Abstract
The radial spoke (RS) is a complex of at least 23 proteins that works as a mechanochemical transducer between the central‐pair apparatus and the peripheral microtubule doublets in eukaryotic flagella and motile cilia. The RS contributes to the regulation of the activity of dynein motors, and thus to flagellar motility. Despite numerous biochemical, physiological and structural studies, the mechanism of the function of the radial spoke remains unclear. Detailed knowledge of the 3D structure of the RS protein complex is needed in order to understand how RS regulates dynein activity. Here we review the most important findings on the structure of the RS, including results of our recent cryo‐electron tomographic analysis of the RS protein complex.
Keywords: axoneme, cilia, cryo‐electron tomography, dynein, flagella, motility, radial spokes
Introduction
Eukaryotic flagella and motile cilia share a common “9 + 2” structure, in which nine peripheral microtubule doublets (MTDs) surround the central‐pair of microtubules (CP) (Fig. 1A). The MTDs and CP are connected by radial spokes (RSs). Genetic, biochemical, and structural analysis indicate that the mechano‐chemical interaction between RSs and CP regulates the activity of dynein motors attached to the MTDs and thus controls the bending motion of the flagellum.
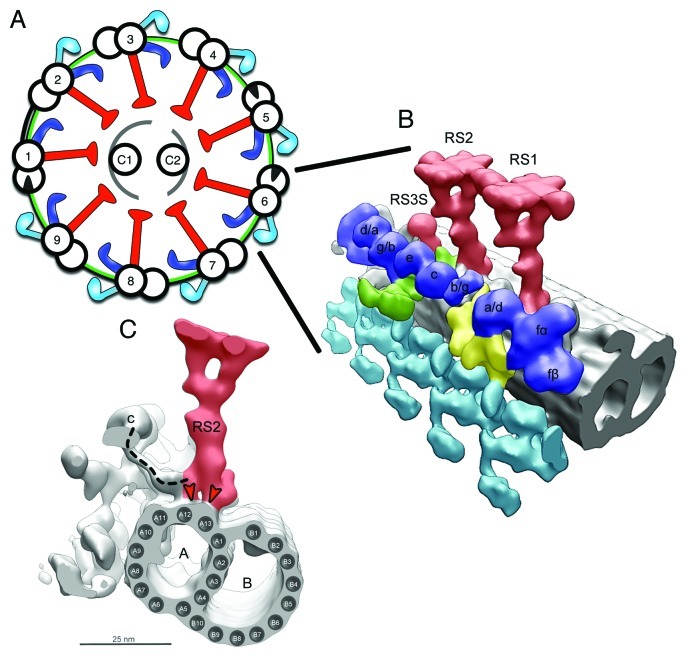
Figure 1. Placement of the RSs in the Axoneme. (A) Scheme of the 9 + 2 axonemal structure, showing the placement of main axonemal components. Radial spokes (red), inner dynein arms (blue), outer dynein arms (turquoise), microtubules (black), N‐DRC (green), central pair complex (gray). (B) Surface renderings of tomographic reconstruction of a 96 nm repeat along one of the MTDs of Chlamydomonas. The microtubules are shown in gray, the rest of the color‐coding is according to (A). RS1, RS2, and RS3 stump (RS3S) are shown. Isoforms of inner arm dyneins are indicated. Dynein b/g is either dynein b or dynein g, but it has not been determined which of the two this dynein is. It is the same case for dynein g/b, a/d, d/a. (C) Side view seen from the proximal end showing RS2, IDA c, ODA, and the microtubule doublet. A, A‐microtubule; B, B‐microtubule. The dashed line indicates the dynein c tail connecting to the RS2 base. The red arrowheads show the binding of the bifurcated base of RS to the protofilaments A12 and A13 of the A‐microtubule. [(B) was modified from ©Pigino et al., 2011. Originally published in JCB. DOI: 10.1083/jcb.201106125].
Morphology
Basic shape and anchoring of the radial spoke
Björn Afzelius, in 1959, was the first to describe the presence of RSs in the axonemes of sea urchin sperm flagella.1 He drew the RSs as slender threads radiating between the CP and the 9 MTDs and revealed that the RS binds only to the A‐microtubule of the MTD.1 In a recent cryo‐electron tomography study of the detailed 3D structure of the RSs in Chlamydomonas reinhardtii flagella and Tetrahymena thermophila cilia we have shown that the RS binds onto the protofilaments A12 and A13 of the A‐microtubule2 (Fig. 1C).
