Abstract
Although histochemical staining has been believed to inhibit DNA amplification reaction, no previous study has systematically evaluated the influence of histochemical staining on downstream molecular assays. To evaluate an influence of hematoxylin and eosin (HE) staining on DNA testing, we isolated DNA from 10 unstained, 10 hematoxylin-stained, 10 eosin-stained or 10 HE-stained tissue sections (ie, 4 groups), from each of 5 colon cancers. Among those 4 groups, we did not observe any significant or appreciable difference in DNA fragmentation by agarose gel electrophoresis; in DNA amplification by real-time PCR; in microsatellite PCR fragment analyses; or in PCR-Pyrosequencing. As a proof-of-principle study, we successfully performed microsatellite instability analysis and sequencing of KRAS and BRAF on over 1300 colorectal cancers using DNA extracted from HE stained tissue sections. Our data provide no evidence for interfering effect of HE staining on DNA testing, suggesting that DNA from HE-stained sections can be effectively used for routine DNA testing.
Keywords: histochemical staining, polymerase chain reaction, molecular diagnostics, mutation detection, microdissection, solid tumor
INTRODUCTION
Screening and identification of genetic alterations in formalin-fixed paraffin-embedded (FFPE) tumor materials have been important in molecular diagnosis and clinical medicine.1-6 Solid tumors are inherently heterogeneous tissues that contain neoplastic cells admixed with non-neoplastic cells including inflammatory cells, vascular and lymphatic endothelial cells, smooth muscle cells, and stromal fibroblasts. Therefore, an enrichment of neoplastic cells is commonly performed before DNA extraction in molecular testing, to avoid false negative results due to low neoplastic cellularity.7
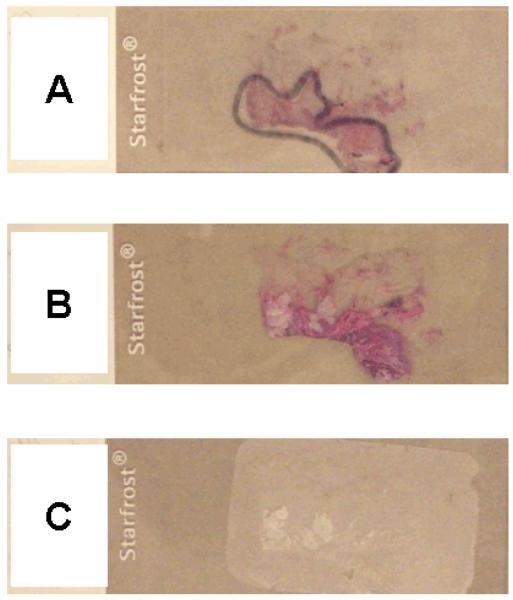
Macrodissection from tissue sections on glass slides has been a widely used method to exclude pure non-neoplastic areas and enrich neoplastic cellularity for molecular testing. For macrodissection of tumor areas, an accurate morphologic identification of tumor areas is essential.8, 9 Thus, macrodissection should be guided by a hematoxylin and eosin (HE)-stained tissue section with tumor areas marked by a pathologist under a microscope (Figure 1A). It would be ideal to dissect tumor tissue from a HE-stained section (without coverslip) because HE stain enables us to easily identify tumor areas (Figure 1B), particularly next to the HE slide with the marked tumor areas (Figure 1A). In contrast, it is difficult to accurately identify tumor areas of an unstained tissue section (Figure 1C) and tumor areas can be missed.
Figure 1.
Examples of HE-stained and unstained tissue sections.
A. A HE-stained tissue section slide to guide tumor tissue dissection. A pathologist marked a tumor area under a microscope.
B. A HE-stained tissue section without coverslip for subsequent DNA extraction. It is easy to identify the tumor area.
C. An unstained tissue section for DNA extraction. It is not as easy to identify the tumor area as in the HE-stained tissue section (B).
HE, hematoxylin and eosin.
With regard to the influence of histochemical staining on downstream molecular assays, some previous studies have shown interfering effects of histochemical staining, especially hematoxylin stain,10-12 while another study failed to show significant difference in PCR amplification between DNA specimens from HE-stained and unstained tissue sections.13 To avoid possible interfering effects of histochemical staining on molecular assays, macrodissection of tumor from unstained tissue sections is commonly performed, despite the difficulty in identifying tumor areas (Figure 1C) and the risk of missing tumor areas.1, 14-17 To our knowledge, no previous study has systematically evaluated the influence of histochemical staining on downstream molecular assays.
