SUMMARY
The secondary metabolome of Basidiomycota represents a largely uncharacterized source of pharmaceutically relevant natural products. Terpenoids are the primary class of bioactive compounds isolated from mushrooms. The Jack O’Lantern mushroom Omphalotus olearius was identified 50 years ago as a prolific producer of anticancer illudin sesquiterpenoids, however to date there have been exceptionally few studies into the biosynthesis of these important compounds. Here we report the draft genome sequence of O. olearius, which reveals a diverse network of sesquiterpene synthases and two metabolic gene clusters associated with illudin biosynthesis. Their biochemical characterization enabled a comprehensive survey of all currently available Basidiomycota genomes, thereby creating a predictive resource for terpenoid natural product biosynthesis in these organisms. Our results will facilitate discovery and biosynthetic production of unique pharmaceutically relevant bioactive compounds from Basidiomycota.
INTRODUCTION
Current estimates of fungal diversity (3 to 5 million species) exceed that of terrestrial plants by an order of magnitude (Blackwell, 2011). To date, only a fraction of all fungal species have been described (about 100,000), and an even smaller number explored (mostly filamentous fungi and yeast: phylum Ascomycota) for the production of pharmacologically relevant secondary metabolites, including important drugs such as β-lactam antibiotics, statins, and cyclosporine (Zhong and Xiao, 2009). Widely used in traditional medicine, mushroom forming fungi (phylum Basidiomycota) are known to be prolific producers of secondary metabolites (Schueffler and Anke, 2009; Wasser, 2011). Despite their rich chemodiversity, very few studies have addressed their secondary metabolic pathways, largely due to their complex life cycle and frequently poor growth under laboratory conditions (Erkel and Anke, 2008). Over the past two years there has been a rapid influx of Basidiomycota genomes; these fungi are mostly genome sequenced because they are pathogens, parasites, mycorrhiza, or wood-rotting lignocellulose degraders. However, their secondary metabolome, unlike that of many Ascomycota such as e.g. Aspergillus species, has received virtually no attention thus far.
Terpenoids are the primary class of secondary metabolites isolated from Basidiomycota, and often have unique structures not observed elsewhere in the natural world (Schueffler and Anke, 2009). Examples include the diterpenoid antibiotic pleuromutilin and the anticancer, antimicrobial, and immunomodulating triterpenoid compounds from Ganoderma species (Novak and Shlaes, 2010; Paterson, 2006). Mushrooms are particularly adept at producing a wide range of structurally diverse sesquiterpenes (Abraham, 2001; Kramer and Abraham, 2012), many of which exhibit antibiotic and cytotoxic activities (Schueffler and Anke, 2009; Zaidman et al., 2005). Sesquiterpene scaffolds are synthesized from the prenyl compound farnesyl diphosphate (FPP) by a class of enzymes known as sesquiterpene synthases (STS) (Christianson, 2006). These enzymes catalyze the release of diphosphate and then guide migration of the reactive carbocation along the prenyl chain, thereby inducing a series of cyclization and rearrangement reactions, until final carbocation quenching. Mechanistic studies have provided insight into the different cyclization pathways catalyzed by diverse STS (Cane and Ikeda, 2011; Davis and Croteau, 2000); Cane and Ikeda, 2011. Two pathways can be distinguished based on whether a synthase catalyzes isomerization of the trans 2,3-double bond of 2E,6E-FPP to generate a secondary carbocation from the resulting isomer (3R)-nerolidyl diphosphate (NPP). Additionally, sesquiterpenoid structures can be distinguished by the first carbon-carbon bond forming ring-closure reaction that can be 1,10 or 1,11 with a 2E,6E-FPP carbocation or one of four (1,6-, 1,7-, 1,10-, 1,11-) closures with a (3R)-NPP carbocation. Biosynthesis of many of the unique sesquiterpene compounds in mushrooms is proposed to proceed through a trans-humulyl cation intermediate (1,11-ring closure, 2E,6E-FPP carbocation), leading to a product that may be modified further to yield the final biologically active compounds (Abraham, 2001; Kramer and Abraham, 2012).
In this study we set out to sequence, for the first time, the genome of a mushroom species known to be a producer of bioactive terpenoid natural products. By combining genomic and biochemical information, we sought to develop a predictive framework that will allow researchers to tap into the vast terpenomes of Basidiomycota, and target specific biosynthetic genes for heterologous pathway engineering in order to produce natural and novel compounds with potential new bioactivities. As our model organism, we chose the Jack O’Lantern mushroom (Omphalotus olearius, taxonomic synonyms: O. illudens, Clitocybe illudens), which was identified decades ago as a prolific producer of anticancer illudin sesquiterpenoids (McMorris, 1999; Morisaki, 1985). Clinical studies showed that illudins are effective against several tumors, including metastatic prostate cancer (McMorris et al., 2010; Schobert et al., 2011; Wiltshire et al., 2007).
Here, we report the draft genome sequence of the O. olearius strain VT-653.13 which enabled the identification of a diverse network of 11 STS, along with two illudin biosynthetic clusters. The O. olearius genome sequence also provides an insight into biosynthetic pathways for non-terpenoid secondary metabolites, such as polyketides and non-ribosomal peptides. Finally, we present a phylogenetic analysis of the fast growing number of available Basidiomycota genome sequences for cross-species analysis of STS diversity, thereby providing a resource for further research into fungal enzymes producing structurally diverse and therapeutically relevant sesquiterpenoids.
RESULTS
O. olearius produces structurally diverse sesquiterpene hydrocarbons
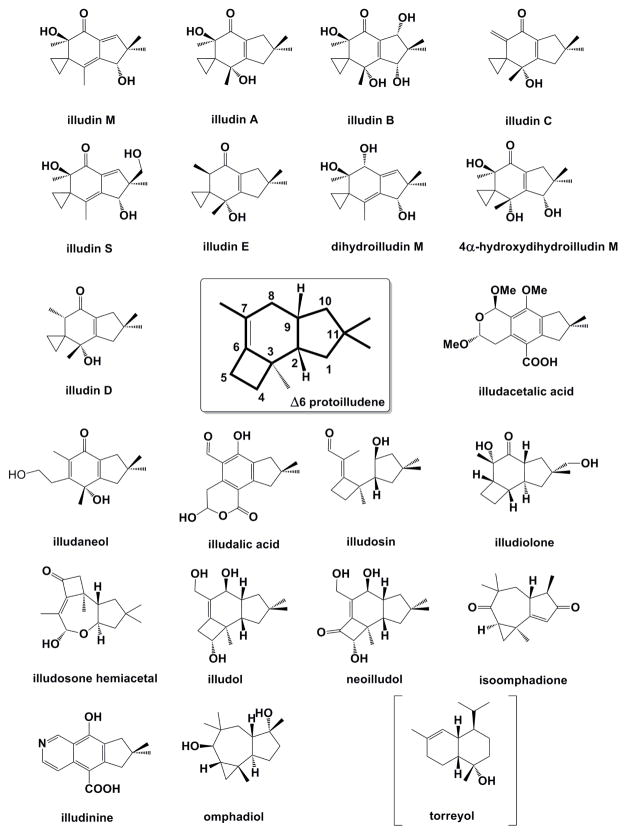
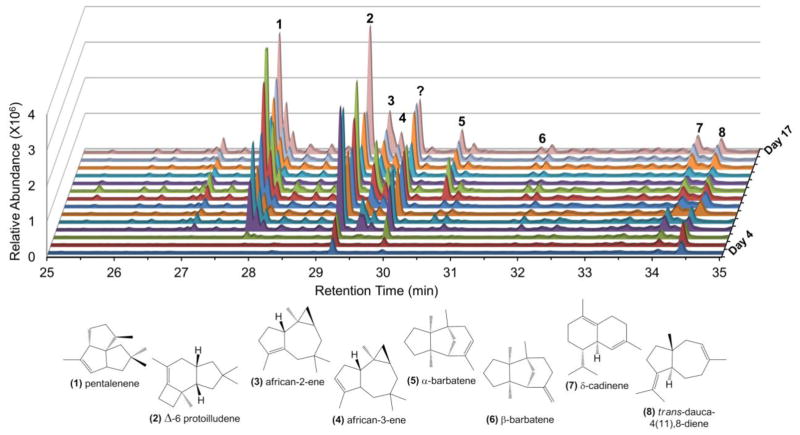
Almost all of the reported O. olearius sesquiterpenoids appear to be derivatives of the Δ-6 protoilludene scaffold (McMorris, 1999; McMorris et al., 2002; McMorris et al., 2000; Morisaki, 1985) (Figure 1). To obtain an estimate of the mushroom’s capacity to create other sesquiterpene hydrocarbon scaffolds as well, we first determined its product profile of unmodified sesquiterpenes. Volatile metabolites produced by liquid cultures of O. olearius were analyzed by Gas Chromatography-Mass Spectrometry (GC-MS). As shown in Figure 2, the illudin precursor Δ-6 protoilludene 2, along with the second-most abundant terpene pentalenene 1, accumulated steadily over the entire sampling period. The two other major compounds were identified as african-2-ene 3 and african-3-ene 4 (see Figure S1 for mass spectra of all compounds identified). Interestingly, all four of these major products are derived from the same initial trans-humulyl carbocation, suggesting that O. olearius possesses STS that favor 1,11 ring closure of the primary (E,E)-farnesyl cation (Tantillo, 2011). Four less prominent products were identified as α-barbatene 5, β-barbatene 6, δ-cadinene 7, and trans-dauca-4(11),8-diene 8. One major sesquiterpene product and many small terpene peaks could not be conclusively identified. While the same major and minor sesquiterpene products were detected in multiple cultures, their relative amounts varied, suggesting secondary metabolite regulation influenced by minor changes in culture conditions (Figure S2).