An RS is a T‐shaped structure, composed of (1) an elongated stalk that is anchored on the A‐microtubule of a peripheral doublet, and (2) an orthogonal head, which is thought to have transient contacts with the inner sheath and the CP.3-5 The RS head was first recognized and described by Warner in the blowfly sperm flagella Sarcophaga bullata and in Chlamydomonas flagella.6 The same T‐shaped structure was later observed in the flagella and cilia of many different species, ranging from protozoa to mammals.7-9
Placement of the RSs in the Axoneme
Groups of RSs repeat every 96 nm along the A‐microtubule of each MTD. In most organisms these groups contain a triplet of spokes, RS1, RS2, and RS3 (Tetrahymena,2,5,7 Paramecium tetraurelia,8 Trypanosoma brucey9 and sperm cells of sea urchin species10,11). However, in some other organisms these groups contain only pairs of spokes, RS1 and RS2 (Chlamydomonas3,5,12 and S. bullata6). Proceeding from the proximal to the distal axonemal end, RS1 is the first spoke in each triplet and RS2 is about 32 nm distal to RS1. RS2 and RS3 are 24 nm apart, and the distance between RS2 and RS1 of the next triplet is the remaining 64 nm.2,5 We have shown that the RS3 structure in Tetrahymena is distinct from RS1 and RS2.2
Strikingly, we have also revealed that the 96 nm repeat in Chlamydomonas contains a fraction of RS3 within the doublet repeat (Figs. 2A and 3A).2 However, the functional difference between triplet and doublet repeats remains to be elucidated.
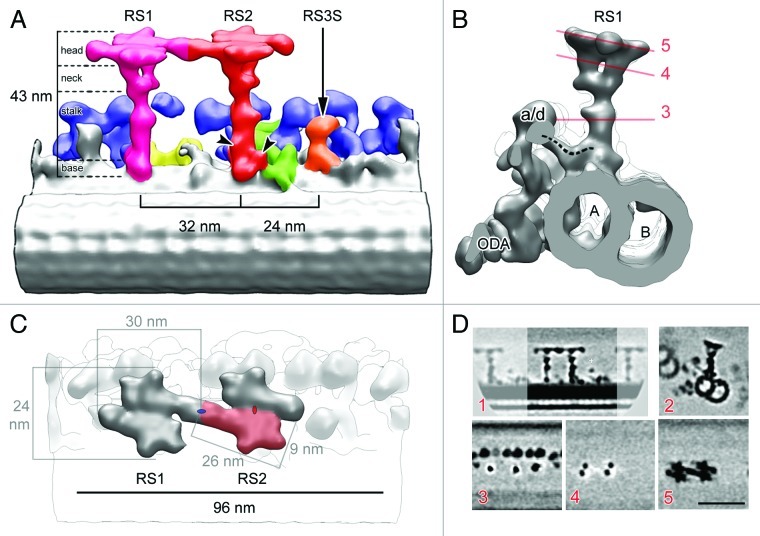
Figure 2. 3D structure of WT RSs in C. reinhardtii. (A–C) Surface renderings of tomographic reconstruction after 3D subtomogram averaging. (A) Longitudinal view showing the B‐ microtubule (foreground), radial spoke 1 (RS1)(purple), radial spoke 2 (RS2)(red), the RS3 stump (RS3S)(orange) (arrow), the IDAs (blue), the intermediate and light chains of IDAs (yellow), the DRC (green). The proximal end of the axoneme points toward the left. Arrowheads indicate densities specific to RS2. The boundaries between the head, neck, stalk, and base domains are shown. (B) Side view seen from the proximal end showing RS1, IDA a or d (a/d), ODA, and the microtubule doublet. A, A‐microtubule; B, B‐microtubule. The dashed line indicates the dynein a/d tail connecting to the RS1 base. The red lines show the position of section planes through the