Abstract
Aim
To determine the pharmacogenetics of platinum-based chemotherapy in Non Small Cell Lung Cancer (NSCLC) patients.
Methods
Publications were selected from PubMed, Cochrane Library and ISI Web of Knowledge. A meta-analysis was conducted to determine the association between genetic polymorphisms and platinum-based chemotherapy by checking odds ratio (OR) and 95% confidence interval (CI).
Results
Data were extracted from 24 publications, which included 11 polymorphisms in 8 genes for meta-analysis. MDR1 C3435T (OR = 1.97, 95% CI: 1.11–3.50, P = 0.02), G2677A/T (OR = 2.61, 95% CI: 1.44–4.74, P = 0.002) and GSTP1 A313G (OR = 0.32, 95% CI: 0.17–0.58, P = 0.0002) were significantly correlated with platinum-based chemotherapy in Asian NSCLC patients.
Conclusion
Attention should be paid to MDR1 C3435T, G2677A/T and GSTP1 A313G for personalized chemotherapy treatment for NSCLC patients in Asian population in the future.
Introduction
Lung cancer is one of the most serious public health problems around the globe. There are two major forms of lung cancer: small cell lung cancer (SCLC) and non small cell lung cancer (NSCLC), with the latter accounting for 85% of total cases [1]. Although intensive effort has been made to improve the efficacy of lung cancer therapy, the 5-year relative survival rate still remains between 11% and 17% [2].
Platinum-based chemotherapy is one of the major therapeutic methods for NSCLC, especially for advanced cancer. However, in clinical practice, the chemotherapy response varies wildly among individuals. Some patients respond to the chemotherapy, while others confer intrinsic or acquired resistance. It is speculated that some genetic polymorphisms may affect drug response, and there are accumulating evidences to support this hypothesis [3]–[5]. Therefore, understanding the association between the polymorphisms and platinum response will be beneficial for individualized chemotherapy. Although a number of studies investigated this issue, there is no congruency for the relationship between genetic polymorphisms and NSCLC platinum-based chemotherapy response. Results from different studies are inconsistent with one another. For example, Su et al. found excision repair cross-complementing 1 (ERCC1) C354T was significantly associated with drug response while other studies presented contradictory results [6]–[8]. The same situation exists for GSTP1 A313G [9]–[11]. A recent review summarized the pharmacogenomics of platinum-based chemotherapy in NSCLC [12]. However, a quantitative evaluation is still lacking.
The aim of this study is to provide a comprehensive assessment on the association between genetic polymorphisms and platinum-based drug response in NSCLC. We collected all available publications on pharmacogenetic studies of platinum-based chemotherapy in NSCLC, and quantitatively studied them using meta-analysis.
Materials and Methods
Literature search: A systematic literature search was performed independently by two authors (J.Y.Y. and Q.H.) in three electronic databases: PubMed database, Cochrane Library and ISI Web of Knowledge. The identified articles were reviewed carefully to find more relevant articles. All search results were reviewed and compared by a third reviewer (Z.Q.L.) and the discrepancies between searchers were discussed and solved with consensus. The time period for literature searching was from the first available article to April 1th 2012. Publications were retrieved using terms related to platinum drugs (“platinum” or “cisplatin” or “carboplatin” or “oxaliplatin”) in combination with keywords related to genetic variation (“polymorphism” or “SNP” or “single nucleotide polymorphism” or “mutation” or “variation”) and “lung cancer”.
Publication selection criteria: Publications meeting the following criteria were included: (a) patients with NSCLC; (b) trials had platinum-based chemotherapy; (c) the data of response rate stratified by polymorphisms could be obtained or derived from the original article or corresponding author. Studies were excluded if any of the following applies: (a) papers written in a language other than English; (b) the data of response rate stratified by polymorphisms could not be provided; (c) repeated publications, abstracts, letters, or review articles.
Data extraction: Data were manually extracted independently by two authors (J.Y.Y. and Q.H.), who were blind to each other, using the same data recording form. After exaction, all results were reviewed and compared by a third reviewer (Z.Q.L.) and the discrepancies between extractors were discussed and solved with consensus. When necessary, data of some studies were obtained directly from corresponding authors. The following information of each study was collected: first author’s name, publication year, ethnicity (country), sample size, polymorphisms, dbSNP number of investigated polymorphisms, genotyping methods, disease stage, chemotherapeutic drugs, and the number of responders and non-responders in different genotypes.
Data analysis: The patients were divided into two groups: responders (including complete responders (CR) and partial responders (PR)) and non-responders (including stable disease (SD) and progressive disease (PD)). All data were loaded into and analyzed by the Cochrane Collaboration software (Review Manager 5, the Cochrane Collaboration, Oxford, UK). The pooled odds ratio (OR) and associated 95% confidence interval (CI) were calculated. The significance of the pooled OR was determined using the Z test. The heterogeneity of publications in each meta-analysis was evaluated by I2 statistical analysis, which describes the proportion of total variation across studies due to heterogeneity rather than chance. As a guide, I2 value ranged from 0 to 100%, and values of 25%, 50% and 75% were considered as low, moderate and high heterogeneity respectively [13]. To further evaluate the extent of heterogeneity between publications, Cochran’s χ2 based Q statistic test was also employed [14]. The threshold significance level of heterogeneity was set at P = 0.10. If P<0.10, the random-effect model using DerSimonian and Laird method was selected to pool the results, yielding wider CIs [15], [16]. Otherwise, the pooled ORs and P values were calculated by fixed-effect model using the Mantel-Haenszel method [17]. To control publication bias, the relationship between OR and SE (log [OR]) was estimated using funnel plot. The symmetry of funnel plots was visually inspected. To further evaluate publication bias, Begg’s test [18] and Egger’s test [19] were also performed using Stata 12.0 software (StataCorp LP, College Station, USA). A P<0.05 was considered statistically significant in all analyses, except for heterogeneity test.