Figure 1.
The following sesquiterpenoid compounds have been isolated from O. olearius and are proposed to be derived from the Δ-6 protoilludene scaffold: Illudin A and B, illudalenol (Arnone et al., 1991); illudin C,D,E (Arnone et al., 1991); illudosin (Arnone et al., 1991); dihydroilludin (Singh et al., 1971); 4α-Hydroxyilludin M (Bradshaw et al., 1982); neoilludol (Nair and Anchel, 1975); illudacetalic acid (Nair and Anchel, 1972); illudol (McMorris et al., 1971); illudalic acid and illudinine (Nair et al., 1969); illudosone hemiacetal, isoomphadione and illudiolone (McMorris et al., 2002), and omphadiol (McMorris et al., 2000). The sesquiterpene alcohol (+) torreyol (Nair and Anchel, 1973) (in square brackets) is not derived from Δ-6 protoilludene, instead it is proposed to be a derivative of α–muurolene.
Figure 2.
Time course of sesquiterpene production by O. olearius. Volatile metabolites produced by liquid cultures of O. olearius were sampled by solid phase microextraction (SPME) and analyzed by Gas Chromatography-Mass Spectrometry (GC-MS). Identified sesquiterpene compounds are numbered and structures are shown for each compound (see Figure S1 for compound mass spectra). One major sesquiterpene product and many small terpene peaks could not be conclusively identified. See also Figure S2. Genome assembly statistics and associated data can be found in Table S1 and Data S1.
The genome and sesquiterpenome of O. olearius
The production of several different sesquiterpene hydrocarbons by O. olearius indicated that this organism expresses a large complement of functional STS, which we sought to identify by sequencing the genome of the haploid O. olearius (DC.) Singer strain VT-653.13. The obtained final assembly length was estimated at 28.15 Mbp, with approximately 89-fold sequencing depth and approximately 94 % genome coverage. Contained within the assembly are 8,172 predicted protein-coding ORFs encoding at least 100 amino acids (Data S1). The genome size and number of predicted genes falls within the range of other Basidiomycota genomes (Martin et al., 2011). Functional annotation of the predicted genes was carried out using BLAST with the following databases: KEGG, KOG, SwissProt, TrEMBL, NR, and GO (Data S1). Additional genome statistics are included in Table S1.
To find putative STS encoding genes, we performed BLAST homology searches with six STS (Cop1-6) previously cloned by our group from the model Basidiomycete Coprinus cinereus (Agger et al., 2009; Lopez-Gallego et al., 2010; Lopez-Gallego et al., 2010). We identified a total of 11 putative STS genes, which were named Omp1-10 (with 5a and 5b denoting two STS genes located adjacent to one another on the same scaffold). These were located on 10 different scaffolds, and were predicted to contain two to six introns each (Table S2 and Data S2). A preliminary phylogenetic analysis showed that the Omp STS clustered with the Cop STS into four major groups, suggesting that the enzymes in each cluster may produce related sesquiterpenes through conserved cyclization pathways. Since functional data for Basidiomycota STS are extremely limited, we sought to clone and individually characterize all 11 putative O. olearius STS.
Predicted O. olearius sesquiterpene synthases catalyze a wide range of cyclization reactions
To facilitate cloning of functional STS, each individual gene model was manually inspected and splicing predictions were confirmed by alignment with the other STS in order to compare conserved residues. Cloned putative STS from O. olearius were then functionally characterized through heterologous expression in E. coli, followed by GC-MS analysis of the sesquiterpene products of the recombinant cultures (Figure 3).
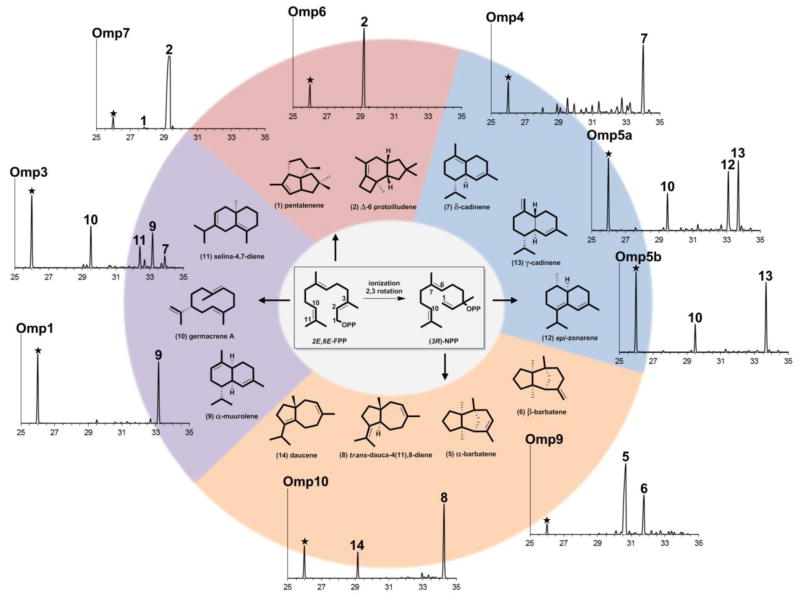
Figure 3.
Sesquiterpenes produced by E. coli expressing STS genes from O. olearius. Volatile sesquiterpene compounds in the culture head space were analyzed by GC-MS. Identified major compound peaks are labeled by numbers corresponding to structures shown below. Indole (★) endogenously produced by E. coli cultures serves as an internal reference point to compare relative amounts of sesquiterpene compounds produced (see Figure S1 for compound mass spectra). Compounds with retention times between 25 and 35 minutes (x-axis) were analyzed. Omp1 synthesizes α-muurolene 9 (84 % of total sesquiterpene products), while Omp3 synthesizes β-elemene 10 (29 %) (the heat induced Cope-rearrangement product of germacrene A) (Faraldos et al., 2007), selina-4,7-diene 11 (16 %), α-muurolene 9 (26 %), and δ-cadinene 7 (9 %). Omp4 synthesizes primarily δ-cadinene 7 (41 %), while Omp5a and Omp5b both produce β-elemene 10 (17 % and 23 %, respectively) and γ-cadinene 13 (36 % and 64 %, respectively) as major products, with Omp5a also producing epi-zonarene 12 (32 %). Omp6 and Omp7 synthesize the illudin precursor Δ-6 protoilludene 2 as their major products (95 % and 96 %, respectively), with Omp7 synthesizing 0.7 % pentalenene 1. Omp9 synthesizes α-barbatene 5 (57 %) and β-barbatene 6 (21 %), while Omp10 synthesizes daucene 14 (21 %) and trans-dauca-4(11),8-diene 8 (71 %). See also Table S2 for gene information for all 11 STS, Figure S3 for expression analysis in E. coli, and Data S2 for STS gene sequences.