original density map used to generate subfigures (shown in D, 3–5). (C) Top view showing the two RS heads. The proximal end points to the left as in A. The pale red area identifies one of the symmetrical subdomains composing the RS head. Two such subdomains build one RS head. The red ellipse indicates the 2-fold rotational symmetry between these subdomains. The two RS heads are also symmetrical, also following a 2-fold rotational symmetry, denoted by a blue ellipse. (D) Sections through the density map of the model shown in A–C. (1) Same orientation as in A; (2) Same orientation as in B; (3–5) Same orientation as in C. The proximal end is pointing toward the left in all sections, except for section 2, where the proximal end is oriented toward the reader. Bar, 50 nm. (Modified from ©Pigino et al., 2011. Originally published in JCB. doi: 10.1083/jcb.201106125)
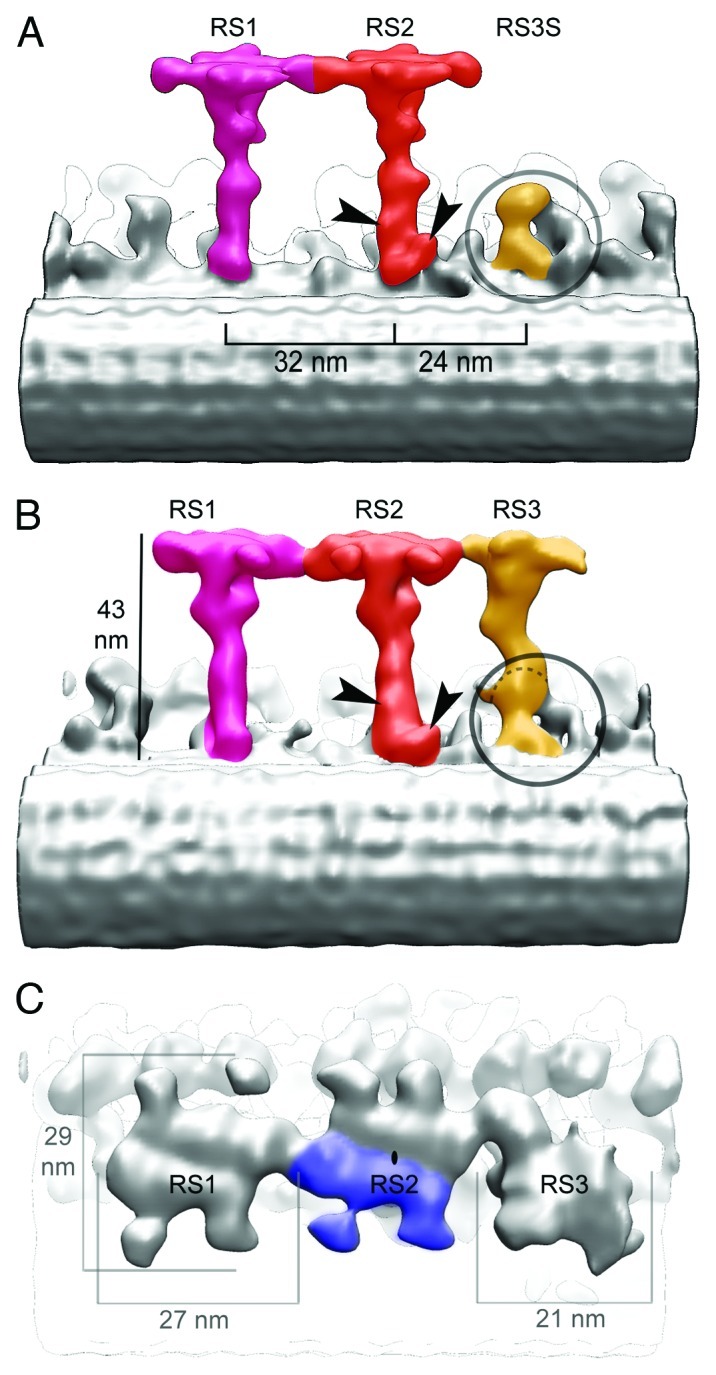
Figure 3. RS3S in C. reinhardtii and 3D structure of RSs in T. thermophila. (A) Longitudinal (proximal end of the axoneme to the left) view of the WT C. reinhardtii RSs. The RS3 stump (RS3S)(orange) is encircled. (B‐C) Surface rendering of the RS triplet in T. thermophila. (B) Longitudinal view with arrowheads pointing to RS2‐specific densities. The base of RS3 is encircled, and a dashed line marks the approximate location corresponding to the upper side of RS3S seen in C. reinhardtii. (C) Top view showing the three RS heads. The heads of RS1 and RS2 share the same structure (2-fold rotational symmetry). Blue area shows one of the two subdomains that assemble in a single head and also has 2-fold rotational symmetry. The structure of the RS3 head differs from that of RS1 and RS2.
Characterization of the Radial Spoke Proteins (RSPs)
Initially, SDS‐PAGE analysis of the WT axonemes and axonemes of paralyzed mutants of Chlamydomonas revealed 17 polypeptide chains that were ascribed to the RS complex.13,14 Following that, a procedure for purification of the RS complex from axonemes15,16 enabled identification of a total number of 23 proteins.16,17 These proteins are called RSP1 to RSP23. The RSPs 2–4, 6, 16, 20, 22 and 23 have been identified and sequenced.18-24 In addition to the 23 RPSs, another complex, known as calmodulin and spoke‐associated complex (CSC), binds to the MTD and is essential for anchoring the RSs (Table 1).25,26
Table 1. Radial spoke proteins.