Results
1. Literature Review, Characteristics of Studies and Publication Bias
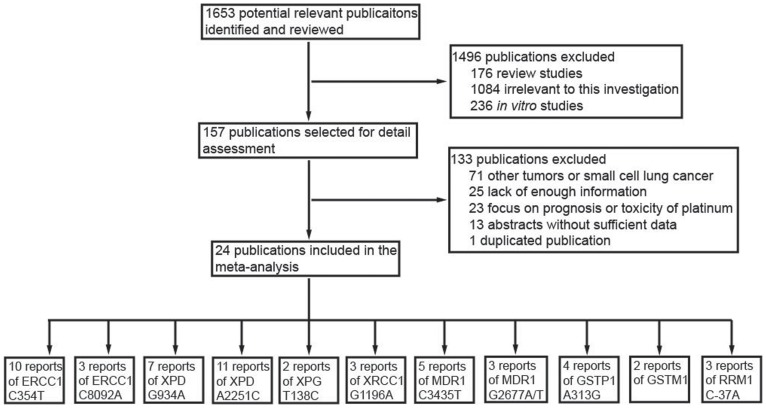
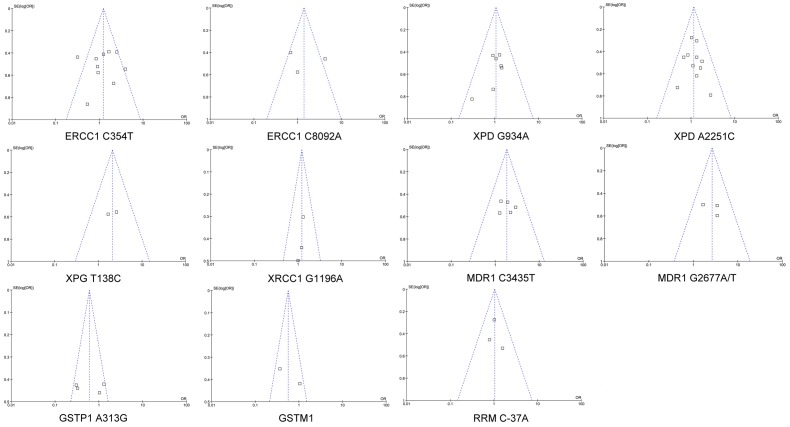
The contents of this meta-analysis were summarized in Table S1. Our initial electronic search retrieved 1653 publications, among which, 176 were review articles, 1084 were obviously not relevant to this investigation, and 236 were in vitro studies. Therefore, 157 studies were included in the next round of review. After reading the full text of these articles, we found that 71 focused on other tumors, 25 didn’t provide enough information, 19 focused on disease prognosis, 4 focused on drug side effect, 13 were conference abstracts without sufficient data, and 1 was duplicated publication. Finally, 24 publications met the inclusion criteria with enough data to be extracted (Figure 1 and S7). These publications included 11 polymorphisms in 8 genes (Table 1). Visual inspection of the funnel plots of all meta-analysis revealed a symmetrical inverted V shape (Figure 2). Begg’s test and Egger’s test also didn’t suggest evidence of publication bias. Thus, there was no publication bias for all meta-analysis in the present study. The characteristics of these studies were summarized in Table 2.
Figure 1. Flow chart of literature selection.
Table 1. Polymorphisms involved in this study and the response rate in different alleles.
| Genes | Polymorphisms | NCBI SNP ID | Allele | Responders | Non-responders | References |
| ERCC1 | C354T | rs11615 | C | 210 | 285 | [6]–[8], [22], [59]–[64] |
| (Asn118Asn) | T | 199 | 308 | |||
| C8092A | rs3212986 | C | 85 | 87 | [9], [22], [65] | |
| A | 85 | 93 | ||||
| XPD | G934A | rs1799793 | G | 114 | 174 | [7]–[9], [59], [63], [66]–[68] |
| (Asp312Asn) | A | 93 | 181 | |||
| A2251C | rs13181 | A | 259 | 528 | [7]–[9], [59], [63], [65]–[70] | |
| (Lys751Gln) | C | 172 | 372 | |||
| XPG | T138C | rs1047768 | C | 13 | 20 | [23], [24] |
| (His46His) | T | 41 | 131 | |||
| XRCC1 | G1196A | rs25487 | G | 60 | 110 | [9], [23], [68], [69] |
| (Arg399Gln) | A | 71 | 152 | |||
| MDR1 | C3435T | rs1045642 | C | 49 | 46 | [8], [32], [60], [63], [71] |
| (Ile1145Ile) | T | 88 | 141 | |||
| G2677T/A | rs2032582 | G | 31 | 22 | [32], [60], [71] | |
| (Ala893Ser/Thr) | T/A | 44 | 67 | |||
| GSTP1 | A313G | rs1695 | A | 64 | 181 | [9]–[11], [72] |
| (Ile105Val) | G | 67 | 113 | |||
| GSTM1 | Deletion | NR | presence | 31 | 63 | [9], [62] |
| deletion | 38 | 40 | ||||
| RRM1 | C-37A | NR | C | 87 | 123 | [8], [36], [63] |
| A | 65 | 92 |
NR: not reported.
Figure 2. Funnel plots of SE (log [OR]) by the OR in meta-analysis.
. SE (log [OR]) is standard error of log [OR]. Each dot represents one article.
Table 2. Characteristics of studies involved in the meta-analysis.
| Authors | Year | Ethnicity (Country) | Number of patients | Disease stage | Genotyping method | Chemotherapeutic drugs | Genes and polymorphism | Reference |
| Camps et al. | 2003 | Caucasian (Spain) | 38 | IIIB-IV | Direct sequencing | Cisplatin/Gemcitabine | XPD: G934A XPD: A2251C | [67] |
| Isla et al. | 2004 | Caucasian (Spain) | 62 | IIIB-IV | TaqMan genotyping assay | Cisplatin/Docetaxel | ERCC1: C354T MDR1: C3435T RRM1: A-37C XPD: G934A XPD: A2251C | [8] |
| Ryu et al. | 2004 | Asian (Korea) | 108 | IIIB-IV | SNaPShot assay | Platinum-based chemotherapy | ERCC1: C354T XPD: G934A XPD: A2251C | [59] |
| Booten et al. | 2006 | Caucasian (UK) | 89 | III-IV | Direct sequencing | Platinum-based chemotherapy | GSTP1:A313G | [11] |
| Booton et al. | 2006 | Caucasian (UK) | 89 | III-IV | PCR-RFLP Direct sequencing | Platinum-based chemotherapy | XPD: G934A XPD: A2251C | [66] |
| Su et al. | 2007 | Asian (China) | 76 | IIIA-IV | TaqMan genotyping assay | Platinum-based chemotherapy | ERCC1: C354T | [6] |
| Giachino et al. | 2007 | Caucasian (Italy) | 188 | IIIA-IV | PCR-RFLP | Platinum-based chemotherapy | XRCC1: G1196A XPD: A2251C | [69] |
| Tibaldi et al. | 2008 | Caucasian (Italy) | 65 | IIIB-IV | TaqMan genotyping assay | Cisplatin/Gemcitabine | ERCC1: C354T XPD: G934A XPD: A2251C | [7] |
| Pan et al. | 2008 | Asian (China) | 69 | IIIB-IV | PCR-RFLP | Cisplatin/Vinorelbine | MDR1: C3435T MDR1: G2677A/T | [71] |
| Yu et al. | 2008 | Asian (China) | 117 | NR | Direct sequencing | Carboplatin/Etoposide | ERCC1: C354T ERCC1: C8092A | [22] |
| Kalikaki et al. | 2009 | Caucasian (Greece) | 119 | IIIA-IV | PCR-RFLP Direct sequencing | Platinum-based chemotherapy | ERCC1: C354T ERCC1: C8092A GSTP1: A313G GSTM1: deletion XPD: G934A XPD: A2251C XRCC1: G1196A | [9] |
| Feng et al. | 2009 | Asian (China) | 214 | IIB-IV | PCR-RFLP | Platinum-based chemotherapy | RRM1: A-37C | [36] |
| Yao et al. | 2009 | Asian (China) | 102 | IIIB-IV | PCR-RFLP | Platinum-based chemotherapy | XRCC1: G1196A | [68] |
| Sun et al. | 2009 | Asian (China) | 90 | IV | 3-D polyacrylamide gel-based DNA microarray | Platinum-based chemotherapy | XRCC1: G1196A | [23] |
| Pan et al. | 2009 | Asian (China) | 54 | IIIB-IV | PCR-RFLP | Cisplatin/Docetaxel | MDR1: C3435T MDR1: G2677A/T | [32] |
| Zhou et al. | 2010 | Asian (China) | 130 | IIIB-IV | TaqMan genotyping assay | Platinum-based chemotherapy | ERCC1: C354T | [61] |
| Chen et al. | 2010 | Asian (China) | 95 | IIIB-IV | Ligase detection reactions (LDR) | Platinum-based chemotherapy | ERCC1: C354T MDR1: C3435T MDR1: G2677A/T | [60] |
| Sun et al. | 2010 | Asian (China) | 113 | IIIA-IV | 3-D polyacrylamide gel-based DNA microarray | Platinum-based chemotherapy | GSTP1: A313G | [10] |
| Li et al. | 2010 | Asian (China) | 115 | IIIB-IV | 3-D polyacrylamide gel-based DNA microarray | Platinum-based chemotherapy | ERCC1: C354T XPD: A2251C | [65] |
| Vinolas et al. | 2011 | Caucasian (Spain) | 94 | IIIB-IV | 5′ nuclease allelic discrimination assay | Cisplatin/Vinorelbine | ERCC1: C354T XPD: G934A XPD: A2251C MDR1: C3435T RRM1: A-37C | [63] |
| Zhou et al. | 2011 | Asian (China) | 111 | IV | Direct sequencing | Platinum-based chemotherapy | GSTP1: A313G | [72] |
| Li et al. | 2012 | Asian (China) | 58 | NR | PCR-RFLP | Platinum-based chemotherapy | GSTM1:deletion | [62] |
| Chen et al. | 2012 | Asian (China) | 355 | IIIB-IV | TaqMan genotyping assay | Platinum-based chemotherapy | XPD: A2251C | [70] |
| Cheng et al. | 2012 | Asian (China) | 142 | IIIB-IV | Direct sequencing | Platinum-based chemotherapy | ERCC1: C354T | [64] |
NR: not reported.