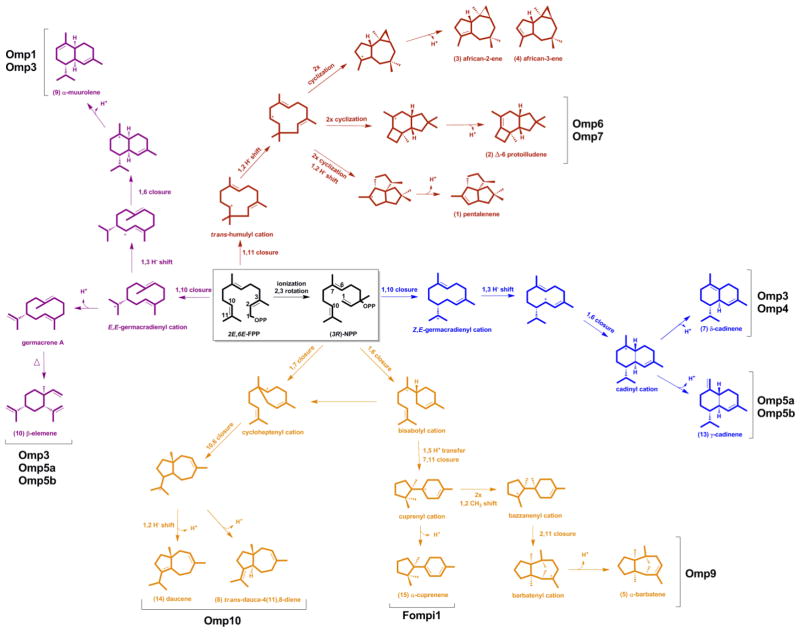
As shown in Figure 4, the identified Omp STS family includes enzymes that catalyze five of the six described ring-closures of the initial (E,E)-farnesyl and secondary (3R)-nerolidyl cations to reach their respective major products (Degenhardt et al., 2009; Tantillo, 2011). Notably, this first cyclization step is conserved among the Omp and Cop STS in each of the four homology groups, supporting our hypothesis that sequence conservation among fungal STS is predictive of their activities. Except for the Δ-6 protoilludene 2 derived illudins and the α-muurolene 9 derived (+)-torreyol (Figure 1), modified products of the other cyclization pathways catalyzed by STS have not been described from O. olearius.
Figure 4.
Proposed cyclization pathways leading to O. olearius sesquiterpene products. Ionization of FPP results in a primary carbocation from (E,E)-FPP that can either undergo two different ring closures (1,10 or 1,11) or is isomerized to a secondary carbocation from (3R)-NPP, which can undergo four different ring closures (1,6; 1,7; 1,10 or 1,11). Shown are pathways leading to major sesquiterpene products identified in the culture headspace of O. olearius (pentalenene 1, Δ-6 protoilludene 2, african-2-ene 3, african-3-ene 4, α-barbatene 5, δ-cadinene 7, and trans-dauca-4(11),8-diene 8) and synthesized by recombinant Omp sesquiterpene synthases (Δ-6 protoilludene 2, α-barbatene 5, δ-cadinene 7, trans-dauca-4(11),8-diene 8, α-muurolene 9, β-elemene 10, γ-cadinene 13, and daucene 14) (see Figure S1 for compound mass spectra). Also shown is the cyclization pathway leading to α-cuprenene 15 synthesized by sesquiterpene synthase Fompi1 cloned from Fomitopsis pinicola.
Three Omp enzymes (Omp1-3) preferentially catalyzed a 1,10 cyclization of (E,E)-FPP to form bicyclic sesquiterpenes with two six-membered rings. Omp1 is a selective α-muurolene 9 synthase (Figure 3), while Omp3 produces three additional major products. Omp2 was not functional, despite soluble protein expression (Figure S3). The Omp4, Omp5a and Omp5b group of STS synthesized major compounds that require 1,10 cyclization of (3R)-nerolidyl diphosphate (NPP). Omp4 is a highly promiscuous enzyme, synthesizing small amounts of at least 16 different sesquiterpenes in addition to its major product, δ-cadinene 7, which is a trend seen in other cadinane-type sesquiterpene synthases (Faraldos et al., 2012; Garms et al., 2010). Both Omp5a and Omp5b synthesize γ-cadinene 13 as their major product, while Omp5a also synthesizes epi-zonarene 12 (a germacrene D rearrangement product (Bulow and Konig, 2000)).
Two new cyclization activities for which no dedicated STS are currently known were identified for Omp9 and Omp10. Omp9 is highly active and generates two major products, α-barbatene 5 and β-barbatene 6, compounds known to be produced by fungi and plants (Dickschat et al., 2011; Tholl et al., 2005). Biosynthesis of these compounds was first proposed to proceed through a bisabolyl cation followed by the formation of a (R)-cuprenyl cation (via 7,11- cyclization and subsequent 1,4-hydride shift) that is then rearranged to the tricyclic barbatenes (Vedula et al., 2008). Recent feeding studies with Fusarium strains (Dickschat et al., 2011), however, corrobate a mechanism shown in Figure 4 that proceeds through a (S)-cuprenyl cation which is obtained from a (R)-bisabolyl cation via 1,5-proton transfer followed by 7,11-cyclization as proposed earlier by quantum chemical calculations (Hong and Tantillo, 2006).
Omp10 produces two carotane sesquiterpenes: daucene 14 and trans-dauca-4(11),8-diene 8, which are antimicrobial terpenes reported to be produced by carrots (Daucus carota) and wood-rotting Basidiomycota (Ghisalberti, 1994; Rosecke et al., 2000). Two cyclization pathways leading to the carotane skeletons can be envisioned: 1,7- or 1,6-cyclization of (3R)-NPP, where in the latter case a subsequent rearrangement yields the cycloheptenyl cation that generates the daucene products of Omp10 (Figure 3 and Figure 4). Omp8, another Omp9/10 homolog, was located at the end of a scaffold and lacked approximately 100 amino acids at its N-terminus and was not functional when expressed in E. coli. Creation of a chimeric gene with the N-terminal region of Omp9, however, failed to generate a functional enzyme.
Absent from the previously characterized Cop STS (Agger et al., 2009) was an enzyme capable of catalyzing a 1,11 cyclization of (E,E)-FPP leading to major groups of bioactive sesquiterpenes in Basidiomycota, including Δ-6 protoilludene and thereof derived illudins. When Omp6 and Omp7 were expressed in E. coli, both enzymes proved to be very active and synthesized the illudin precursor Δ-6 protoilludene 2; thereby identifying the first step of illudin biosynthesis in O. olearius. Surprisingly, both enzymes were very specific in E. coli and only recombinant Omp7 generated minute amounts of the trans-humulyl derived pentalenene 1. None of the Omp STS generated the africanenes 3, 4 detected in O. olearius cultures (Figure 2).
Omp6 and Omp7 are highly active and specific Δ-6 protoilludene synthases
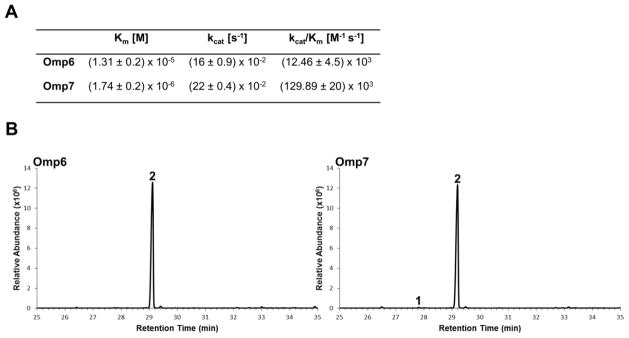
To determine whether alternative reactions catalyzed by Omp6 and Omp7 under different reaction conditions could account for the synthesis of the africanenes 3, 4 and of pentalenene 1 by the fungus; we characterized their activities in vitro. Both Δ-6 protoilludene synthases, Omp6 and Omp7, were overexpressed and purified from E. coli, and the kinetic parameters of the purified proteins were calculated (Figure 5A). The two enzymes display similar catalytic turnover rates (kcat) with (E,E)-FPP. However, Omp7 binds the substrate with ~10-fold greater affinity (Km) resulting in a 10-fold increase in catalytic efficiency.