| RSP | Molecular weight (kDa) | Motif | Position |
|---|---|---|---|
| RSP1 |
78.6 |
MORN |
head |
| RSP2 |
77.4 |
GAF/calmodulin binding |
neck |
| RSP3 |
56.8 |
A-kinase anchoring |
stalk |
| RSP4 |
49.8 |
unknown |
head |
| RSP5 |
55.9 |
Aldo-keto reductase |
stalk |
| RSP6 |
48.8 |
unknown |
head |
| RSP7 |
55 |
RII α/EF hand |
stalk |
| RSP8 |
40.5 |
Armadillo |
stalk |
| RSP9 |
29.5 |
unknown |
head |
| RSP10 |
23.5 |
MORN |
head |
| RSP11 |
21.5 |
RII α |
stalk |
| RSP12 |
19.7 |
peptidl-prolyl isomerase |
stalk |
| RSP13 |
~98 |
|
stalk |
| RSP14 |
28.3 |
Armadillo |
stalk |
| RSP15 |
~38 |
leucine-rich repeat |
stalk |
| RSP16 |
39 |
DnaJ/DnaJ-C |
neck |
| RSP17 |
98.5 |
GAF |
stalk |
| RSP18 |
~210 |
|
stalk |
| RSP19 |
~140 |
|
stalk |
| RSP20 |
18.3 |
EF hand (calmodulin) |
stalk |
| RSP21 |
16 |
|
stalk |
| RSP22 |
10.3 |
LC8 |
stalk |
| RSP23 |
61 |
NDK/IQ motif |
neck |
| CaM- IP2 |
~183 |
AKAP/AAT-I |
CSC |
| CaM- IP3 |
|
pyridine-disulfide oxidoreductase |
CSC |
| CaM- IP4 | 97 | WD repeat | CSC |
To date there has been little structural or functional information to elucidate the mechanism of function of RSPs. Nevertheless, sequence analysis of RSPs suggests the roles of individual components of the complex. For example, several functional domains were found within the sequences of the RSPs 2 and 23 that suggest a role of signal transduction via cyclic nucleotides. These two RSPs contain a cyclic GMP‐binding domain, an adenylyl cyclase domain and a nucleotide diphosphate kinase domain. The RSPs 2 and 23 also contain an FHA domain, which mediates interactions with phosphorylated proteins.17,21,23,24 Indeed, the sliding velocity of axonemes has been shown to be dependent on the activity of phosphate kinases, such as PKA and CK1.27,28 Thus, phosphorylation likely plays an essential role in the RS function. Furthermore, RSPs 2 and 23 contain calmodulin‐binding motifs.17 Additionally, sequences of RSPs 7 and 20 reveal a role of these RSPs in calcium signal transduction.17 Their sequences contain multiple EF‐hand motifs, which are structural domains present in calcium‐binding proteins. RSP20 belongs to the calmodulin family.16 Consequently, RSPs 2 and 23 are likely to interact with the RSP20. However, detailed structural information of the RS complex, along with functional studies of the individual components, will be needed to understand how RSs regulate axonemal dyneins.
Function of Radial Spokes
The role of RSs in the beating motion seems to be a signal transduction between the CP and the dyneins.29 RSs are essential for flagellar motility under physiological ATP concentration (0.2–1 mM ATP). Accordingly, Chlamydomonas mutants that lack RSs show no flagellar beating.30-32 Nevertheless, flagellar beating can be restored by low ATP concentrations (< 0.1 mM ATP) in these mutants.30,33 This shows that flagellar motility is not abolished by the absence of RSs. Rather, RSs act as a motility regulator. Thus, the interaction between CP and dynein motors, mediated by RSs, seems to be critical for motility. However, in the low concentration of ATP, dynein activity is independent of the regulation by RSs. The mechanism of this regulation remains to be elucidated.
The waveform of Chlamydomonas and sea urchin sperm flagella is dependent on the concentration of calcium ions.34,35 As mentioned in the previous chapter, RS proteins contain sequences of domains involved in calcium binding and signaling (Table 1).17 Thus, RS are likely to be involved in the control of flagellum waveform. However, Chlamydomonas mutants that lack RSs also show the calcium‐dependent waveform inversion.33 This suggests a distinct or additional mechanism of calcium regulation to the one mediated by RSs. For example, the outer dynein arms contain calcium‐binding proteins and thus can be involved in waveform regulation.36
In the following chapters, we discuss the interactions between the RSs with central‐pair and microtubule doublets, as well as the interaction between the RS proteins. Furthermore, we point out possible mechanisms of signal transduction by RSs from the CP to the MT doublets.
Interaction between RS Head and Central‐Pair
It is known that the RS head extends toward the CP, but the interaction between them has not yet been studied. It is not known whether all the RSs interact contemporaneously with the asymmetric structure of the CP complex, or if the interaction itself is asymmetric (although all nine RS‐heads face the CP) and only some sets of RSs connect with the CP complex depending on the bending motion phase. The sequences of the RS head proteins (RSPs 1, 4, 6, 9 and 10) do not show any motifs of secondary messenger signal transduction.17 RSP1 and RSP10 are predicted to contain a series of MORN (membrane occupation and recognition nexus) domains.17 The presence of MORN sequences is puzzling. However, this seems to suggest that the interaction between CP and RS heads is likely different from the one in the stalk.
Interaction between RS Head and Stalk
The activity of individual axonemal dyneins must be coordinated in response to the bending of flagella in order to maintain a regular motion (theoretically37-39 and experimentally40). RSs are likely to participate in the differential dynein regulation during the bending of axoneme. In a hypothetical model, the RS base is strongly attached to the MT while the rest of the structure shifts upon bending, resulting in the tilt of RSs. As discussed in the previous paragraph, the tilt could result in the detachment of RSs from the CP followed by reattachment during the bending of the flagellum. Indeed, tilt of RSs was observed by electron microscopy and it was proposed to be the regulation mechanism of bending.3,41 This hypothesis, however, remains to be investigated.