We therefore conducted this study to evaluate various molecular assay results on DNA extracted from HE-stained, hematoxylin-stained, and eosin-stained FFPE tissue sections, in comparison to DNA extracted from unstained tissue sections. We compared results of agarose gel genomic DNA fragment analysis (without PCR), quantitative real-time PCR assay, PCR-Pyrosequencing assay, and PCR-capillary electrophoresis fragment analyses. We did not observe an appreciable difference in any of the results on DNA specimens from stained tissue. Furthermore, as a proof-of-principle study, we successfully utilized DNA from HE-stained tissue sections, and obtained DNA analysis data from over 1300 colorectal cancers. Our data support feasibility of DNA extraction from HE-stained tissue sections for routine clinical laboratory workflow.
MATERIALS AND METHODS
FFPE Tissue Specimens
Specimen collection and analysis in this study were approved by the Harvard School of Public Health, Brigham and Women’s Hospital, and Dana-Farber Cancer Institute Institutional Review Boards. Formalin-fixed paraffin-embedded (FFPE) colorectal cancer tissue specimens (5 cases) were anonymized after collection from the archival file of the Department of Pathology, Brigham and Women’s Hospital, to assess the effects of histochemical staining on downstream molecular testing (Figure 2).
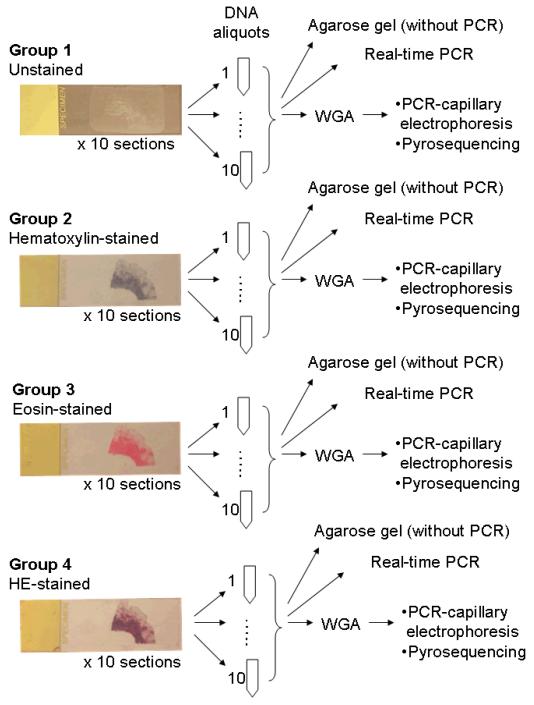
Figure 2.
Our overall strategy to assess influence of tissue staining on downstream DNA testing. DNA was extracted from unstained tissue (Group 1), hematoxylin-stained tissue (Group 2), eosin-stained tissue (Group 3), and HE-stained tissue (Group 4).
HE, hematoxylin and eosin; PCR, polymerase chain reaction; WGA, whole genome amplification.
As a proof-of-principle, feasibility study, we tested DNA extraction from HE-stained tissue sections in a large cohort of colorectal cancer cases. We utilized two U.S. nationwide prospective cohort studies, the Nurses’ Health Study (N=121,701 women followed since 1976) and the Health Professionals Follow-up Study (N=51,529 men followed since 1986).18, 19 We collected FFPE tissue blocks from hospitals and pathology laboratories throughout the U.S., where cohort participants with colorectal cancer underwent cancer resections.18, 19 HE-stained tissue sections were reviewed by a pathologist (S.O.). Genomic DNA was extracted from HE-stained tissue sections using another HE-stained section with marked tumor areas as a guide (as in Figure 1A-1B), for downstream KRAS, BRAF, and MSI testing, and results were available in 1313, 1312, and 1293 cases, respectively.
Overall Study Strategy, DNA Extraction and Agarose Gel Electrophoresis
Deparaffinized and hydrated 10 μm sections from the same tissue block were stained by hematoxylin (Harris hematoxylin; Surgipath Inc., Richmond, IL) alone for 3 minutes (Group 2), by eosin (Surgipath Inc.) alone for 15 seconds (Group 3), or by both hematoxylin and eosin (HE) (Group 4) (Figure 2). Unstained slides were prepared as controls (Group 1). Figure 2 illustrates our overall strategy of experiments to assess the effects of hematoxylin and/or eosin staining on DNA integrity and downstream molecular assays.