2. Nucleotide Excision Repair (NER) Pathway
DNA repair was one of the most important pathways involved in platinum response. Therefore, polymorphisms of genes in DNA repair pathway may affect patients’ response to platinum chemotherapy. There were four major DNA repair pathways: nucleotide excision repair (NER), base excision repair (BER), double-strand break repair (DSB) and mismatch repair (MMR) [20]. NER was the major repair system for removal of platinum-caused DNA lesions [20], [21]. Thus it was closely related with platinum response. Our meta-analysis contained 3 genes in NER pathway: ERCC1, xeroderma pigmentosum group D (XPD) and xeroderma pigmentosum group G (XPG).
2.1 ERCC1
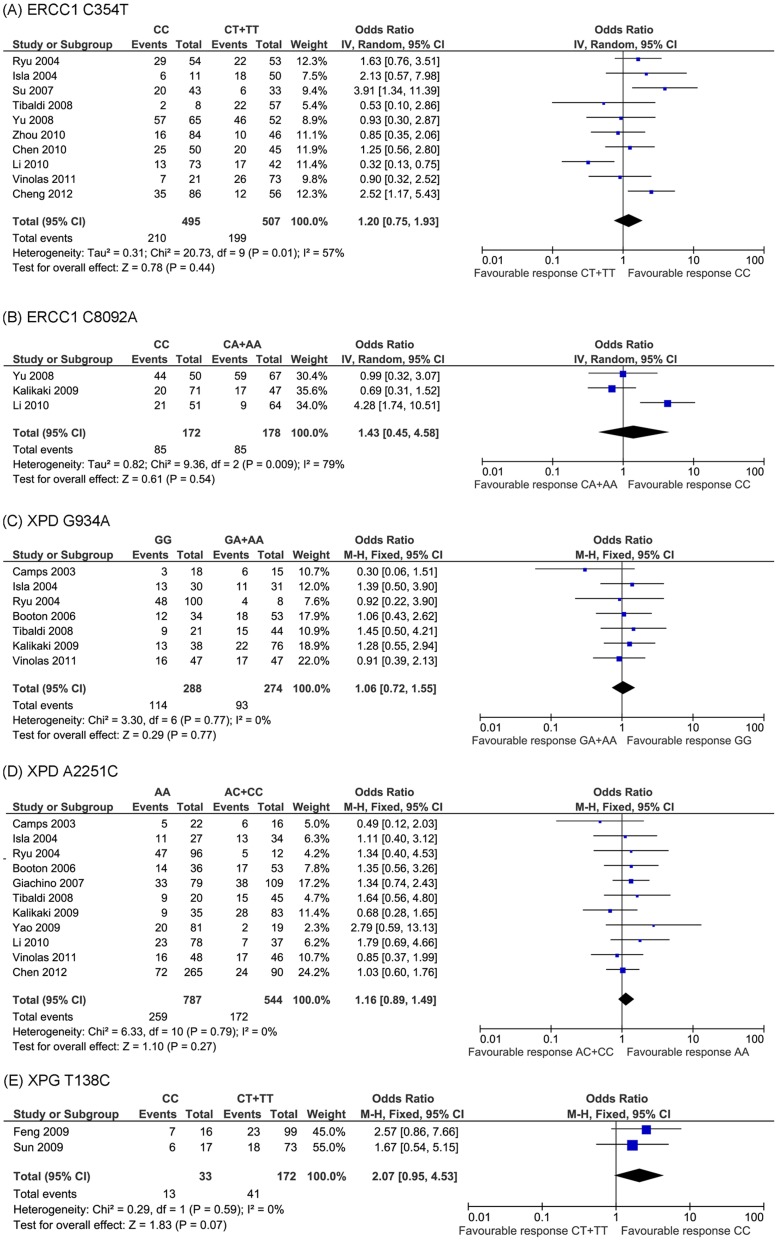
The most extensively studied polymorphism was ERCC1 C354T. Ten studies examined the association between this polymorphism and platinum-based drug response in NSCLC patients. We conducted a meta-analysis on these studies, which included 1002 subjects (495 CC genotype and 507 CT+TT genotype carriers). Because there was heterogeneity across the studies (P = 0.01, I2 = 57%), we chose random-effect model. Pooled data from these investigations showed 42.4% and 39.3% overall response rate in CC genotype and CT+TT genotype group, respectively. No significant relationship was detected between ERCC1 C354T and drug response (OR = 1.20, 95% CI: 0.75−1.93, P = 0.44) (Figure 3A). We further analyzed their relationship stratified by ethnic populations. The result showed that ERCC1 C354T was not correlated with drug response in either Asian or Caucasian population (Figure S1).
Figure 3. Meta-analysis of association polymorphisms in NER DNA repair pathway genes and platinum-based chemotherapy in NSCLC patients.
No significant association was found for these polymorphisms.
C8092A was another polymorphism of ERCC1 that was included in this study [9], [22]. Since heterogeneity was also identified among the three studies (P = 0.009, I2 = 79%), random-effect model was selected. Again, no significant association between ERCC1 C8092A and drug response was detected (OR = 1.43, 95% CI: 0.45−4.58, P = 0.54) (Figure 3B).