Interaction between RSs and Dyneins
RSs interact extensively with the inner arm dyneins (IDAs) (see Fig. 1B for IDAs isoforms identification).2 As mentioned in the beginning of this chapter, mutants lacking RSs are paralyzed under physiological concentration, but the motility is restored in low concentration of ATP.30 A possible explanation to this phenomenon would be intramolecular regulation of dynein activity, proposed by Inoue and Shingyoji.42 In such a model, the motor activity of certain dyneins would be inhibited by physiological concentration of ATP. RSs would suppress this inhibition, promoting the dynein activity even at physiological concentration of ATP. Thus, in mutants lacking RS, the flagellar motility would be impaired at high ATP concentrations due to the inhibition of dynein, but would be restored at low ATP concentrations. This hypothesis is strongly corroborated by the finding that in Chlamydomonas double mutants, lacking CP or RSs, as well as inner dynein arm components, the flagellar motility is recovered.43,44 Thus, suppression of ATP‐inhibition of inner arm dyneins by RSs is a possible mechanism of regulation of flagellar motility.
Detailed Structure of Radial Spokes
All three radial spokes within one repeat have been shown by freeze-fracture deep-etch electron microscopy to have the same T‐shaped structure.5 However, detailed analysis of Chlamydomonas and Tetrahymena RSs by cryo‐electron tomography proved that the three RSs are structurally not identical.2 Our 3D reconstructions show that RS3 has a unique morphology, but that significant differences are also present between RS1 and RS2.
Localization of RS Proteins and Structural Differences between RS1 and RS2
Our 3D reconstructions of Chlamydomonas and Tetrahymena RSs show that RS1 and RS2 share the same overall architecture (Figs. 2 and 3). Their structure can be divided into 4 domains: (1) a very short base that is anchored to the MT, (2) an elongated stalk, (3) a bifurcated neck, and (4) an orthogonal head (Fig. 2).2
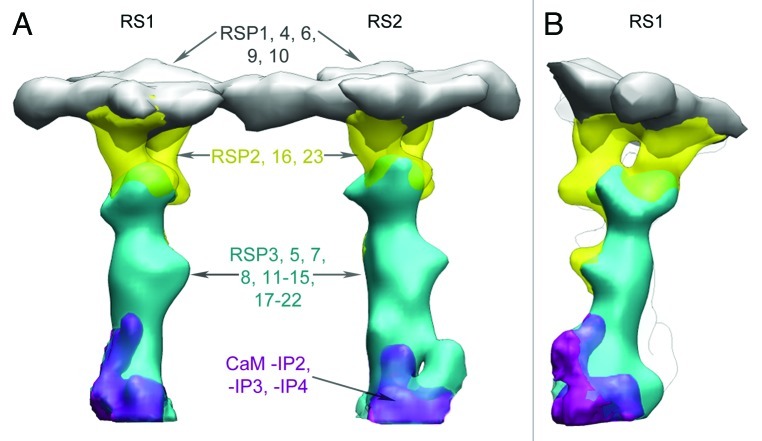
In Chlamydomonas, RS1 and RS2 were subdivided in domains not only on purely morphological grounds, but also based on the protein composition of those four domains (Fig. 4). We were able to locate groups of RS proteins by comparing the RS structure in Chlamydomonas mutants lacking specific subsets of RSPs and specific isoforms of the inner dynein arms (see Table 2 for details about mutants).2 The mutant ida4 lacks dyneins a, c and d, and retain the complete RS complex. The mutant pf14 is deficient in RSP3 and biochemical data show that all the 23 RSPs are missing in pf14 flagella.13 The comparative analysis of axonemes of WT, pf14, and ida4 mutants showed that the RS base might contain adaptor complexes (Fig. 4) such as the calmodulin spoke complex (CSC).25 The CSC, which was shown to be present in pf14 axonemes,25 is important for anchoring the RS stalk to the MT, and it is further known to be involved in the dynein regulatory pathway. Indeed, the 3D reconstructions show that the base of RS1 and the base of RS2 are connected to the tails of the inner dynein arm a/d and the inner dynein arm c, respectively (Figs. 1B and 2B).
Figure 4.C. reinhardtii RS mutants and RS domains. Differential maps of pf14, pf24, and pf1 RS mutants show the boundaries between the various domains of the RS structure (gray, heads; yellow, necks; blue, stalks; purple, adaptor protein complexes). (A) Longitudinal view with proximal end toward the left. (B) Side view (RS1 in the front) seen from the proximal end. RSPs localized in each domain are indicated using the same color code. CaM and CaM‐ IP2, ‐IP3 and ‐IP4 of CSC are present in RS2; the composition of the adaptor complex in RS1 is unknown. (Modified from ©Pigino et al., 2011. Originally published in JCB. DOI: 10.1083/jcb.201106125).
Table 2. Chlamydomomas mutants mentioned in this review.
| Mutants | Missing gene | Missing protein |
|---|---|---|
|
pf1 |
RSP4 |
RSP1, 4, 6, 9 and 10 |
|
pf14 |
RSP3 |
RSP1–23 |
|
pf24 |
RSP2 |
1, 2, 4, 6, 9, 10, 16 and 23 reduced |
| ida4 | IDA4 | dynein a,c,d |
Although similar, the morphology of RS1 and RS2 base differs in some detailed aspects. This is in agreement with the effects of inhibiting the expression of specific CSC proteins, which affects only the assembly of RS2. The CSC might therefore be associated with RS2, but not with RS1.26 The protein composition of the RS1 base is still unknown.