Figure 5.
Activities of purified Δ-6 protoilludene synthases Omp6 and Omp7. (a) Kinetic parameters for Omp6 and Omp7 were determined using (E,E)-FPP as the substrate in a coupled spectrophotometric assay. (b) GC-MS analysis of in vitro reactions containing purified Omp6 or Omp7 incubated with (E,E)-FPP. Both enzymes produce greater than 99 % Δ-6 protoilludene. Omp7 also produces a very small amount of pentalenene 1.
Purified Omp6 and Omp7 were further characterized by GC-MS analysis of their product profiles after in vitro incubation with (E,E)-FPP. Both enzymes maintained their selectivity and produced greater than 99 % Δ-6 protoilludene (Figure 5B). We attempted to alter the enzymes’ product profiles by varying pH, temperature, and buffer conditions, however under all reaction conditions tested the enzymes remained highly selective. Therefore, the missing STS activities may either not be included in the genome assembly, or different splicing isoforms of Omp6 or Omp7 (alternative splicing is well-documented in fungi (Chang and Muddiman, 2011)) or yet other unknown factors that influence the enzymes’ activity in the fungus could be responsible for the synthesis of these compounds.
Omp1, Omp6, and Omp7 are located in biosynthetic gene clusters
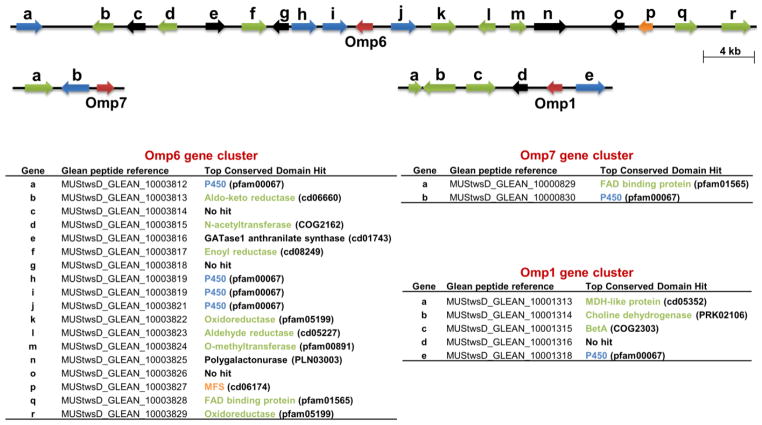
In fungi, secondary metabolite biosynthetic genes are typically located in contiguous clusters (Keller et al., 2005). Anticipating that one or both of the characterized Δ-6 protoilludene synthases Omp6 and Omp7 are involved in the biosynthesis of the cytotoxic illudins, we surveyed the genomic regions surrounding each STS for potential biosynthetic genes. Although they share significant amino acid sequence identity (62 %), Omp6 and Omp7 are located on two separate scaffolds in the assembly (Table S2). As shown in Figure 6, Omp6 is located in a well-defined cluster spanning roughly 25 kb, and including four P450 enzymes and fourteen additional putative genes that include at least five putative oxidoreductases, two putative transferases, a putative multiple drug transporter and two seemingly unrelated enzymes, a GATase1 anthranilate synthase and a polygalactonurase. Omp7, on the other hand, is within a cluster containing only one P450 enzyme and a FAD binding protein (Figure 6). The orientation and homology of Omp7 and the adjacent P450-b to Omp6 and P450-j suggests that a gene duplication event led to the formation of the smaller gene cluster.
Figure 6.
Organization of sesquiterpene biosynthetic gene clusters in O. olearius. Three putative biosynthetic clusters surrounding Omp6, Omp7, and Omp1 were identified after manual annotation. Predicted ORFs are colored according to their putative function, with blue representing P450 enzymes, green representing enzymes with a potential role in sesquiterpene scaffold modification, orange representing a drug transporter, and red representing the respective STS in each cluster. Corresponding peptide reference numbers (Data S1) are included for each putative gene. The top conserved domain hit (CDD) at NCBI is listed in parentheses next to each ORFs putative function. The Omp6 cluster contains four putative P450 monooxygenases, five reductases, a FAD-binding protein, a methyltransferase, an N-acetyltransferase, and a multiple drug transporter. A GATase1 anthranilate synthase and a polygalactonurase were also identified. Omp7 is part of a small gene cluster with a single P450 monooxygenase located less than 1 kb upstream and a FAD-binding protein approximately 4 kb upstream of the STS gene. The biosynthetic cluster containing Omp1 consists of a single P450 enzyme and three additional genes with possible roles in the modification of α-muurolene 9. See also Table S3 for gene information and Figure S4 for transcriptional analysis. Putative PKS and NRPS clusters, and overall P450 monooxygenase content were also examined in Table S5 and Table S6, respectively. Putative P450 monooxygenase sequences are contained in Data S3.
Among all the other identified Omp STS, only Omp1 appears to be located within a biosynthetic cluster containing a single P450 enzyme, and three genes that may be involved in subsequent modification of α-muurolene 9 to its reported alcohol (+)-torreyol (Nair and Anchel, 1973) and other, as yet unreported α-muurolene derivatives (Figure 1 and Figure 6).
To examine whether our cluster predictions for Omp6 and Omp7 are of functional significance, we sought to amplify select genes from each cluster. Five properly spliced ORFs from the Omp6 cluster (6-h, 6-i, 6-j, 6-k, 6-l, Figure 6) and one gene from the Omp7 (7-b, Figure 6) cluster were readily cloned from O. olearius cDNA, indicating expression of the modifying enzymes surrounding Omp6 and Omp7, and confirming preliminary splicing predictions (Table S3). Additionally, examination of transcript levels by qRT-PCR affirmed the co-regulation of Omp6 and Omp7 with the putative modifying genes in their respective clusters (Figure S4). These results showed that both clusters are of functional importance in O. olearius, which is further supported by the high activity and exceptional product selectivity of Omp6 and Omp7. The finding that Omp7 has a 10-fold higher catalytic efficiency than Omp6 suggests that the small Omp7 cluster is a gene duplication to boost rate-limiting steps in the biosynthesis of illudin compounds.
Analysis of Basidiomycota genomes uncovers an extensive sesquiterpenome
In order to establish a predictive framework for sesquiterpene natural product biosynthesis in Basidiomycota, we used the sequence and functional data gained for O. olearius and C. cinereus STS and analyzed the currently available genome data from 40 Basidiomycota (Table S4). We uncovered a total of 542 putative STS sequences in all inspected genomes. Interestingly, genomes without any STS belonged to pathogenic or parasitic fungi. Fungi containing STS generally have an average of 16 and a maximum of 41 putative STS homologs. The large STS gene complements identified in O. olearius and other Basidiomycota genomes confirms that sesquiterpenoids represent a major class of secondary metabolites produced by these fungi.
For phylogenetic analysis, the Omp amino acid sequences were aligned against the other 531 putative STS sequences. The output was manually inspected for erroneous protein predictions resulting in incomplete sequences that deviated from the expected protein length (~250-425 aa) or were lacking the conserved metal-binding DDxxD and NSE/DTE motifs of STS. The final alignment contained 393 putative STS representing 35 basidiomycota species (Table S4 and Data S2).