To avoid bias that could be introduced by tumor-directed macrodissection, a whole tissue section (including tumor and adjacent normal tissue) was entirely scraped off from a glass slide by a sterile needle. Thus, DNA yields were very similar between the Groups. The scraped tissue was collected into a microtube, and genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Valencia, CA). As a result, each of ten tissue sections was aliquoted to one tube, to yield 10 aliquoted DNA specimens for each Group (Figure 2). The 260/280 nm absorbance ratio of extracted DNA was approximately 1.8 (nanodrop; Thermo Scientific, Waltham, MA). Extracted DNA from unstained tissue and HE-stained tissue from all of the five cases was analyzed by electrophoresis in 0.8% agarose gel (Figure 3). Three cases were used for real-time PCR and PCR-Pyrosequencing, and the other two cases were used for microsatellite PCR-fragment analysis.
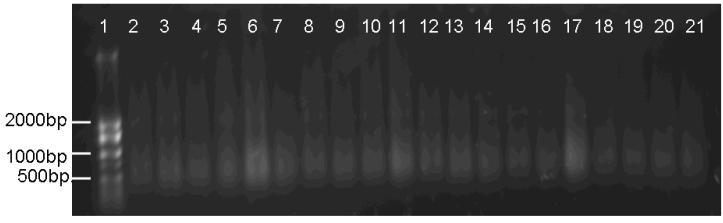
Figure 3.
Agarose gel (0.8%) electrophoresis to assess integrity of DNA from unstained and HE-stained tissue sections. There was no appreciable difference in DNA integrity between DNA specimens from unstained tissue and HE-stained tissue sections. The representative case (10 sections) was shown.
Lane 1: size marker. Lanes 2-11: Group 1 (unstained tissue). Lanes 12-21: Group 4 (HE-stained tissue)
Real-Time PCR Assay for GAPDH
To assess PCR amplification efficiency, we performed real-time PCR using the primers for GAPDH and SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) by ABI 7300 Real-Time PCR system (Applied Biosystems). Primer sequences were 5′-GTCATGGGTGTGAACCATGAGAA-3′ and 5′-TGGTCATGAGTCCTTCCACGAT-3′. PCR reaction was repeated 4 times on each of the 10 DNA aliquots for Groups 1 through 4 and cycle threshold (Ct) values were compared.
Microsatellite PCR-Fragment Analysis
To assess the effects of hematoxylin and/or eosin staining on PCR-fragment analysis, we performed PCR for dinucleotide markers, D2S123 and D5S346,20 after PCR-based whole genome amplification on genomic DNA as previously described.21 Forward primers are labeled with fluorescence and PCR product sizes were 180 bp (D2S123) and 129 bp (D5S346). PCR products were electrophoresed and analyzed by ABI 3730 DNA Analyzer (Applied Biosystems). PCR-fragment analysis was repeated twice on each of the 10 DNA aliquots.
For the large feasibility cohort, we performed microsatellite instability (MSI) analysis using a 10-marker panel (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487).20 MSI-high was defined as the presence of instability in 30% or more of the markers, and MSI-low/microsatellite stability (MSS) as instability in less than 30% of the markers.20
PCR-Pyrosequencing for KRAS and BRAF
To assess the effect of hematoxylin and/or eosin staining on Pyrosequencing, we performed PCR-Pyrosequencing for KRAS (codons 12 and 13) which was previously developed and validated.21 The size of PCR product was 82 bp, and 10 μl of each was sequenced by Pyrosequencing PSQ96 HS System (Qiagen). The PCR-Pyrosequencing reaction was repeated 3 times on each of the 10 aliquots. In addition, in the feasibility cohort, PCR-Pyrosequencing for BRAF (codon 600) was performed as previously described.22
Statistical Analysis
For all statistical analyses, we used SAS program (Version 9.1, SAS Institute, Cary, NC). All p values were two-sided. The cycle threshold (Ct) in real-time PCR, which reflected amplification efficiency of each PCR reaction given similar amounts of scraped tissue and extracted DNA, was compared by ANOVA (analysis of variance) test, adjusting for case, spot of each reaction and plate. We performed ANOVA test for comparing peak heights of fluorescence signal on capillary electropherograms in microsatellite analysis. The distribution of the peak height values (median 3028.5, range 0 to 10618) was normalized by logarithmic transformation after adding 0.5 to each value (in order to log-transform the value of “0”). A deviation from the null hypothesis in any of the comparisons might imply interfering effect of histochemical staining on the DNA testing process.