2.2 XPD
Another intensively investigated gene in the NER pathway was XPD. Two polymorphisms (G934A and A2251C) were widely studied. Meta-analysis of G934A and A2251C included 7 and 11 studies, respectively. No heterogeneity was found across these studies (P = 0.77, I2 = 0% and P = 0.79, I2 = 0% for G934A and A2251C, respectively), thus we chose fixed-effect model. For XPD G934A, pooled data contained 562 patients. Response rates for the GG genotype carriers and GA+AA genotype carriers were 39.6% and 33.9%, respectively. However, the OR (1.06) and 95% CI (0.72−1.55) values indicated that there was no significant correlation between this polymorphism and platinum-based chemotherapy (P = 0.77) (Figure 3C). In terms of XPD A2251C, 11 studies included 1331 individuals in total. The response rate for AA genotype was 32.9%, while that for AC+CC genotype was 31.6%. No significant association was found between this polymorphism and drug response (OR = 1.16, 95% CI: 0.89−1.49, P = 0.27) (Figure 3D). Analysis stratified by different populations was further investigated, with no significant correlation detected for polymorphism in either Asian or Caucasian population (Figures S3 and S4). Taken together, neither polymorphism of XPD was significantly related with platinum-based chemotherapy in NSCLC patients.
2.3 XPG
Another genetic polymorphism in the XP family involved in this study was XPG T138C. Sun et al. showed that it was significantly associated with platinum-based chemotherapy (P = 0.047) [23] while, another study didn’t find significant correlation between this SNP and chemotherapeutic drug response (OR = 2.57, 95% CI: 0.86−7.66, P = 0.083) [24]. T138C was a synonymous mutation. Our meta-analysis contained these two contradictory investigations, and no heterogeneity was found across these two studies (P = 0.59, I2 = 0%). As a result, no significant correlation was found between this polymorphism and drug response (OR = 2.07, 95% CI: 0.95−4.53, P = 0.07) (Figure 3E).
3. Other Pathways
Although NER was the most important pathway for platinum drug resistance, other genetic polymorphisms were also widely studied, including other DNA repair pathways, detoxification system and drug transporters.
3.1 X-Ray Cross-Complementing Group 1 (XRCC1)
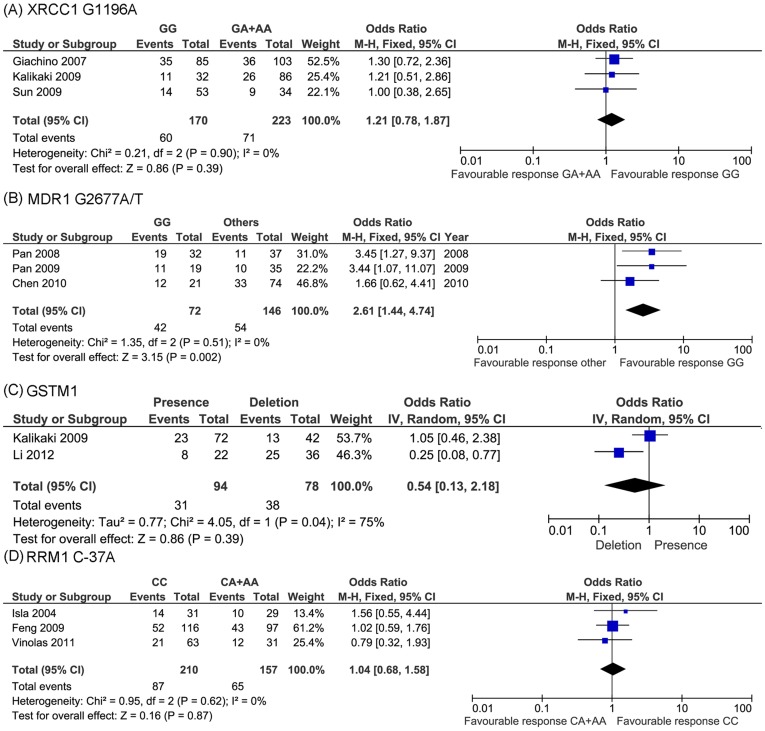
Another extensively studied DNA repair gene is XRCC1, which belongs to BER pathway. In this meta-analysis, three publications investigated G1196A of XRCC1. No heterogeneity was found across the three studies (P = 0.90, I2 = 0%). Thus fixed-effect model was selected. The pooled data showed that no significant association was found between this polymorphism and drug response (OR = 1.21, 95% CI: 0.78−1.87, P = 0.39) (Figure 4A).
Figure 4. Meta-analysis of association between other pathways gene polymorphisms and platinum-based chemotherapy in NSCLC patients.
No significant association was found for XRCC1 G1196A (A), GSTM1 (C) and RRM C-37A (D), while significant correlation was identified for MDR1 G2677A/T (P = 0.002) (B).
3.2 Multidrug resistance 1 (MDR1)
The decreased accumulation of drug in the cell has been proved to be a common mechanism of drug resistance [25]. There were two major causes of reduced intracellular drug accumulation: decreased drug influx and increased drug efflux. In terms of platinum resistance, polymorphisms of MDR1 were extensively studied.
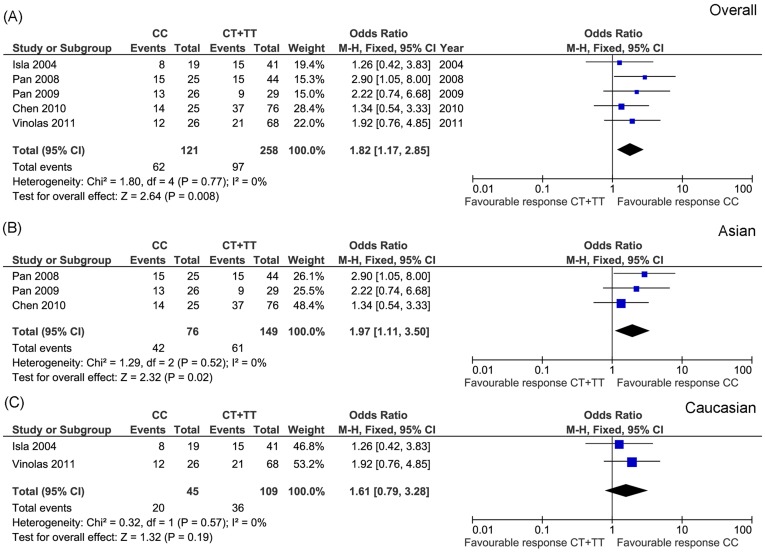
Five studies investigated MDR1 C3435T, including a total of 379 patients, the response rates in the CC and CT+TT genotype group were 51.2% and 37.6%, respectively. No heterogeneity across the studies was found (P = 0.77, I2 = 0%). We thus selected fixed-effect model. The result showed that this polymorphism was significantly correlated with drug response (OR = 1.82, 95% CI: 1.17−2.85, P = 0.008) (Figure 5A). CC genotype carrier showed significant increased drug response. Considering that ethnic differences between populations may contribute to the drug response and our included five studies comprise two different ethnic populations, we further conducted separate analyses in Asian and Caucasian population, respectively. It was interesting to note that MDR1 C3435T was only significantly correlated with platinum response in Asian population (P = 0.02) (Figure 5B). No association was detected in Caucasian population (P = 0.19) (Figure 5C).
Figure 5. Meta-analysis of association between MDR1 C3435T and platinum-based chemotherapy in overall (A), Asian (B) and Caucasian (C) NSCLC patients.
Significant association was identified in overall (P = 0.008) and Asian (P = 0.02) populations.