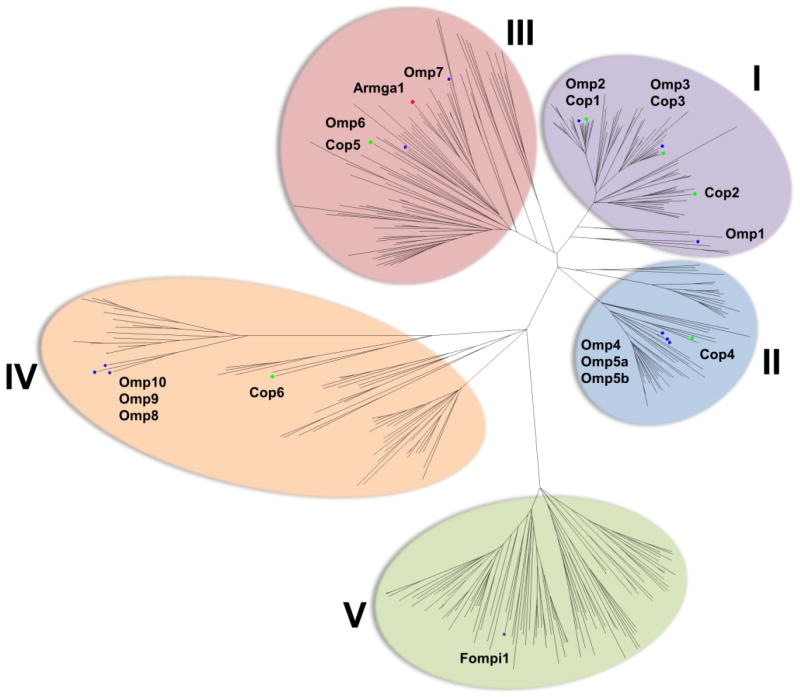
A phylogenetic tree was constructed, revealing five distinct STS clades (Figure 7). Interestingly, the trans-humulyl-type synthases clustered together (Omp6, Omp7; clade III), while STS responsible for 1,6 or 1,7 cyclization (Omp9, Omp10) of (3R)-NPP were segregated in another group (clade IV). STS that catalyze the 1,10 cyclization of either (E,E)-FPP or (3R)-NPP formed two clades around Omp1-3 (clade I) and Omp4-5a/b homologs (clade II). One clade (clade V) did not contain any characterized Omp STS but possessed significant sequence similarity to clade IV. Knowledge of sequence conservation for each distinct clade of STS provides a basis for more effective site-directed mutagenesis to modulate activity and develop a mechanistic understanding of the distinct cyclization activities exhibited by each STS group.
Figure 7.
Unrooted Neighbor-Joining phylogram of STS homologs identified in 42 Basidiomycota genomes. Five clades are highlighted with colors as in Figure 4. STS from C. cinereus (Cop), O. olearius (Omp), F. pinicola (Fompi1), and the single STS described from Armillaria gallica (Armga1) (Engels et al., 2011) are labeled. Refer to the methods for tree construction parameters. Sequences used in the final alignment and the resultant phylogram data can be found in Data S2. See also Table S4 for a breakdown of total number of putative STS and number of STS sequences used in tree building after manual inspection of alignment. See Figure S5 for product analysis of the STS from F. pinicola.
Using the predictive framework to uncover a new STS activity in Fomitopsis pinicola
We next sought to not only validate the use of our framework for identification of additional STS from other Basidiomycota, but also assign biochemical data to clade V, which contained no STS characterized from O. olearius, but shared high sequence similarity with clade IV. Fomitopsis pinicola was chosen because 7 of its 17 STS are found in clade V and 2 additional STS cluster with sequences in clade IV. In addition, F. pinicola has been reported to produce several volatile sesquiterpenes, including the Omp9 (Clade IV) products α-barbatene 5 and β-barbatene 6 (Rosecke et al., 2000) (Figure 4). Upon examination of the genomic region surrounding the seven STS, only one, Fompi84944 (hereby referred to to Fompi1), was contained within a small biosynthetic cluster including two putative P450 monooxygenases (Figure S5A). This STS was selected for subsequent cloning and functional characterization.
Interestingly, expression of Fompi1 in E. coli revealed it to be a highly active and specific α-cuprenene 15 synthase (Figure S5B), a sesquiterpene synthase activity previously reported by us for Cop6 (Agger et al., 2009) which is present in Clade IV (Figure 7). This finding suggests conservation of the 1,6 cyclization mechanism between clades IV and V, leading to the barbatene 5, 6 (Omp9), α-cuprenene 15 (Cop6 and Fompi1), and possibly carotane 8, 14 (Omp10) sesquiterpenes. By using genomic context and the framework presented above, we successfully identified a new STS activity in F. pinicola, responsible for the production of α-cuprenene 15, which was previously not reported (Rosecke et al., 2000).
Insights into other secondary metabolic capabilities of O. olearius
Non-terpenoid secondary metabolites of O. olearius include the nematicidal omphalotins (A-F) (Liermann et al., 2009) and the siderophore ferrichrome A (Welzel et al., 2005). Omphalotins are cyclopeptides that are likely made by a non-ribosomal peptide synthase (NRPS). A set of genes including a NRPS (fso1), L-ornithine-N5-monoosuxtenase (omo1) and acyltransferase (ato1) were previously cloned from O. olearius and presumed to be involved in ferrichrome A biosynthesis (Welzel et al., 2005). To gain further insight into the secondary metabolome of O. olearius, SMURF (Khaldi et al., 2010) was used to identify metabolic clusters in the genome assembly. Five putative polyketide synthase genes (PKS) and eight putative NRPS genes, each located on a different scaffold, were identified (Table S5). The biosynthetic gene cluster predictions include fso1 and a previously described fatty acid synthase (Antelo et al., 2009).
Another measure of the metabolic diversity of an organism is its complement of P450 enzymes (Grogan, 2011) (CYPome). P450s have undergone extensive diversification in plants and in some fungal species (Cresnar and Petric, 2011; Park et al., 2008), where they play roles in biosynthesis and detoxification/degradation of xenobiotic compounds, including plant defense compounds (Lah et al., 2011). We therefore conducted a comparative analysis of the CYPome of O. olearius with that of other studied Basidiomycota using the Fungal Cytochrome P450 Database (FCPD) (Park et al., 2008) (Table S6 and Data S3). Notably, many of the pathogenic and parasitic Basidiomycota which possess no STS are also significantly lacking in P450 enzymes, with the ratio of P450 enzymes to total ORFs not exceeding 0.30 %. Conversely, there is strong positive correlation between fungi possessing STS and the number of P450 enzymes present, with most species surveyed containing a greater than 1.00 % ratio, and P. placenta containing the highest ratio at 2.06 %. O. olearius contains 121 putative cytochrome P450 enzymes, accounting for 1.48 % of its total ORFs, further substantiating its use as a model for the study of secondary metabolism in Basidiomycota.
DISCUSSION
O. olearius is known for its production of anticancer illudin compounds (Figure 1), and with this draft genome sequence we have identified two Δ-6 protoilludene synthases, Omp6 and Omp7, each located within a gene cluster likely responsible for the biosynthesis of these and related illudin compounds (Figure 6). Both STS genes and neighboring, putative biosynthetic genes were readily cloned and shown to be co-regulated during fungal growth (Table S3 and Figure S4), suggesting that the two gene clusters are functional and expressed in O. olearius. The purified, recombinant Δ-6 protoilludene synthases proved to be highly active and possessed exceptional product selectivity. Surprisingly, Omp7 has a 10-fold higher catalytic efficiency than Omp6, bringing into question their specific roles in illudin biosynthesis. Omp6 and its adjacent P450 monooxygenase (Figure 6, 6-i) share high sequence homology with Omp7 and its adjacent P450 monooxygenase (Figure 6, 7-b), which leads us to believe that gene duplication occurred, likely in order to boost rate-limiting steps in illudin biosynthesis. Common to all of the identified illudin structures shown in Figure 1 is an oxidation at position 8 of the six-carbon ring, which may suggest that these monooxygenase enzymes are responsible for this reaction. Subsequent oxidation reactions then may lead to a α,β-unsaturated ketone, which is important for illudin cytotoxicity (McMorris et al., 1990), and oxidation at position 3 and/or 7 may trigger ring contraction, forming the reactive cyclopropyl-ring found in the illudins.
Synthesis of the anticancer illudins M and S appears to involve at least five oxidation steps which may be catalyzed by the core group of P450s and oxidoreductases surrounding Omp6 (6-h to 6-l) that are all expressed by the fungus (Table S3). Additional biosynthetic enzyme present in the Omp6 cluster likely are responsible for the synthesis other illudanes made by O.olearius (Figure 1). For example, the putative GATase1 anthranilate synthase (6-e) may install the nitrogen in the illudinine ring, while the putative O-methyltransferase (6-m) is likely involved in the synthesis of illudacetalic acid (Figure 1). Biosynthesis of several other known illudane compounds (i.e. illudalic acid, Figure 1) appear to involve a Baeyer-Villiger monooxygenase (BVM) that catalyzes lactonization (Leisch et al., 2011). Two putative enzymes (6-q and 7-a) in the Omp6 and Omp7 cluster are pfam01565 family proteins that utilize FAD as co-factor, which indicates that they may encode enzymes similar to BVMs (Kharel et al., 2009).