RESULTS
Integrity of DNA from Unstained and Stained Tissue
Our overall strategy to assess influence of hematoxylin and eosin (HE) staining on subsequent DNA testing for formalin-fixed paraffin-embedded (FFPE) tissue is shown in Figure 2.
First, we assessed the influence of HE staining on DNA integrity. We extracted DNA specimens of Group 1 (unstained) and Group 4 (HE-stained) were loaded onto 0.8% agarose gel (Figure 3). There was no appreciable difference in DNA integrity between the DNA specimens from HE-stained and unstained tissue sections from all of the five cases.
Comparison of Cycle Threshold (Ct) Values in Real-Time PCR
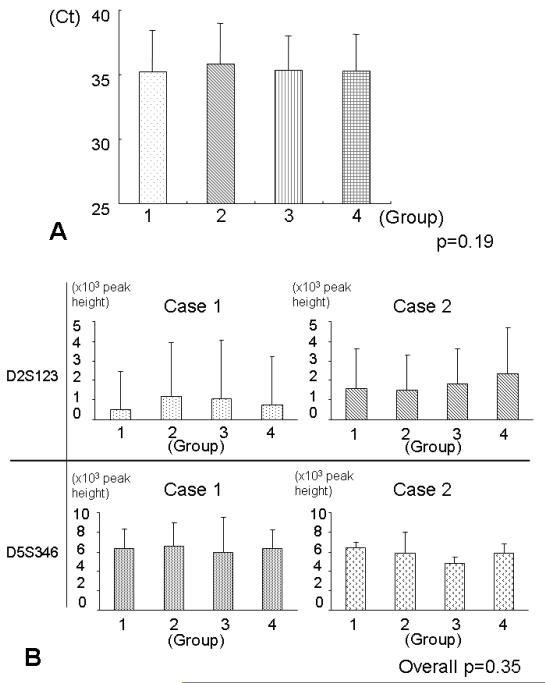
We compared Ct values in real-time PCR for GAPDH between the four groups using three cases (× 10 sections × 4 repeated runs), by ANOVA test which adjusted for case, spot of each reaction, and plate (Figure 4A). There was no significant difference in Ct values between the groups (p=0.19).
Figure 4.
Tissue staining and subsequent PCR analysis.
A. Mean Ct values in quantitative real-time PCR for GAPDH on DNA specimens (3 cases × 10 sections × 4 repeated runs) in each group. The vertical bar indicates standard deviation. There was no significant difference in the mean Ct values between the groups (p=0.19).
Ct, cycle threshold.
B. Mean peak heights in electropherograms in PCR-fragment analysis of the microsatellite markers (D2S123 and D5S346) on DNA specimens (2 cases × 10 sections × 2 repeats) from each group. The vertical bar indicates standard deviation. There was no significant difference in the mean peak heights between the four groups (Overall p=0.35).
Microsatellite PCR-Fragment Analysis
We performed PCR-fragment analysis for the two microsatellite markers, D2S126 and D5S346, and assessed potential influence of tissue staining, using two cases (× 10 sections × 2 repeats). We measured the peak height in electropherograms, and compared the mean peak height value between the four groups. There was no significant difference in the mean peak height between the four groups (overall p=0.35) (Figure 4B).
PCR-Pyrosequencing for KRAS
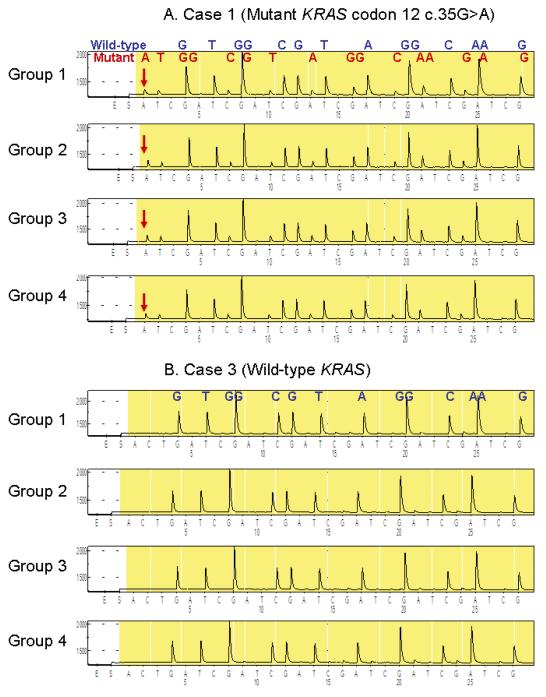
We performed PCR-Pyrosequencing for KRAS codons 12 and 13 on three cases (× 10 sections × 3 repeats), and compared results between the four groups. Two cases had c.35G>A (p.G12D) mutation, and one case showed only wild-type sequence (Figure 5). All Pyrograms for KRAS codons 12 and 13 showed clear peaks and little background, and there was no appreciable difference between the four groups.