Another MDR1 polymorphism involved in this meta-analysis was G2677A/T. Heterogeneity test showed that fixed-effect model should be selected (P = 0.51, I2 = 0%). It was interesting to found that this SNP was also significantly associated with platinum response (OR = 2.61, 95%CI: 1.44−4.74, P = 0.002), patients with GG genotype had better response to platinum based chemotherapy (Figure 4B). Furthermore, it is noteworthy that all studied included for this SNP were conducted in the Asian population. Thus, taken together, MDR1 polymorphisms were correlated with NSCLC patients’ platinum-based chemotherapy in Asian population.
3.3 Glutathione S-transferase P1 (GSTP1)
Once platinum enters the cells, it could be conjugated by glutathione (GSH). The formed complex reduces the toxicity of platinum and can be more easily transported out of the cells. The conjugation is determined by the cellular GSH level. Therefore, some GSH synthesis enzymes are correlated with platinum response. One of the most important enzymes is glutathione S-transferase (GST), which includes several members such as GSTP1, GSTM1 and GSTT1.
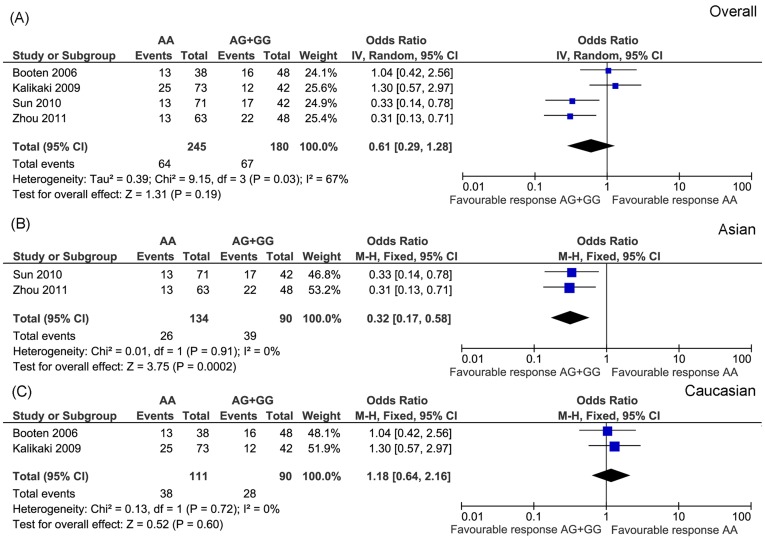
GSTP1 A313G was the most widely studied polymorphism. We included four publications and conducted a meta-analysis. These studies included 425 patients. Response rates of the AA and AG+GG genotype group were 26.1% and 37.2%, respectively. Because heterogeneity was detected across these studies (P = 0.03, I2 = 67%), we selected random-effect model. The result showed no association between GSTP1 A313G and platinum-based chemotherapy (OR = 0.61, 95% CI: 0.29−1.28, P = 0.19) (Figure 6A). To avoid the additional heterogeneity introduced by different ethnic population, we further analyzed the correlation stratified by different populations. As showed in Figure 6 B and C, GSTP1 A313G was only significantly associated with drug response in Asian population.
Figure 6. Meta-analysis of association between GSTP1 A313G and platinum-based chemotherapy in overall (A), Asian (B) and Caucasian (C) NSCLC patients.
Significant association was identified in the Asian (P = 0.0002) populations.
One deletion polymorphism of GSTM1 was also investigated in this study, heterogeneity was detected across involved two studies (P = 0.04, I2 = 75%), random-effect model was used to evaluate correlation between GSTM1 polymorphism and drug response. However, no significant association was detected (OR = 0.54, 95% CI: 0.13−2.18, P = 0.39) (Figure 4C).
3.4 Ribonucleotide reductase M1 (RRM1)
The last enrolled gene was RRM1. Three studies investigated the polymorphism of C-37A. No heterogeneity was detected across the studies (P = 0.62, I2 = 0%). Thus fixed-effect model was selected. We didn’t find significant correlation between this SNP and drug response (OR = 1.04, 95% CI: 0.68−1.58, P = 0.87) (Figure 4D).
Discussion
In this study, we investigated the phamacogenetics of platinum-based chemotherapy in NSCLC patients. We conducted meta-analysis for 11 polymorphisms in 8 genes. The results showed that MDR1 C3435T, G2677A/T and GSTP1 A313G were significantly correlated with platinum-based chemotherapy in the Asian NSCLC patients.
Platinum is an effective chemotherapeutic drug for lung cancer patients. However, the mechanisms for individual difference on drug response remain unknown. Gene polymorphisms have been proved to play an important role in the drug therapeutic efficacy [5], [25]–[27]. Therefore, researchers focused on the polymorphisms of the genes in the platinum pathways [12]. In the present study, it is interesting that two polymorphisms of MDR1 were significantly correlated with platinum response. MDR1 encodes P-glycoprotein (P-gp), which is responsible for transporting a broad spectrum of drugs out of cells [28]. It was widely reported to be implicated in platinum resistance due to its activity of drug efflux [29]. Thus it is not surprising that MDR1 polymorphisms were associated with platinum response. However, the mechanisms of their correlation remain unclear. One proposal is that these polymorphisms affect P-gp protein expression or function which, in turn, alter drug efflux activity. A number of publications reported the association between MDR1 C3435T and G2677A/T and P-gp expression or function. The mutated MDR1 had altered P-gp expression level or aberrant protein function [30], [31]. It was previously reported that C3435T and G2677A/T were in linkage disequilibrium (LD) with each other [32]. Thus their haplotype may also have significant contribution to platinum response. However, existing literatures do not provide enough data for a meta-analysis for this hypothesis. Therefore, their correlation awaits further investigations. Another polymorphism found to be significantly correlated with drug response was GSTP1 A313G, which is a non-synonymous SNP located in exon 5 [33]. GSTs are phase II metabolic enzymes involved in the platinum detoxification mediated by glutathione (GSH) conjugation [34]. GSTP1 is one of the major isoforms of GST and its activity level is associated with platinum detoxification and subsequent drug response. Previous study showed that GSTP1 A313G resulted in reduced glutathione conjugating ability [33]. Thus, patients harboring this polymorphism may have decreased platinum detoxification capacity, which is consistent with our result that mutated genotype carriers were more sensitive to platinum-based chemotherapy.
Except for drug transporters and detoxification, DNA repair was also one of the classical platinum resistance mechanisms [34], We proposed that genetic polymorphisms in DNA repair pathways were correlated with drug response prior to this study. However, we failed to find the significant correlation between genetic polymorphisms in any DNA repair pathway and platinum chemotherapy response in this study. To further verify this result, we examined more DNA repair pathway polymorphisms, which were not included in this study because they didn’t meet our including criteria. Among these, ERCC1 T262G [22], hMSH2 IVS12-6T>C [35], RRM1 C-524T [36], XPA A23G [24] and XRCC1 T194C [23] were claimed to be significantly associated with drug response by other authors. However, after careful examination on these studies, we found several problems in these studies. For example, adjusted OR values were not calculated for RRM1 C-524T. Besides, the results of hMSH2 IVS12-6T>C and XRCC1 T194C were inconsistent between different genotypes. We did not find similar problems in the analyses of ERCC1 T262G and XPA A23G. However, due to the small sample size, we think these results need to be further validated. Taken both our and these studies together, the association between genetic polymorphisms in DNA repair pathway and platinum-based chemotherapy in NSCLC is not supported by experimental evidence.