Unfortunately, O. olearius is not a genetically tractable fungus that allows testing of biosynthetic functions by creating gene knockouts. Delineation of its illudane pathways will require stepwise reconstitution of the biosynthetic reactions sequences in another fungal host such as S. cerevisae for functional expression of the many P450 enzymes present in the Omp6 and Omp7 clusters. Since illudin M and S have poor therapeutic indices, the development of semi-synthetic derivatives with better pharmacological properties has been pursued (Schobert et al., 2011). The large set of putative Δ-6 protoilludene tailoring enzymes identified in this study will enable the development of combinatorial biosynthetic approaches to obtain new illudin derivatives with improved bioactivities.
In addition to the two Δ-6 protoilludene synthases, O. olearius possesses at least nine additional STS. Two of these enzymes, Omp9 and Omp10, encode sesquiterpene synthases with new cyclization activities. We found similar or even larger STS gene complements in other Basidiomycota genomes, confirming that sesquiterpenoids represent a major class of secondary metabolites produced by higher fungi. The number of putative terpene synthases (531 putative sequences) identified in only 40 available genome sequences from Basidiomycota by far exceeds the number of terpene synthases present in bacteria. In fact, a recent study surveying the publicly available genomes of all bacteria (presently, 1750 bacterial genomes are listed at MicrobesOnline (www.microbesonline.org)) identified only 179 putative terpene synthase genes, mostly from actinomycetes (Citron et al., 2012). Basidiomycota will soon prove to be a valuable resource for genome mining of STS, PKS and NRPS biosynthetic enzymes. The rapid influx of new Basidiomycota genomes (12 genomes in the last year alone at the Joint Genome Institute) provides an unmatched opportunity for biosynthetic gene discovery. However, most of the recent genome sequencing projects focused on wood-rotting lignocellulose degraders. To our knowledge, this is the first published sequencing initiative centered solely on biosynthetic gene elucidation in higher fungi.
EXPERIMENTAL PROCEDURES
Chemicals
(E,E)-FPP was purchased from Sigma-Aldrich (St. Louis, MO). Other chemicals were from suppliers as indicated or from Sigma-Aldrich.
Fungal strains and growth conditions
The haploid monokaryotic strain O. olearius (DC.) Singer VT-653.13 and the F. pinicola (Sw.) P. Karst. strain CS-1 (USDA, Forest Mycology Center, Madison, WI) were grown on potato dextrose agar (PDA) at room temperature for 2–3 weeks in the dark prior to inoculating liquid cultures with an agar plug containing mycelium. Liquid media consisted of 1 g L−1 of potassium phosphate (monobasic), 3 g L−1 of sodium nitrate, 0.5 g L−1 of potassium chloride, 0.5 g L−1 of magnesium sulfate (heptahydrate), 5 g L−1 of corn steep powder and 40 g L−1 of glucose and was adjusted to pH 4.0. Cultures were grown in 250 mL flasks containing 100 mL of media at 28 °C and 225 rpm for 2–3 weeks in the dark.
Preparation of fungal genomic DNA for sequencing
O. olearius was grown by placing an agar plug containing mycelium on to a nitrocellulose membrane covering a PDA plate. After 3 weeks of growth in the conditions described above the mycelium was collected from the plate, frozen in liquid N2, and ground to a fine powder. Roughly 0.5 g of crushed tissue was placed in an Eppendorf tube to which 600 μl of pre-warmed (60 °C) CTAB buffer (2% CTAB, 100 mM TrisHCl pH 8.0, 20 mM EDTA, 1.4 M NaCl, 0.1 mg/mL proteinase K) was added, followed by 66 μl of 20% SDS. The solution was incubated at 65 °C for 2 hrs. The resultant mixture was extracted using an equal volume of phenol:chloroform:isoamyl alcohol mix (25:24:1). The aqueous layer was treated with RNase A for 2 hrs at 37 °C. Genomic DNA was precipitated by adding 0.6 volume of room temperature isopropyl alcohol, pelleted by centrifugation, and washed with cold 70% ethanol. DNA was resuspended in 1x TE buffer (pH 8.0)
Genome sequencing, assembly, annotation, and data deposition
De novo genome sequencing, assembly and initial annotation were carried out by the Beijing Genomics Institute (BGI) at Shenzhen, China. Paired-end (90 bp reads) Illumina sequencing of a 500 bp fragment library yielded 2,500 Mb of clean data that were assembled into genomic sequences using SOAPdenovo (Li et al., 2010). The optimal kmer length identified after assembly with SOAPdenovo was k = 15 and a pkdepth = 28 after processing 836,889,213 15-mers. The assembly yielded 868 scaffolds with a N50 of 199,357 bp and a N90 of 22,541 bp. The total assembly size was 28.15 Mbp. Genome coverage based on kmer analysis is estimated at 94.16 %. Augustus (Stanke et al., 2004) was used for gene prediction. Functional annotation was performed using BLAST (Altschul et al., 1990) for alignment of gene predictions with the following databases: KEGG, COG, SwissProt, TrEMBL, NR and GO.
Analysis of sesquiterpenes produced by recombinant E. coli cultures
Omp STS sequences were each cloned into our in-house vector pUCBB (Vick et al.) by for expression under the control of a constitutive lac promoter (Schmidt-Dannert et al., 2000). Single colonies of E. coli BL21 transformants were used to inoculate 4 mL seed cultures of LB supplemented with 100 μg/ml ampicillin. These cultures were used to inoculate 50 mL of LB media supplemented with 100 μg/ml ampicillin and allowed to grow for 18 hrs. The culture headspace was sampled for 15 min by solid phase microextraction (SPME) using a 100 μm polydimethylsiloxane fiber (Supelco, Bellefonte, PA). The fiber was inserted through a tin foil seal into the gas phase of the flask and, after adsorption, inserted into the injection port of the GC-MS for thermal desorption.
Gas chromatography-mass spectrometry (GC-MS) analysis
GC-MS analysis was conducted on an HP GC 7890A coupled to an anion-trap mass spectrometer HP MSD triple axis detector (Agilent Technologies, Santa Clara, CA). Separation of compounds was performed using a HP-5MS capillary column (30 m by 0.25 mm by 1.0 μm) with an injection port temperature of 250 °C and helium for the carrier gas. Mass spectra were recorded in electron impact ionization mode. Volatile compounds adsorbed on the fiber were allowed to desorb for 35 min in the injection port. The oven temperature started at 60 °C and was ramped up 4 °C min−1 to a final oven temperature of 250 °C. Mass spectra were scanned in the range of 5 to 300 atomic mass units at 1 s intervals.
Structural identification of sesquiterpene compounds
Compounds produced were identified by first calibrating the GC-MS with a C8-C20 alkane mix. Retention indices and mass spectra of compound peaks were compared to the published reference spectra in MassFinder’s (software version 3) terpene library (Joulain and König, 1998) and in the National Institute of Standards and Technology (NIST) standard reference database 08 (http://www.nist.gov/srd/nist1a.cfm). Major STS products were confirmed, when possible, by using essential oils with known terpene compositions and authentic standards as described previously (Agger et al., 2009; Lopez-Gallego et al., 2010; Lopez-Gallego et al., 2010). In addition, carrot seed oil was used for the identification of daucene 14 and calamus root oil was used for the identification of γ-cadinene 13. α-cuprenene was confirmed using the previously characterized Cop6 from C. cinereus. No essential oil or standard could be obtained for α-barbatene 5, β-barbatene 6, trans-dauca-4(11),8-diene 8, selina-4,7-diene 11, or epi-zonarene 12.
Kinetic parameters for Omp6 and Omp7
The Km and kcat of Omp6 and Omp7 were determined by incubation of the purified proteins with a range of concentrations of (E,E)-FPP (1 – 100 uM). Terpene synthase activity was monitored using an enzyme coupled spectrophotometric assay to measure the release of pyrophosphate (PPi) (PiPer™ Pyrophosphate Assay Kit, Invitrogen, Carlsbad, CA). Known concentrations of PPi were used to obtain a standard curve. Microplate assays were performed as previously described (Lopez-Gallego et al., 2010). Control reactions without enzyme and without substrate were performed simultaneously, and all reactions were carried out in triplicate. The Km and Vmax values were determined by non-linear regression analysis of the data in SigmaPlot 11.