Figure 5.
Tissue staining and subsequent PCR-Pyrosequencing for KRAS
A. Case 1 with mutant KRAS codon 12 (c.35G>A) admixed with wild-type sequence. The assay was repeated 3 times on 120 DNA aliquot specimens (3 cases × 4 groups × 10 sections), and representative results are shown. There was no appreciable difference between DNA specimens from the four groups.
B. Case 3 with wild-type KRAS. The assay was repeated 3 times on 120 DNA aliquot specimens (3 cases × 4 groups × 10 sections), and representative results are shown. There was no appreciable difference between DNA specimens from the four groups.
MSI Testing and Pyrosequencing for KRAS and BRAF in the Large Feasibility Cohort
Finally, we performed a proof-of-principle, feasibility study on FFPE tissue blocks which were fixed and processed in numerous different pathology laboratories throughout the U.S., utilizing the two U.S. nationwide prospective cohort studies.18 We dissected tumor tissue from HE-stained sections (without coverslip) using a guide HE slide with marked tumor areas (as in Figure 1A-1B), and extract DNA for subsequent MSI analysis and PCR-Pyrosequencing for KRAS and BRAF. KRAS and BRAF mutations were detected in 470 (36%) of 1313 cases and 186 (14%) of 1312 cases, respectively (Table 1). MSI-high was present in 197 (15%) of 1293 cases. These frequencies are compatible with the published data on these molecular features in colorectal cancers,23-27 particularly in population-based studies.28-33 There was no evidence for interfering effect of HE staining on these molecular assays. These data suggest that DNA from HE-stained sections can be used for routine tumor DNA testing on FFPE tissue.
Table 1.
Molecular features of colorectal cancer by analysis of DNA from HE-stained tissue in two U.S. nationwide prospective cohort studies, the Nurses’ Health Study and the Health Professionals Follow-up Study.
| Molecular features | N | |
|---|---|---|
| KRAS | 1314 | |
| Codon 12 or 13 mutation | 470 (36%) | |
| Wild-type | 844 (64%) | |
| BRAF | 1313 | |
| Codon 600 mutation | 186 (14%) | |
| Wild-type | 1127 (86%) | |
| MSI | 1293 | |
| MSI-high | 197 (15%) | |
| MSS/MSI-low | 1096 (85%) | |
HE, hematoxylin and eosin; MSI, microsatellite instability; MSS, microsatellite stable.
DISCUSSION
We conducted this study to evaluate potential influence of HE staining on subsequent DNA testing. Solid tumor tissue is inherently heterogeneous tissue with admixture of neoplastic cells and non-neoplastic cells including inflammatory, stromal, endothelial, and smooth muscle cells, even within tumor tissue areas. Macrodissection from HE-stained tissue sections enables accurate identification and dissection of tumor areas, to obtain DNA specimens with a high content of neoplastic cell DNA. Especially for small tumor nests or metastatic tumor foci which must be carefully and precisely dissected, it is very useful to stain and clearly visualize the tissue section. HE staining is a common and simple method to visualize the tumor area of interest. Although hematoxylin staining has been believed to inhibit DNA amplification reaction, only a small number of studies have been performed to assess potential influence of tissue staining on amplification of DNA by PCR.10, 11, 13 Notably, no previous study has systematically evaluated the influence of HE staining on downstream molecular assays. Our current study has provided no evidence for interfering effects of HE staining on subsequent DNA assays. Our data support routine clinical use of HE staining of tissue sections for subsequent tumor tissue dissection, DNA extraction and molecular analyses.