Considering the importance of these genes in platinum resistance, especially for NER DNA repair pathway genes, it is surprising that no correlation was detected. We think one possibility is that cancer genetics is different from somatic genetics. Although a number of studies showed that polymorphisms in these genes affect their expression or function [37], [38], they may have no effect on cancer patients’ chemotherapeutic efficiency. We understand that many other cancer genetic factors also affect the therapeutic efficiency of platinum. Thus, the polymorphisms in DNA repair pathway genes may not play a central role in platinum drug response in NSCLC. This is supported by a large sample size study conducted by Shiraishi et al [39], who genotyped 30 SNPs in 27 DNA repair genes in 640 NSCLC patients. Some of the polymorphisms were also investigated in the present study such as XPD A2251C and XRCC1 G1196A. Their results were in agreement with ours in that, most of polymorphisms in the DNA repair pathway genes were not significantly involved in platinum drug response. Our results are also supported by several recent large genome wide association studies (GWAS) in NSCLC [40]–[42]. Wu et al. conducted a GWAS (including 307,260 SNPs) in a total of 1062 NSCLC patients, they identified that the rs1878022 in the chemokine-like receptor 1 (CMKLR1) was statistically associated with poor overall survival [41]. No polymorphisms in the DNA repair pathway genes were identified. Another GWAS study performed by Yang group also didn’t identify the correlation between DNA repair gene polymorphisms and patients overall survival in NSCLC [42], either. They explored 894 SNPs in 70 genes in four pathways related to chemotherapy drug actions, including GSH, DNA repair, cell cycle and EGFR pathways. However, this 1076 patients study concluded that only genetic variations in EGFR and glutathione pathways were associated with overall survival in NSCLC patients receiving platinum based chemotherapy. Although the endpoints in these studies were overall survival, considering the close association between drug response and survival, they may also provide some clues for the correlation between these SNPs and drug response. Taken together, compared with DNA repair pathway genetic polymorphisms, other gene polymorphisms may have more contribution to chemotherapy response in NSCLC patients receiving platinum-based treatment.
Lung cancer is a heterogeneous disease. Thus pathological and ethnic difference may affect drug response. To clarify this concern, we analyzed the correlation between genetic polymorphisms and drug response stratified by different ethnic populations (Figure 5, 6 and S1, S2, S3, S4, S5, and S6). Although most of the separately analyzed results were consistent with overall population, two polymorphisms (MDR1 C3435T and GSTP1 A313G) showed ethnic difference. Both polymorphisms were only significantly associated with drug response in Asian population, not in Caucasian population (Figure 5 and 6). This result indicates that the outcomes of platinum-based chemotherapy differ between Asian and Caucasian population. Thus, ethnic factor should be considered if personalized chemotherapy treatment is conducted in the future.
Pathology is another factor to be considered when pooled data was analyzed, because NSCLC includes adenocarcinoma and squamous cell carcinoma. Although both types of tumor are treated with the same regimen, the outcomes may be completely different. However, studies included in this meta-analysis didn’t provide the data of response rate stratified by both polymorphisms and pathological types. Thus it is impossible to do the meta-analysis separately on adenocarcinoma and squamous cell carcinoma based on existing studies. This is one of the limitations of our study. To address this issue, future studies are needed when enough data are available.
We also identified some flaws in existing studies. One common flaw is that the authors didn’t perform comprehensive analysis. As our result indicated, a single polymorphism has little contribution to chemotherapy efficacy. Thus comprehensive analysis was needed. One restriction for the past comprehensive study is shortage of high-throughput genotyping methods. With the advancement of technology, more and more methods have been developed for performing high-throughput genotyping, which can be used in the future study. For example, in our included studies, one group used 3D polyacrylamide gel-based DNA microarray to determine genotypes [10], [23], [24], [35]. This is a powerful high-throughput genotyping assay, which is frequently utilized by this group. As gene sequencing technology develops rapidly, another available method is genome-wide approach. One group identified some variants that were related with platinum response using this method [43]–[46]. However, all these studies are in vitro investigations and the results need to be verified in the patients. On the other hand, GWAS have been successfully used in disease susceptibility and pharmacogenetic research [47]–[50]. Thus, we believe GWAS is a powerful tool to investigate the pharmacogenetics of platinum-based chemotherapy in the future. Another problem is inadequate sample size. The sample sizes of involved studies were from 33 to 355, and none of these studies did a sample size calculation. In some situation, especially for the low frequency polymorphisms, small sample size cannot generate solid conclusion. Furthermore, as described previously, some studies didn’t calculate adjusted OR [36], [51]. They just used χ2 test to assess the distribution of different genotypes in responders and non-responders. Thus, these studies can’t exclude the effect of age, sex, smoking habit, tumor histology, and tumor stages. To make the result more reliable, future studies should avoid these pitfalls.
The cellular process of platinum is complicated, which includes a lot of pathways. However, existing studies didn’t cover all platinum pathways. Therefore, some most important polymorphisms affecting platinum chemotherapy may still be elusive. For instance, compared with efflux, drug influx is also crucial for platinum accumulation in the tumor cells. Copper transporter-1 (CTR1) and ATPases ATP7 have a substantial role in platinum influx [52]–[54]. However, no polymorphisms of these genes were investigated. Other important pathways that didn’t attract attention include DNA damage signal transduction pathways. These pathways are, but not limited to, AKT, c-ABL, p53, MAPK/JNK/ERK and p38 MAPK [55]. We didn’t find publications investigating polymorphisms of genes in these pathways. Therefore, we propose that another possible direction in the future is to investigate genes in these pathways.
Prior to this study, another two groups performed meta-analysis to detect the correlation of ERCC1 and MDR1 polymorphisms and platinum-based chemotherapies in advanced NSCLC [56], [57]. Our results are in agreement with these two studies. However, a study conducted by Wei et al. had different results. They investigated the association between ERCC1 and XPD polymorphisms and platinum-based chemotherapy in advanced NSCLC patients [58]. They studied ERCC1 C354T, XPD G934A and A2251C, and found that ERCC1 C354T was significantly correlated with drug response in patients treated with platinum-based chemotherapy. Apparently, their result is inconsistent with our study. We think that the discrepancy comes from the difference in enrolled studies. Our meta-analysis included six more studies, five of which are published after Wei’s meta-analysis. In the other one, data were obtained directly from the author. Besides, our investigation didn’t include two studies, which were in Wei’s investigation. One study was not published in English, and the other had errors in genotype classification. As a result, our meta-analysis included 1002 patients, while Wei’s study included 556 patients. Therefore, the difference in patients pool possibly caused inconsistency in results.
In conclusion, we identified that MDR1 C3435T, G2677A/T and GSTP1 A313G were significantly correlated with platinum-based chemotherapy in Asian NSCLC patients. These polymorphisms should be considered for personalized chemotherapy treatment for NSCLC patients in Asian population in the future.
Supporting Information
Meta-analysis of association between ERCC1 C354T and platinum-based chemotherapy in NSCLC patients stratified by ethnic population. No significant association was found in either Asian (A) or Caucasian (B) population.