The RS stalk is the part of the spoke that carries the most evident morphological differences between RS1 and RS2. Two additional densities are visible at the base of the RS2 stalk (Figs. 2A and 3A amd B). One of these occurs at the microtubule‐anchoring area and connects the RS2 to the nexin‐dynein regulatory complex (N‐DRC) (Figs. 2A and 3A and B), which is a component of the dynein activity regulatory system.43 The additional density at the base of the RS2 stalk disappears in the mutant pf14, thereby confirming that it is a component of RS, but that it must not be confused with the CSC.2
The mutant pf24 is deficient in RSP2, and lacks RSP1–2, RSP4, RSP6, RSP9–10, RSP16 and RSP23. The differential analysis of the 3D structure of WT, pf24, and pf14 mutant flagella showed that the RS stalk is composed of RSP3, RSP5, RSP7‐8, RSP11‐15, and RSP17‐22 (Fig. 4).2 With the exception of RSP3, which is a component of both RS1 and RS2, it is not yet known (1) which of the other RSP stalk proteins are contained in the additional RS2 densities, and (2) which of them are shared between RS1 and RS2. The morphological and structurally inferred biochemical heterogeneity between RS1 and RS2 was also studied in Tetrahymena2 (Fig. 3A), and the sea urchin.45 The morphology of the stalks is very similar in all those species.
3D reconstructions of Chlamydomonas and Tetrahymena RSs showed that the neck is a bifurcated area that connects the spoke stalk to the head (Figs. 2B–D2 and 4B).2 In the mutant pf1 the RSP1, RSP4, RSP6 and RSP9–10 are missing. The morphological analysis of Chlamydomonas WT, pf24 and pf1 flagella revealed that the radial spoke neck is composed of multiple copies (presumably two) of RSP2, RSP16 and RSP23 (Fig. 4).2 The head of the spoke contains more than two copies of RSP1, RSP4, RSP6, RSP9 and RSP10.2 In contrast to the stalk, the head and the neck of RS1 and RS2 appear identical.
Another important finding is that the heads of RS1 and RS2 are symmetrical structures that consist of two identical sub‐domains (Figs. 2C and 3C). In Chlamydomonas, each of them is 26 nm long and 9 nm wide, and in the head of the spoke exhibits a 2-fold rotational symmetry (Fig. 2C). Although the two head sub‐domains are tightly interconnected, each one of them is independently attached to one of the branches of the bifurcated neck (Fig. 2B and C). Interestingly, RS1 and RS2 heads are also interconnected with a 2-fold translational symmetry. These findings, which were also found in Tetrahymena (Fig. 3C), allow a proposal of new theories about the interaction of RS with the CP and the RS assembly process at the flagellar tip.
RS Pairs and Triplets, and Peculiarities of RS3
As mentioned before, the 3D analysis of RSs in Tetrahymena and Chlamydomonas revealed significant structural differences between the three radial spokes in a triplet. The RS3 in Tetrahymena can still be seen as a T‐shaped structure at low resolution.5 In our cryo‐tomographic studies it is easy to see that the morphology and orientation of head, neck, and stalk are very different compared with RS1 and RS2 (Fig. 3B and C). Rather than extending perpendicularly from the MTD, as both RS1 and RS2 do, the RS3 is slanted toward RS2 and the B‐microtubule. A similar orientation of RS3 is also seen in the sperm flagellum of Gallus domesticus.46 As RS1 and RS2, RS3 also binds to protofilaments A12 and A13, but its bifurcated base appears, with respect to the orientation of the RS1 and RS2 stalk bases, to be rotated by roughly 45°. The slanted base of RS3 is clearly visible (Fig. 3A and B). Further evident structural differences are that the RS3 neck is not bifurcated and the head doesn’t show any evidence of a symmetrical arrangement. The morphological peculiarities of RS3 suggest a substantially different protein composition compared with RS1 and RS2. It is therefore likely that the role of RS3 in regulating the flagellum bending motion is different than the roles of RS1 and RS2.
Maybe the most surprising discovery of our cryo‐tomographic analysis of Chlamydomonas RSs was a short structure located at precisely the same place where RS3 is located in Tetrahymena (Figs. 2A, 3A and 5). The positioning and the morphological similarity between this short structure in Chlamydomonas and the reconstructed base of RS3 in Tetrahymena indicate that the Chlamydomonas repeat contains, in addition to the doublet of RS1 and RS2, a portion of the RS3.2 This finding was further confirmed by a comparison of the structure of Chlamydomonas with the structure of RS3 in sea urchin sperm flagella.45 We introduced the name RS3 stump (RS3S) for the portion of RS3 that is present in Chlamydomonas.