Supplementary Material
SIGNIFICANCE.
Except for the lignolytic white rot fungus Stereum, which is a producer of bioactive sesquiterpenoids (many with unique ring-structures derived from Δ-6 protoilludene (Liermann et al., 2010)), none of the Basidiomycota with sequenced genomes surveyed in this work have been studied primarily for their abilities to produce terpenes. Yet, our analysis reveals that almost all harbor extensive STS gene families, suggesting that they have the ability to produce diverse and potentially novel terpene scaffolds. Moreover, many of the STS genes identified here appear to be located in biosynthetic gene clusters, thus providing access to a large number of terpene modifying enzymes. Our study shows that the genomes of Basidiomycota present an unprecedented opportunity for the discovery and heterologous engineering of novel terpene biosynthetic routes leading to compounds with new bioactivities. The phylogenetic tree described here presents a road-map for exploring the exciting and as yet untraversed terpenome of this fascinating class of organisms.
Acknowledgments
This research was supported by the National Institute of Health Grant GM080299 (to C.S-D.). G.T.W. was supported by a predoctoral National Institute of Health traineeship (GM08700).
Footnotes
ACCESSION NUMBERS
The sequencing reads for the genome sequence of O. olearius VT-653.13 have been deposited in the NCBI sequence read archive. The contigs used for whole genome assembly, and the scaffolds from the resultant assembly have been deposited at NCBI and can be accessed through BioProject ID: 79063, Accession: PRJNA79063. Additional sequence data are also provided in the Supplementary Information in Data S1 and S2.
Supplemental Information includes five figures, six tables, three data sets, and Supplemental Experimental Procedures.
References
- Abraham WR. Bioactive sesquiterpenes produced by fungi: are they useful for humans as well? Curr Med Chem. 2001;8:583–606. doi: 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- Agger S, Lopez-Gallego F, Schmidt-Dannert C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol. 2009;72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger S, Lopez-Gallego F, Schmidt-Dannert C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol. 2009;72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antelo L, Schlipp A, Hof C, Eisfeld K, Berg H, Hornbogen T, Zocher R, Anke H. The fatty acid synthase of the basidiomycete Omphalotus olearius is a single polypeptide. Z Naturforsch (C) 2009;64:244–250. doi: 10.1515/znc-2009-3-416. [DOI] [PubMed] [Google Scholar]
- Arnone A, Cardillo R, Dimodugno V, Nasini G. Secondary mold metabolites: 33. Structure elucidation of illudin C, illudin D and illudin E, novel illudane sesquiterpenoids isolated from Clitocybe illudens, using one-dimensional and two-dimensional NMR-spectroscopy. Gazz Chim Ital. 1991;121:345–348. [Google Scholar]
- Arnone A, Cardillo R, Nasini G, Depava OV. Secondary mold metabolites: 31. Isolation and structure elucidation of illudins A and illudins B, and illudalenol, new sesquiterpenoids from Clitocybe illudens. J Chem Soc Perk T. 1991;1:733–737. [Google Scholar]
- Arnone A, Cardillo R, Nasini G, Depava OV. Secondary Mold Metabolites: 34. Isolation and structure elucidation of illudosin, a novel sesquiterpene from Clitocybe illudens using one and two-dimensional NMR Techniques. J Chem Soc Perk T. 1991;1:1787–1791. [Google Scholar]
- Blackwell M. The Fungi: 1, 2, 3 … 5.1 million species? Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Bradshaw A, Peter W, Hanson JR, Hitchcock PB. 4α-Hydroxydihydroilludin M. A new sesquiterpenoid metabolite of Clitocybe illudens. Phytochemistry. 1982;21:942–943. [Google Scholar]
- Bulow N, Konig WA. The role of germacrene D as a precursor in sesquiterpene biosynthesis: investigations of acid catalyzed, photochemically and thermally induced rearrangements. Phytochemistry. 2000;55:141–168. doi: 10.1016/s0031-9422(00)00266-1. [DOI] [PubMed] [Google Scholar]
- Cane DE, Ikeda H. Exploration and mining of the bacterial terpenome. Acc Chem Res. 2011 doi: 10.1021/ar200198d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Muddiman DC. Identification of alternative splice variants in Aspergillus flavus through comparison of multiple tandem MS search algorithms. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Citron CA, Gleitzmann J, Laurenzano G, Pukall R, Dickschat JS. Terpenoids are widespread in actinomycetes: A correlation of secondary metabolism and genome data. Chem Bio Chem. 2012;13:202–214. doi: 10.1002/cbic.201100641. [DOI] [PubMed] [Google Scholar]
- Cresnar B, Petric S. Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta. 2011;1814:29–35. doi: 10.1016/j.bbapap.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Davis EM, Croteau R. Biosynthesis: Aromatic Polyketides, Isoprenoids, Alkaloids. Berlin: Springer-Verlag Berlin; 2000. Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes; pp. 53–95. [Google Scholar]
- Degenhardt J, Kollner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70:1621–1637. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Dickschat JS, Brock NL, Citron CA, Tudzynski B. Biosynthesis of sesquiterpenes by the fungus Fusarium verticillioides. Chem Bio Chem. 2011;12:2088–2095. doi: 10.1002/cbic.201100268. [DOI] [PubMed] [Google Scholar]
- Engels B, Heinig U, Grothe T, Stadler M, Jennewein S. Cloning and characterization of an Armillaria gallica cDNA encoding protoilludene synthase, which catalyzes the first committed step in the synthesis of antimicrobial melleolides. J Biol Chem. 2011;286:6871–6878. doi: 10.1074/jbc.M110.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkel G, Anke T. Biotechnology. Wiley-VCH Verlag GmbH; 2008. Products from Basidiomycetes; pp. 489–533. [Google Scholar]
- Faraldos JA, Miller DJ, González V, Yoosuf-Aly Z, Cascón O, Li A, Allemann RK. A 1,6-ring closure mechanism for (+)-δ-cadinene synthase? J Am Chem Soc. 2012;134:5900–5908. doi: 10.1021/ja211820p. [DOI] [PubMed] [Google Scholar]
- Faraldos JA, Wu S, Chappell J, Coates RM. Conformational analysis of (+)-germacrene A by variable temperature NMR and NOE spectroscopy. Tetrahedron. 2007;63:7733–7742. doi: 10.1016/j.tet.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garms S, Köllner TG, Boland W. A multiproduct terpene synthase from Medicago truncatula generates cadalane sesquiterpenes via two different mechanisms. J Org Chem. 2010;75:5590–5600. doi: 10.1021/jo100917c. [DOI] [PubMed] [Google Scholar]
- Ghisalberti EL. The daucane (carotane) class of sesquiterpenes. Phytochemistry. 1994;37:597–623. [Google Scholar]
- Grogan G. Cytochromes P450: exploiting diversity and enabling application as biocatalysts. Curr Opin Chem Biol. 2011;15:241–248. doi: 10.1016/j.cbpa.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Hong YJ, Tantillo DJ. Which is more likely in trichodiene biosynthesis: Hydride or proton transfer? Org Lett. 2006;8:4601–4604. doi: 10.1021/ol061884f. [DOI] [PubMed] [Google Scholar]
- Joulain D, König WA. The Atlas of Spectral Data of Sesquiterpene Hydrocarbons. Hamburg, Germany: E.B. Verlag; 1998. [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharel MK, Pahari P, Lian H, Rohr J. GilR, an unusual lactone-forming enzyme involved in gilvocarcin biosynthesis. Chem Bio Chem. 2009;10:1305–1308. doi: 10.1002/cbic.200900130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R, Abraham WR. Volatile sesquiterpenes from fungi: what are they good for? Phytochem Rev. 2012;11:15–37. [Google Scholar]