(TIF)
Meta-analysis of association between ERCC1 C8092A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was identified.
(TIF)
Meta-analysis of association between XPD G934A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was detected.
(TIF)
Meta-analysis of association between XPD A2251C and platinum-based chemotherapy in NSCLC patients stratified by ethnic population. No significant association was found in either Asian (A) or Caucasian (B) populations.
(TIF)
Meta-analysis of association between XRCC1 G1196A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was found.
(TIF)
Meta-analysis of association between RRM1 C-37A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was detected.
(TIF)
PRISMA flow diagram
(DOC)
PRISMA checklist
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Natural Science Foundation of China (81173129) (Z.Q.L.), National High-Tech R&D Program of China (863Program) (2009AA022703) (Z.Q.L. and J.Y.Y.), Specialized Research Fund for the Doctoral Program of Higher Education (20113420120006) (Q.H.) and Grants for Scientific Research of BSKY (XJ201021) from Anhui Medical University (Q.H.). J.Y.Y. was supported in part by Scholarship Award for Excellent Doctoral Student granted by the Ministry of Education of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Kweekel DM, Gelderblom H, Guchelaar HJ. Pharmacology of oxaliplatin and the use of pharmacogenomics to individualize therapy. Cancer Treat Rev. 2005;31:90–105. doi: 10.1016/j.ctrv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Yu M, Xu XJ, Yin JY, Wu J, Chen X, et al. KCNJ11 Lys23Glu and TCF7L2 rs290487(C/T) polymorphisms affect therapeutic efficacy of repaglinide in Chinese patients with type 2 diabetes. Clin Pharmacol Ther. 2010;87:330–335. doi: 10.1038/clpt.2009.242. [DOI] [PubMed] [Google Scholar]
- 5.Huang Q, Yin JY, Dai XP, Wu J, Chen X, et al. Association analysis of SLC30A8 rs13266634 and rs16889462 polymorphisms with type 2 diabetes mellitus and repaglinide response in Chinese patients. Eur J Clin Pharmacol. 2010;66:1207–1215. doi: 10.1007/s00228-010-0882-6. [DOI] [PubMed] [Google Scholar]
- 6.Su D, Ma S, Liu P, Jiang Z, Lv W, et al. Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer. 2007;56:281–288. doi: 10.1016/j.lungcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Tibaldi C, Giovannetti E, Vasile E, Mey V, Laan AC, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14:1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 8.Isla D, Sarries C, Rosell R, Alonso G, Domine M, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 9.Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 10.Sun N, Sun X, Chen B, Cheng H, Feng J, et al. MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2010;65:437–446. doi: 10.1007/s00280-009-1046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booton R, Ward T, Heighway J, Ashcroft L, Morris J, et al. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. 2006;1:679–683. [PubMed] [Google Scholar]
- 12.Hildebrandt MA, Gu J, Wu X. Pharmacogenomics of platinum-based chemotherapy in NSCLC. Expert Opin Drug Metab Toxicol. 2009;5:745–755. doi: 10.1517/17425250902973711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 15.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 18.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Campelo R, Alonso-Curbera G, Anton Aparicio LM, Rosell R. Pharmacogenomics in lung cancer: an analysis of DNA repair gene expression in patients treated with platinum-based chemotherapy. Expert Opin Pharmacother. 2005;6:2015–2026. doi: 10.1517/14656566.6.12.2015. [DOI] [PubMed] [Google Scholar]
- 22.Yu D, Zhang X, Liu J, Yuan P, Tan W, et al. Characterization of functional excision repair cross-complementation group 1 variants and their association with lung cancer risk and prognosis. Clin Cancer Res. 2008;14:2878–2886. doi: 10.1158/1078-0432.CCR-07-1612. [DOI] [PubMed] [Google Scholar]
- 23.Sun X, Li F, Sun N, Shukui Q, Baoan C, et al. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65:230–236. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Feng J, Sun X, Sun N, Qin S, Li F, et al. XPA A23G polymorphism is associated with the elevated response to platinum-based chemotherapy in advanced non-small cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 2009;41:429–435. doi: 10.1093/abbs/gmp027. [DOI] [PubMed] [Google Scholar]
- 25.Yin JY, Huang Q, Yang Y, Zhang JT, Zhong MZ, et al. Characterization and analyses of multidrug resistance-associated protein 1 (MRP1/ABCC1) polymorphisms in Chinese population. Pharmacogenet Genomics. 2009;19:206–216. doi: 10.1097/FPC.0b013e328323f680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J, Zhang J. Multidrug resistance-associated protein 1 (MRP1/ABCC1) polymorphism: from discovery to clinical application. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011;36:927–938. doi: 10.3969/j.issn.1672-7347.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin JY, Han LF, Huang Q, Xu XJ, Zhou HH, et al. ABCC1 polymorphism Arg723Gln (2168G> A) is associated with lung cancer susceptibility in a Chinese population. Clin Exp Pharmacol Physiol. 2011;38:632–637. doi: 10.1111/j.1440-1681.2011.05571.x. [DOI] [PubMed] [Google Scholar]
- 28.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 29.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 30.Woodahl EL, Ho RJ. The role of MDR1 genetic polymorphisms in interindividual variability in P-glycoprotein expression and function. Curr Drug Metab. 2004;5:11–19. doi: 10.2174/1389200043489108. [DOI] [PubMed] [Google Scholar]
- 31.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 32.Pan JH, Han JX, Wu JM, Huang HN, Yu QZ, et al. MDR1 single nucleotide polymorphism G2677T/A and haplotype are correlated with response to docetaxel-cisplatin chemotherapy in patients with non-small-cell lung cancer. Respiration. 2009;78:49–55. doi: 10.1159/000158454. [DOI] [PubMed] [Google Scholar]
- 33.Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 34.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Cheng H, Sun N, Sun X, Chen B, Li F, et al. Polymorphisms in hMSH2 and hMLH1 and response to platinum-based chemotherapy in advanced non-small-cell lung cancer patients. Acta Biochim Biophys Sin (Shanghai) 2010;42:311–317. doi: 10.1093/abbs/gmq023. [DOI] [PubMed] [Google Scholar]
- 36.Feng JF, Wu JZ, Hu SN, Gao CM, Shi MQ, et al. Polymorphisms of the ribonucleotide reductase M1 gene and sensitivity to platin-based chemotherapy in non-small cell lung cancer. Lung Cancer. 2009;66:344–349. doi: 10.1016/j.lungcan.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Yu JJ, Lee KB, Mu C, Li Q, Abernathy TV, et al. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555–560. doi: 10.3892/ijo.16.3.555. [DOI] [PubMed] [Google Scholar]
- 38.Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, et al. A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8658. [PubMed] [Google Scholar]
- 39.Shiraishi K, Kohno T, Tanai C, Goto Y, Kuchiba A, et al. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol. 2010;28:4945–4952. doi: 10.1200/JCO.2010.30.5334. [DOI] [PubMed] [Google Scholar]