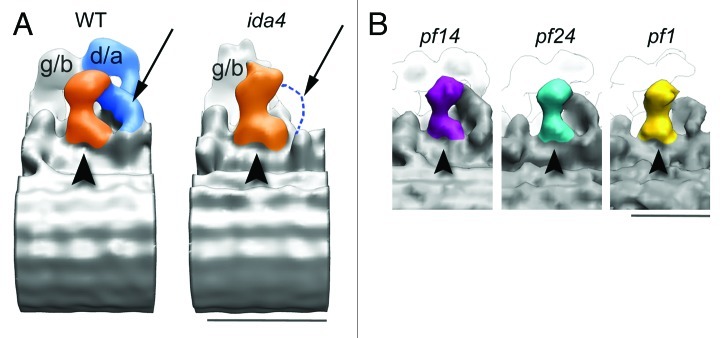
Figure 5. 3D structure of Chlamydomonas RS3 stump (RS3S). (A) RS3S in WT (left) and ida4 (right) flagella. The RS3S is colored in orange and IDA d/a in blue. The IDA d/a is missing in ida4 flagella. The arrows show the approximate position of the dynein d/a tail in WT and its corresponding location in ida4. In ida4, no densities are visible at this location as indicated by the blue dashed line. (View from the adjacent doublet microtubules. The proximal end of the axoneme is to the left.) (B) The structure of RS3S is unchanged in pf14 (purple), pf24 (turquoise), and pf1 (yellow) RS mutants. This indicates a different protein composition between RS3S and the other RSs. Bar, 25 nm.
RS3S is connected to dynein d/a (Fig. 5A). The comparative analysis in wild type (WT) and ida4 mutant flagella shows that the arc‐like structure that is connected to RS3 is a portion of the dynein d/a tail and not a portion of the RS3 structure.2 In Tetrahymena the RS3 stalk is connected also to dynein g/b.2
Further, we compared the 3D structure of RSs in Chlamydomonas mutants pf1, pf24 and pf14.2 These mutants lack specific RS proteins and can therefore be used to visualize the structural changes associated with these missing proteins. Although the structures of RS1 and RS2 in these mutants lack big portions of head and stalk, the RS3 stump maintained its wild‐type morphology in all mutants (Fig. 5B). RS3S remains unaltered even in pf14, which lacks the RS1 and RS2 structures almost entirely. Our comparative analysis between Chlamydomonas mutants and WT is the evidence that the RS3 has a very different protein composition.2
The identity of the RS3 proteins, though, remains largely unknown. Extraction of spokes from Tetrahymena cilia by KI, purification, and molecular characterization of the RS proteins followed by a comparative analysis with the RS proteomics in Chlamydomonas might be a first step toward understanding of the composition of RS3 and its functional role.
Assembly of the Radial Spokes: the Dimerization Hypothesis
In Chlamydomonas the head of the radial spoke is a symmetrical structure (Fig. 2C), with two identical, elongated domains positioned in a 2-fold rotational symmetry.2 This is consistent with the “hypothesis of dimerization” of the 12S intermediate complex during 20S RS assembly.47 The 12S RSs are “7”‐ or “L”‐shaped structures, made of a long rod (~28 nm) and a head (the projection) (~20 nm).47 Their size and shape suggest that two of these L‐shaped 12S intermediates combine in a 2-fold symmetrical way to form one RS. In the cryo‐electron tomography structure, each of the two identical head domains binds to one of the two branches of the bifurcated neck (Fig. 2B). The RS stalk doesn’t appear symmetrical, although its base has a bifurcated structure that anchors on adjacent protofilaments (Fig. 1C). The stalk might contain a big coil‐coiled-like structure and additional proteins might attach subsequently to an initial dimerization process, thereby masking its initially symmetric shape. RSPs 13‐20 are not part of the 12S complex and their contribution to the final RS 20S axonemal complex is not yet resolved. They might create the asymmetry in the stalk structures of RS1 and RS2.
Each symmetrical domain of the head could contain two copies of the RSPs 1, 4, 6, 9 and 10, which have been identified to form the head. Whether these proteins form dimers remains to be elucidated, as well as the possibility of heterodimeric formation. Only RSP10, however, has been shown to form dimers in vitro.48 Kohno et al. also showed that RSP4 and RSP6 interact with RSP9 and RSP10, but not with each other.48 RSP1 was shown to interact with RSP4, but not with the other RS head proteins.49 Although the protein interactions within the head of RS have started to emerge, high‐resolution structural studies and in vitro reconstitution are needed to understand the assembly, structure and function of the head.
The symmetrical arrangement of the head is a conserved feature. Thus, even though the shape of the head differs significantly between Tetrahymena and Chlamydomonas spokes, the symmetrical organization remains the same (Fig. 3).2 Homologs of RSP4 and RSP6 were identified in Tetrahymena,49 and sequence analysis shows that homologs of RSP1, RSP9, and RSP10 are also present in Tetrahymena. The structural difference between Chlamydomonas and Tetrahymena RS heads could be explained by a different organization of the five head proteins or by the presence of additional proteins.
Functional implications of symmetrical RS heads are not known. While MTs are known to be unidirectional, the RS head is symmetrical. Whether the signal transduction between the CP and the RS is performed by a specific protein‐protein interaction remains obscure. The signal could also be propagated in another way, for example, by mechanical interaction.
The reconstruction of 3D models of RSs by cryo‐electron tomography has revealed features of the structure of the individual RSs as never seen before. Important information about the organization of RSPs was also revealed. The new findings support the hypothesis of the RS assembly by dimerization of the 12S RS precursors.