Previous studies evaluated potential influence of tissue staining on DNA assays.10, 11, 13 Burton et al.10 and Chen et al.34 showed that hematoxylin staining inhibited DNA amplification by PCR, and Diss et al.11 reported that DNA from HE-stained tissue was amplified less efficiently than DNA from unstained tissue. On the other hand, Murase et al.13 reported no significant difference in PCR amplification between DNA specimens from unstained and HE-stained tissue. However, none of the previous studies10, 11, 13 has quantitatively evaluated efficiencies of PCR amplification on DNA specimens from unstained and stained tissues. In our current study, there was no statistically significant difference in GAPDH amplification by quantitative real-time PCR between DNA specimens from unstained tissue, hematoxylin-stained tissue, eosin-stained tissue and HE-stained tissue.
Pyrosequencing is a nonelectrophoretic sequencing by nucleotide extension. Pyrosequencing is useful for sequencing of relatively short length of nucleotides (up to 40 to 50). Therefore, Pyrosequencing has been applied for single nucleotide polymorphism genotyping,35 bacterial strain typing,36 mutation detection in tumors,21, 37, 38 quantitative promoter CpG island methylation analysis,39, 40 and measurement of LINE-1 methylation.41 We evaluated the influence of HE staining for PCR-Pyrosequencing for KRAS, and showed no appreciable difference in results between DNA specimens from unstained and HE-stained tissues.
Capillary electrophoresis is commonly used for microsatellite analysis in FFPE clinical tissue specimens (e.g., MSI and loss of heterozygosity analyses).42-45 We showed no significant difference in peak heights of electropherogram of two microsatellite markers (D2S126 and D5S346) between DNA specimens from unstained and stained tissues.
Murphy et al.46 reported an artifact (a peak at 71 bp) in capillary electrophoresis due to autofluorescence from eosin which was probably contaminated by non-stringent DNA purification. A notable difference from our study was that Murphy et al.46 experienced the artifact after biopsy tissue (not a tissue section on a slide) was stained with eosin “to enable identification of small tissue fragments during sectioning”. We have not experienced this problem despite the fact that we have performed microsatellite PCR-fragment analysis on over 1300 cases using DNA from HE-stained tissue. Nonetheless, we agree with Murphy et al.46 that artifacts caused by eosin should be avoided by using stringent DNA purification steps, and that artifacts from histochemical staining should be considered when peaks of the same product size (e.g., 71 bp) are present in multiple specimens or primary peaks contain additional underlying peaks of other colors.
As a potential limitation of our study, we used relatively small number of tissue blocks (N=5) to systematically compare the effects of histochemical staining in downstream molecular assays. Nonetheless, we used as many as 10 sections and 10 DNA aliquots per each condition to achieve a precise measurement estimate. Moreover, as the proof-of-principle, feasibility study, we used DNA extracted from HE-stained tissue of over 1300 colorectal cancers in two U.S. nationwide prospective cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study) for MSI analysis and Pyrosequencing of KRAS and BRAF (Table 1). Those FFPE tissue specimens were retrieved from numerous pathology laboratories throughout the U.S. Despite the diversity of laboratories which processed and stored tissues, we have been successful in performing those molecular assays in over 95% of cases tested, and the frequencies of MSI-high and mutations of KRAS and BRAF were compatible with published data on colorectal cancer,23-27 particularly in population-based studies.28-33 In addition, using DNA from HE-stained tissue sections, we have previously shown that both BRAF mutation and MSI-high in colon cancer are associated with the CpG island methylator phenotype (CIMP20, 28, 47-52) and patient outcome,20, 53 and KRAS mutation is associated with CIMP-low in our cohort studies.22 Our data indicate that HE-staining is useful for DNA analysis on FFPE from numerous pathology laboratories in multicenter clinical trials or population-based cohort studies,42, 54 which can contribute to improvement of both patient care and public heath.
In conclusion, our current study represents the first systematic investigation on the effects of HE staining in downstream molecular assays. Our results provide no evidence to support the common conception that HE staining interferes molecular assays on DNA from FFPE tissue. Our data support routine clinical use of HE staining on tumor sections for subsequent tissue dissection and DNA extraction. HE staining enables easy and accurate identification of tumor areas to obtain DNA with a high content of neoplastic cell DNA. HE staining can be easily incorporated into routine pathology workflow, so that laboratory personnel can easily identify and dissect tumor areas from HE-stained tissue sections, guided by a HE slide with marked tumor areas. Thus, our data may have considerable impacts on clinical molecular diagnostics and routine tumor molecular testing towards personalized medicine.