- Lah L, Podobnik B, Novak M, Korosec B, Berne S, Vogelsang M, Krasevec N, Zupanec N, Stojan J, Bohlmann J, et al. The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification. Mol Microbiol. 2011;81:1374–1389. doi: 10.1111/j.1365-2958.2011.07772.x. [DOI] [PubMed] [Google Scholar]
- Leisch H, Morley K, Lau PCK. Baeyer-villiger monooxygenases: More than just green chemistry. Chem Rev. 2011;111:4165–4222. doi: 10.1021/cr1003437. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liermann JC, Opatz T, Kolshorn H, Antelo L, Hof C, Anke H. Omphalotins E–I, five oxidatively modified nematicidal cyclopeptides from Omphalotus olearius. Eur J Org Chem. 2009:1256–1262. [Google Scholar]
- Liermann JC, Schuffler A, Wollinsky B, Birnbacher J, Kolshorn H, Anke T, Opatz T. Hirsutane-type sesquiterpenes with uncommon modifications from three basidiomycetes. J Org Chem. 2010;75:2955–2961. doi: 10.1021/jo100202b. [DOI] [PubMed] [Google Scholar]
- Lopez-Gallego F, Agger SA, Abate-Pella D, Distefano MD, Schmidt-Dannert C. Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: Catalytic promiscuity and cyclization of farnesyl pyrophosphate geometric isomers. Chem Bio Chem. 2010;11:1093–1106. doi: 10.1002/cbic.200900671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gallego F, Wawrzyn GT, Schmidt-Dannert C. Selectivity of fungal sesquiterpene synthases: Role of the active site’s H-1 alpha loop in catalysis. Appl Environ Microbiol. 2010;76:7723–7733. doi: 10.1128/AEM.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Cullen D, Hibbett D, Pisabarro A, Spatafora JW, Baker SE, Grigoriev IV. Sequencing the fungal tree of life. New Phytol. 2011;190:818–821. doi: 10.1111/j.1469-8137.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- McMorris TC. Discovery and development of sesquiterpenoid derived hydroxymethylacylfulvene: a new anticancer drug. Bioorg Med Chem. 1999;7:881–886. doi: 10.1016/s0968-0896(99)00016-4. [DOI] [PubMed] [Google Scholar]
- McMorris TC, Chimmani R, Alisala K, Staake MD, Banda G, Kelner MJ. Structure-activity studies of urea, carbamate, and sulfonamide derivatives of acylfulvene. J Med Chem. 2010;53:1109–1116. doi: 10.1021/jm901384s. [DOI] [PubMed] [Google Scholar]
- McMorris TC, Kashinatham A, Lira R, Rundgren H, Gantzel PK, Kelner MJ, Dawe R. Sesquiterpenes from Omphalotus illudens. Phytochemistry. 2002;61:395–398. doi: 10.1016/s0031-9422(02)00205-4. [DOI] [PubMed] [Google Scholar]
- McMorris TC, Kelner MJ, Wang W, Moon S, Taetle R. On the mechanism of toxicity of illudins - the role of glutathione. Chem Res Toxicol. 1990;3:574–579. doi: 10.1021/tx00018a013. [DOI] [PubMed] [Google Scholar]
- McMorris TC, Lira R, Gantzel PK, Kelner MJ, Dawe R. Sesquiterpenes from the basidiomycete Omphalotus illudens. J Nat Prod. 2000;63:1557–1559. doi: 10.1021/np9904760. [DOI] [PubMed] [Google Scholar]
- McMorris TC, Nair MS, Singh P, Anchel M. Metabolites of Clitocybe illudens VII Structure of illudol. Phytochemistry. 1971;10:16115–11615. [Google Scholar]
- Morisaki N, Furukawa J, Kobayashi H, Iwasaki S, Nozoe S, Okuda S. Conversion of 6-protoilludene into illudin-M and -S by Omphalotus olearius. Tetrahedron Lett. 1985;26:4755–4758. [Google Scholar]
- Nair MS, Anchel M. Metabolic products of Clitocybe illudens IX Structure of illudacetalic acid and its conversion to illudinine. Tetrahedron Lett. 1972;27:2753–2754. [Google Scholar]
- Nair MS, Anchel M. Metabolic products of Clitocybe illudens. X (+)-Torreyol Lloydia. 1973;36:106. [Google Scholar]
- Nair MS, Takeshita H, McMorris TC, Anchel M. Metabolites of Clitocybe illudens IV Illudalic acid, a sesquiterpenoid, and illudinine, a sesquiterpenoid alkaloid. J Org Chem. 1969;34:240–243. doi: 10.1021/jo00838a058. [DOI] [PubMed] [Google Scholar]
- Nair MSR, Anchel M. Metabolic Products of Clitocybe illudens.11. Structure of Neoilludol. Tetrahedron Letters. 1975:1267–1268. [Google Scholar]
- Novak R, Shlaes DM. The pleuromutilin antibiotics: A new class for human use. Curr Opin Investig Drugs. 2010;11:182–191. [PubMed] [Google Scholar]
- Park J, Lee S, Choi J, Ahn K, Park B, Kang S, Lee YH. Fungal cytochrome P450 database. BMC Genomics. 2008;9:402. doi: 10.1186/1471-2164-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson RR. Ganoderma - a therapeutic fungal biofactory. Phytochemistry. 2006;67:1985–2001. doi: 10.1016/j.phytochem.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Rosecke J, Pietsch M, Konig WA. Volatile constituents of wood-rotting basidiomycetes. Phytochemistry. 2000;54:747–750. doi: 10.1016/s0031-9422(00)00138-2. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nat Biotechnol. 2000;18:750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- Schobert R, Knauer S, Seibt S, Biersack B. Anticancer active illudins: recent developments of a potent alkylating compound class. Curr Med Chem. 2011;18:790–807. doi: 10.2174/092986711794927766. [DOI] [PubMed] [Google Scholar]
- Schueffler A, Anke T. Secondary metabolites of basidiomycetes. In: Anke T, Weber E, editors. The Mycota: Physiology and Genetics XV. Berlin, Heidelberg: Springr; 2009. pp. 2009–2231. [Google Scholar]
- Singh P, Nair MS, McMorris TC, Anchel M. Metabolites of Clitocybe illudens VI Isolation of dihydroilludin M from Clitocybe illudens. Phytochemistry. 1971;10:2229–22230. [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantillo DJ. Biosynthesis via carbocations: theoretical studies on terpene formation. Nat Prod Rep. 2011;28:1035–1053. doi: 10.1039/c1np00006c. [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 2005;42:757–771. doi: 10.1111/j.1365-313X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- Vedula LS, Jiang JY, Zakharian T, Cane DE, Christianson DW. Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: Probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg-B(2+) motif. Arch Biochem Biophys. 2008;469:184–194. doi: 10.1016/j.abb.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick JE, Johnson ET, Choudhary S, Bloch SE, Lopez-Gallego F, Srivastava P, Tikh IB, Wawrzyn GT, Schmidt-Dannert C. Optimized compatible set of BioBrick vectors for metabolic pathway engineering. Appl Microbiol Biotechnol. 92:1275–1286. doi: 10.1007/s00253-011-3633-4. [DOI] [PubMed] [Google Scholar]
- Wasser SP. Current findings, future trends, and unsolved problems in studies of medicinal mushrooms. Appl Microbiol Biotechnol. 2011;89:1323–1332. doi: 10.1007/s00253-010-3067-4. [DOI] [PubMed] [Google Scholar]
- Welzel K, Eisfeld K, Antelo L, Anke T, Anke H. Characterization of the ferrichrome A biosynthetic gene cluster in the homobasidiomycete Omphalotus olearius. FEMS Microbiol Lett. 2005;249:157–163. doi: 10.1016/j.femsle.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, Senft J, Wang Y, Konat GW, Wenger SL, Reed E, Wang W. BRCA1 contributes to cell cycle arrest and chemoresistance in response to the anticancer agent irofulven. Mol Pharmacol. 2007;71:1051–1060. doi: 10.1124/mol.106.029504. [DOI] [PubMed] [Google Scholar]
- Zaidman BZ, Yassin M, Mahajna J, Wasser SP. Medicinal mushroom modulators of molecular targets as cancer therapeutics. Appl Microbiol Biotechnol. 2005;67:453–468. doi: 10.1007/s00253-004-1787-z. [DOI] [PubMed] [Google Scholar]
- Zhong JJ, Xiao JH. Secondary metabolites from higher fungi: discovery, bioactivity, and bioproduction. Adv Biochem Eng Biotechnol. 2009;113:79–150. doi: 10.1007/10_2008_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.