- 40.Tan XL, Moyer AM, Fridley BL, Schaid DJ, Niu N, et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17:5801–5811. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Ye Y, Rosell R, Amos CI, Stewart DJ, et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J Natl Cancer Inst. 2011;103:817–825. doi: 10.1093/jnci/djr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Sun Z, Cunningham JM, Aubry MC, Wampfler JA, et al. Genetic variations in multiple drug action pathways and survival in advanced stage non-small cell lung cancer treated with chemotherapy. Clin Cancer Res. 2011;17:3830–3840. doi: 10.1158/1078-0432.CCR-10-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 44.Shukla SJ, Duan S, Badner JA, Wu X, Dolan ME. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet Genomics. 2008;18:253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol Cancer Ther. 2008;7:3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 49.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Link E, Parish S, Armitage J, Bowman L, Heath S, et al. SLCO1B1 variants and statin-induced myopathy–a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 51.Gautschi O, Hugli B, Ziegler A, Bigosch C, Bowers NL, et al. Cyclin D1 (CCND1) A870G gene polymorphism modulates smoking-induced lung cancer risk and response to platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients. Lung Cancer. 2006;51:303–311. doi: 10.1016/j.lungcan.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 52.Ishida S, Lee J, Thiele DJ, Herskowitz I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci U S A. 2002;99:14298–14302. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katano K, Kondo A, Safaei R, Holzer A, Samimi G, et al. Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res. 2002;62:6559–6565. [PubMed] [Google Scholar]
- 54.Safaei R, Holzer AK, Katano K, Samimi G, Howell SB. The role of copper transporters in the development of resistance to Pt drugs. J Inorg Biochem. 2004;98:1607–1613. doi: 10.1016/j.jinorgbio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 56.Yu D, Shi J, Sun T, Du X, Liu L, et al. Pharmacogenetic role of ERCC1 genetic variants in treatment response of platinum-based chemotherapy among advanced non-small cell lung cancer patients. Tumour Biol. 2012. [DOI] [PubMed]
- 57.Wei HB, Lu XS, Shang LH, Xu G, Hu J, et al. Polymorphisms of ERCC1 C118T/C8092A and MDR1 C3435T predict outcome of platinum-based chemotherapies in advanced non-small cell lung cancer: a meta-analysis. Arch Med Res. 2011;42:412–420. doi: 10.1016/j.arcmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 58.Wei SZ, Zhan P, Shi MQ, Shi Y, Qian Q, et al. Predictive value of ERCC1 and XPD polymorphism in patients with advanced non-small cell lung cancer receiving platinum-based chemotherapy: a systematic review and meta-analysis. Med Oncol. 2010. [DOI] [PubMed]
- 59.Ryu JS, Hong YC, Han HS, Lee JE, Kim S, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44:311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Chen S, Huo X, Lin Y, Ban H, Li W, et al. Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients. Int J Hyg Environ Health. 2010;213:140–145. doi: 10.1016/j.ijheh.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Zhou C, Ren S, Zhou S, Zhang L, Su C, et al. Predictive effects of ERCC1 and XRCC3 SNP on efficacy of platinum-based chemotherapy in advanced NSCLC patients. Jpn J Clin Oncol. 2010;40:954–960. doi: 10.1093/jjco/hyq071. [DOI] [PubMed] [Google Scholar]
- 62.Li W, Yue W, Zhang L, Zhao X, Ma L, et al. Polymorphisms in GSTM1, CYP1A1, CYP2E1, and CYP2D6 are associated with susceptibility and chemotherapy response in non-small-cell lung cancer patients. Lung. 2012;190:91–98. doi: 10.1007/s00408-011-9338-8. [DOI] [PubMed] [Google Scholar]
- 63.Vinolas N, Provencio M, Reguart N, Cardenal F, Alberola V, et al. Single nucleotide polymorphisms in MDR1 gen correlates with outcome in advanced non-small-cell lung cancer patients treated with cisplatin plus vinorelbine. Lung Cancer. 2011;71:191–198. doi: 10.1016/j.lungcan.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 64.Cheng J, Ha M, Wang Y, Sun J, Chen J, et al. A C118T polymorphism of ERCC1 and response to cisplatin chemotherapy in patients with late-stage non-small cell lung cancer. J Cancer Res Clin Oncol. 2012;138:231–238. doi: 10.1007/s00432-011-1090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li F, Sun X, Sun N, Qin S, Cheng H, et al. Association between polymorphisms of ERCC1 and XPD and clinical response to platinum-based chemotherapy in advanced non-small cell lung cancer. Am J Clin Oncol. 2010;33:489–494. doi: 10.1097/COC.0b013e3181b9cedc. [DOI] [PubMed] [Google Scholar]
- 66.Booton R, Ward T, Heighway J, Taylor P, Power F, et al. Xeroderma pigmentosum group D haplotype predicts for response, survival, and toxicity after platinum-based chemotherapy in advanced nonsmall cell lung cancer. Cancer. 2006;106:2421–2427. doi: 10.1002/cncr.21885. [DOI] [PubMed] [Google Scholar]
- 67.Camps C, Sarries C, Roig B, Sanchez JJ, Queralt C, et al. Assessment of nucleotide excision repair XPD polymorphisms in the peripheral blood of gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Lung Cancer. 2003;4:237–241. doi: 10.3816/clc.2003.n.004. [DOI] [PubMed] [Google Scholar]
- 68.Yao CY, Huang XE, Li C, Shen HB, Shi MQ, et al. Lack of influence of XRCC1 and XPD gene polymorphisms on outcome of platinum-based chemotherapy for advanced non small cell lung cancers. Asian Pac J Cancer Prev. 2009;10:859–864. [PubMed] [Google Scholar]
- 69.Giachino DF, Ghio P, Regazzoni S, Mandrile G, Novello S, et al. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13:2876–2881. doi: 10.1158/1078-0432.CCR-06-2543. [DOI] [PubMed] [Google Scholar]
- 70.Chen X, Sun H, Ren S, Kim Curran V, Zhang L, et al. Association of XRCC3 and XPD751 SNP with efficacy of platinum-based chemotherapy in advanced NSCLC patients. Clin Transl Oncol. 2012;14:207–213. doi: 10.1007/s12094-012-0785-3. [DOI] [PubMed] [Google Scholar]
- 71.Pan JH, Han JX, Wu JM, Sheng LJ, Huang HN, et al. MDR1 single nucleotide polymorphisms predict response to vinorelbine-based chemotherapy in patients with non-small cell lung cancer. Respiration. 2008;75:380–385. doi: 10.1159/000108407. [DOI] [PubMed] [Google Scholar]
- 72.Zhou F, Yu Z, Jiang T, Lv H, Yao R, et al. Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients. Swiss Med Wkly. 2011;141:w13275. doi: 10.4414/smw.2011.13275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Meta-analysis of association between ERCC1 C354T and platinum-based chemotherapy in NSCLC patients stratified by ethnic population. No significant association was found in either Asian (A) or Caucasian (B) population.
(TIF)
Meta-analysis of association between ERCC1 C8092A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was identified.
(TIF)
Meta-analysis of association between XPD G934A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was detected.
(TIF)
Meta-analysis of association between XPD A2251C and platinum-based chemotherapy in NSCLC patients stratified by ethnic population. No significant association was found in either Asian (A) or Caucasian (B) populations.
(TIF)
Meta-analysis of association between XRCC1 G1196A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was found.
(TIF)
Meta-analysis of association between RRM1 C-37A and platinum-based chemotherapy in Caucasian NSCLC patients. No significant association was detected.
(TIF)
PRISMA flow diagram
(DOC)
PRISMA checklist
(DOC)