ACKNOWLEDGEMENTS
We deeply thank the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS) cohort participants who have agreed to provide us with information through questionnaires and biological specimens; hospitals and pathology departments throughout the U.S. for generously providing us with tissue specimens; the staff of the NHS and the HPFS for their valuable contributions; and all of the U.S. State Cancer Registries for their help.
This work was supported by U.S. National Institute of Health (NIH) grants P01 CA87969 (SE Hankinson), P01 CA55075 (WC Willett), P50 CA127003 (to CSF), R01 CA118553 (CSF), R01 CA124908 (CSF), and R01 CA151993 (to SO). The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
TM, KS and AK contributed equally
No conflicts of interest exist.
Contributor Information
Teppei Morikawa, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Kaori Shima, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Aya Kuchiba, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Mai Yamauchi, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Noriko Tanaka, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Yu Imamura, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Xiaoyun Liao, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Zhi Rong Qian, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Mohan Brahmandam, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA.
Janina A. Longtine, Department of Pathology, Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA.
Neal I. Lindeman, Department of Pathology, Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA.
Charles S. Fuchs, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA; Department of Medicine, Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA.
Shuji Ogino, Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA; Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA; Department of Pathology, Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA.
REFERENCES
- 1.Ausch C, Buxhofer-Ausch V, Oberkanins C, et al. Sensitive detection of KRAS mutations in archived formalin-fixed paraffin-embedded tissue using mutant-enriched PCR and reverse-hybridization. J Mol Diagn. 2009;11:508–513. doi: 10.2353/jmoldx.2009.090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng S, Cummings OW, Saxena R, et al. Clonality and TP53 mutation analysis of focal nodular hyperplasia of the liver. Am J Clin Pathol. 2010;134:65–70. doi: 10.1309/AJCPCIAH79EABQKM. [DOI] [PubMed] [Google Scholar]
- 3.Battochio A, Mohammed S, Winthrop D, et al. Detection of c-KIT and PDGFRA gene mutations in gastrointestinal stromal tumors: comparison of DHPLC and DNA sequencing methods using a single population-based cohort. Am J Clin Pathol. 2010;133:149–155. doi: 10.1309/AJCP1FNW7RGZFTYU. [DOI] [PubMed] [Google Scholar]
- 4.Hostein I, Faur N, Primois C, et al. BRAF mutation status in gastrointestinal stromal tumors. Am J Clin Pathol. 2010;133:141–148. doi: 10.1309/AJCPPCKGA2QGBJ1R. [DOI] [PubMed] [Google Scholar]
- 5.Ross JS, Torres-Mora J, Wagle N, et al. Biomarker-based prediction of response to therapy for colorectal cancer: current perspective. Am J Clin Pathol. 2010;134:478–490. doi: 10.1309/AJCP2Y8KTDPOAORH. [DOI] [PubMed] [Google Scholar]
- 6.Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monzon FA, Ogino S, Hammond ME, et al. The role of KRAS mutation testing in the management of patients with metastatic colorectal cancer. Arch Pathol Lab Med. 2009;133:1600–1606. doi: 10.5858/133.10.1600. [DOI] [PubMed] [Google Scholar]
- 8.Shibata D, Hawes D, Li ZH, et al. Specific genetic analysis of microscopic tissue after selective ultraviolet radiation fractionation and the polymerase chain reaction. Am J Pathol. 1992;141:539–543. [PMC free article] [PubMed] [Google Scholar]
- 9.Lukish JR, Muro K, DeNobile J, et al. Prognostic significance of DNA replication errors in young patients with colorectal cancer. Ann Surg. 1998;227:51–56. doi: 10.1097/00000658-199801000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton MP, Schneider BG, Brown R, et al. Comparison of histologic stains for use in PCR analysis of microdissected, paraffin-embedded tissues. Biotechniques. 1998;24:86–92. doi: 10.2144/98241st01. [DOI] [PubMed] [Google Scholar]
- 11.Diss TC, Pan L, Peng H, et al. Sources of DNA for detecting B cell monoclonality using PCR. J Clin Pathol. 1994;47:493–496. doi: 10.1136/jcp.47.6.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lang A, Wilander E. Sensitivity of HPV tests on stained vs. unstained cervical smears. Acta Cytol. 2005;49:595–599. doi: 10.1159/000326245. [DOI] [PubMed] [Google Scholar]
- 13.Murase T, Inagaki H, Eimoto T. Influence of histochemical and immunohistochemical stains on polymerase chain reaction. Mod Pathol. 2000;13:147–151. doi: 10.1038/modpathol.3880028. [DOI] [PubMed] [Google Scholar]
