacteria. Biosynthetic pathways for natural antibiotics are ancient, and numerous mechanisms for antibiotic resistance and tolerance are likely to have evolved over the last few million years25. Unraveling these additional mechanisms will require concerted efforts by chemical biologists, microbiologists, and clinicians. These efforts will benefit from metabolic models, and other network biology approaches, to guide additional investigations into processes that modulate antibiotic susceptibility. Critically, these models may be used to identify common points of vulnerability and highlight key differences between pathogens, leading to the development of effective adjuvants, novel antibiotics and new antimicrobial strategies.
There is also a crucial need to better understand how bacteria within a population cooperate to overcome antibiotic treatments. Such investigations may benefit from novel chemical probes and experimental techniques to interrogate the physiology and functional dynamics of natural microbial communities. Insights gained from these studies will augment metagenomic models that can be used to identify biomolecules responsible for these cooperative strategies. Further studies of microbial environments by leveraging chemical biology methodologies and systems biology approaches may reveal a wealth of untapped targets for the development of novel compounds to counter the growing threat of resistant and tolerant bacterial infections.
Contributor Information
Henry H. Lee, Howard Hughes Medical Institute, Department of Biomedical Engineering and Center for BioDynamics, Boston University, Boston, MA 02215, USA
James J. Collins, Howard Hughes Medical Institute, Department of Biomedical Engineering and Center for BioDynamics, Boston University and in the Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02215, USA. jcollins@bu.edu
References
- 1.Clatworthy AE, Pierson E, Hung DT. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 2.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drlica K, Malik M, Kerns RJ, Zhao X. Antimicrob Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 5.Oethinger M, Podglajen I, Kern WV, Levy SB. Antimicrob Agents Chemother. 1998;42:2089–2094. doi: 10.1128/aac.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helling RB, Kukora JS. J Bacteriol. 1971;105:1224–1226. doi: 10.1128/jb.105.3.1224-1226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imlay JA, Chin SM, Linn S. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 10.Kohanski MA, DePristo MA, Collins JJ. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro JA. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 12.Lee HH, Molla MN, Cantor CR, Collins JJ. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler MM, et al. Antimicrob Agents Chemother. 2010;54:3974–3977. doi: 10.1128/AAC.00484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernier SP, Letoffe S, Delepierre M, Ghigo JM. Mol Microbiol. 2011;81:705–716. doi: 10.1111/j.1365-2958.2011.07724.x. [DOI] [PubMed] [Google Scholar]
- 15.Barry CE, 3rd, et al. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin BR, Rozen DE. Nat Rev Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 17.Mulcahy LR, Burns JL, Lory S, Lewis K. J Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison KR, Brynildsen MP, Collins JJ. Curr Opin Microbiol. 2011;14:593–598. doi: 10.1016/j.mib.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison KR, Brynildsen MP, Collins JJ. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JS, et al. Antimicrob Agents Chemother. 2011;55:5380–5383. doi: 10.1128/AAC.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dantas G, Sommer MO, Oluwasegun RD, Church GM. Science. 2008;320:100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 22.Brogden KA, Guthmiller JM, Taylor CE. Lancet. 2005;365:253–255. doi: 10.1016/S0140-6736(05)17745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornton RB, et al. BMC Pediatr. 2011;11:94. doi: 10.1186/1471-2431-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 25.D'Costa VM, et al. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]