- 14.Rowe LR, Bentz BG, Bentz JS. Detection of BRAF V600E activating mutation in papillary thyroid carcinoma using PCR with allele-specific fluorescent probe melting curve analysis. J Clin Pathol. 2007;60:1211–1215. doi: 10.1136/jcp.2006.040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Puijenbroek M, Nielsen M, Tops CM, et al. Identification of patients with (atypical) MUTYH-associated polyposis by KRAS2 c.34G > T prescreening followed by MUTYH hotspot analysis in formalin-fixed paraffin-embedded tissue. Clin Cancer Res. 2008;14:139–142. doi: 10.1158/1078-0432.CCR-07-1705. [DOI] [PubMed] [Google Scholar]
- 16.Kristensen LS, Wojdacz TK, Thestrup BB, et al. Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer. 2009;9:453. doi: 10.1186/1471-2407-9-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messick CA, Church JM, Liu X, et al. Stage III colorectal cancer: molecular disparity between primary cancers and lymph node metastases. Ann Surg Oncol. 2010;17:425–431. doi: 10.1245/s10434-009-0783-z. [DOI] [PubMed] [Google Scholar]
- 18.Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–1694. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan AT, Ogino S, Fuchs CS. Aspirin and the Risk of Colorectal Cancer in Relation to the Expression of COX-2. New Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 20.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 24.Figueiredo JC, Grau MV, Wallace K, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1041–1049. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 26.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 27.Zlobec I, Bihl M, Foerster A, et al. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- 28.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Hughes LA, Simons CC, van den Brandt PA, et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP) PloS one. 2011;6:e18571. doi: 10.1371/journal.pone.0018571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limsui D, Vierkant RA, Tillmans LS, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. Journal of the National Cancer Institute. 2010;102:1012–1022. doi: 10.1093/jnci/djq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozek LS, Herron CM, Greenson JK, et al. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:838–843. doi: 10.1158/1055-9965.EPI-09-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brink M, de Goeij AF, Weijenberg MP, et al. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–710. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 33.English DR, Young JP, Simpson JA, et al. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev. 2008;17:1774–1780. doi: 10.1158/1055-9965.EPI-08-0091. [DOI] [PubMed] [Google Scholar]
- 34.Chen JT, Lane MA, Clark DP. Inhibitors of the polymerase chain reaction in Papanicolaou stain. Removal with a simple destaining procedure. Acta Cytol. 1996;40:873–877. doi: 10.1159/000333994. [DOI] [PubMed] [Google Scholar]
- 35.Uhlmann K, Brinckmann A, Toliat MR, et al. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- 36.Jordan JA, Butchko AR, Durso MB. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J Mol Diagn. 2005;7:105–110. doi: 10.1016/s1525-1578(10)60015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia CA, Ahmadian A, Gharizadeh B, et al. Mutation detection by pyrosequencing: sequencing of exons 5-8 of the p53 tumor suppressor gene. Gene. 2000;253:249–257. doi: 10.1016/s0378-1119(00)00257-2. [DOI] [PubMed] [Google Scholar]
- 38.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colella S, Shen L, Baggerly KA, et al. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 40.Mikeska T, Bock C, El-Maarri O, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Earle JS, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433–440. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Chapelle A, Hampel H. Clinical Relevance of Microsatellite Instability in Colorectal Cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–129. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy KM, Berg KD, Geiger T, et al. Capillary electrophoresis artifact due to eosin: implications for the interpretation of molecular diagnostic assays. J Mol Diagn. 2005;7:143–148. doi: 10.1016/S1525-1578(10)60021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2011 doi: 10.1101/gr.117523.110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka N, Huttenhower C, Nosho K, et al. Novel application of structural equation modeling to correlation structure analysis of CpG island methylation in colorectal cancer. Am J Pathol. 2010;177:2731–2740. doi: 10.2353/ajpath.2010.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: Progress and problems. Biochim Biophys Acta. 2011;1825:77–85. doi: 10.1016/j.bbcan.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Patholog Res Int. 2011;2011 doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogino S, Galon J, Fuchs CS, et al. Cancer immunology-analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]