Nevertheless, detailed structural and biochemical studies of the interaction between RSs and the CP are still needed in order to fully understand the role of RSs in dynein regulation. Investigations of the atomic structure of RSPs complexes combined with the analysis of conformational changes of the RS 3D structure during regulation of flagellum motility will hopefully reveal more about the structure and function of the radial spokes.
Acknowledgments
Our work mentioned here was done in collaboration with Khanh Huy Bui, Dennis Diener, Aditi Maheshwari and Pietro Lupetti, and was funded by grants from the Swiss National Science Foundation, Swiss-Japan Cooperative Research Fund, and ETH Independent Investigators’ Research Awards (T.I.) and by a European Molecular Biology Organization fellowship (G.P.). We thank Matthew Cook and Bara Malkova for the English proofreading.
Footnotes
Previously published online: www.landesbioscience.com/journals/BioArchitecture/article/20394
References
- 1.Afzelius B. Electron microscopy of the sperm tail; results obtained with a new fixative. J Biophys Biochem Cytol. 1959;5:269–78. doi: 10.1083/jcb.5.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–87. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warner FD, Satir P. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J Cell Biol. 1974;63:35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol. 1978;76:729–47. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100:2008–18. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warner FD. New observations on flagellar fine structure. The relationship between matrix structure and the microtubule component of the axoneme. J Cell Biol. 1970;47:159–82. doi: 10.1083/jcb.47.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sale WS, Satir P. Splayed Tetrahymena cilia. A system for analyzing sliding and axonemal spoke arrangements. J Cell Biol. 1976;71:589–605. doi: 10.1083/jcb.71.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen J, Barkalow K, Hamasaki T, Satir P. Structural and functional characterization of paramecium dynein: initial studies. J Protozool. 1991;38:55–61. doi: 10.1111/j.1550-7408.1991.tb04801.x. [DOI] [PubMed] [Google Scholar]
- 9.Bastin P, Pullen TJ, Moreira-Leite FF, Gull K. Inside and outside of the trypanosome flagellum:a multifunctional organelle. Microbes Infect. 2000;2:1865–74. doi: 10.1016/S1286-4579(00)01344-7. [DOI] [PubMed] [Google Scholar]
- 10.Baccetti B, Porter KR, Ulrich M. High voltage electron microscopy of sperm axoneme. J Submicrosc Cytol. 1985;17:171–6. [PubMed] [Google Scholar]
- 11.Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci U S A. 2005;102:15889–94. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilula NB, Satir P. The ciliary necklace. A ciliary membrane specialization. J Cell Biol. 1972;53:494–509. doi: 10.1083/jcb.53.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–9. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–8. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams BD, Mitchell DR, Rosenbaum JL. Molecular cloning and expression of flagellar radial spoke and dynein genes of Chlamydomonas. J Cell Biol. 1986;103:1–11. doi: 10.1083/jcb.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153:1315–26. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–74. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J Cell Biol. 1989;109:235–45. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curry AM, Williams BD, Rosenbaum JL. Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol Cell Biol. 1992;12:3967–77. doi: 10.1128/mcb.12.9.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C, Compton MM, Yang P. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol Biol Cell. 2005;16:637–48. doi: 10.1091/mbc.E04-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmer WE, Schloss JA, Silflow CD, Youngblom J, Watterson DM. Structural organization, DNA sequence, and expression of the calmodulin gene. J Biol Chem. 1988;263:19370–83. [PubMed] [Google Scholar]
- 22.King SM, Patel-King RS. Identification of a Ca(2+)-binding light chain within Chlamydomonas outer arm dynein. J Cell Sci. 1995;108:3757–64. doi: 10.1242/jcs.108.12.3757. [DOI] [PubMed] [Google Scholar]
- 23.Patel-King RS, Gorbatyuk O, Takebe S, King SM. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol Biol Cell. 2004;15:3891–902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang P, Yang C, Sale WS. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot Cell. 2004;3:72–81. doi: 10.1128/EC.3.1.72-81.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179:515–26. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dymek EE, Heuser T, Nicastro D, Smith EF. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell. 2011;22:2520–31. doi: 10.1091/mbc.E11-03-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–92. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang P, Sale WS. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J Biol Chem. 2000;275:18905–12. doi: 10.1074/jbc.M002134200. [DOI] [PubMed] [Google Scholar]
- 29.Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omoto CK, Yagi T, Kurimoto E, Kamiya R. Ability of paralyzed flagella mutants of Chlamydomonas to move. Cell Motil Cytoskeleton. 1996;33:88–94. doi: 10.1002/(SICI)1097-0169(1996)33:2<88::AID-CM2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 31.Frey E, Brokaw CJ, Omoto CK. Reactivation at low ATP distinguishes among classes of paralyzed flagella mutants. Cell Motil Cytoskeleton. 1997;38:91–9. doi: 10.1002/(SICI)1097-0169(1997)38:1<91::AID-CM8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Yagi T, Kamiya R. Vigorous beating of Chlamydomonas axonemes lacking central pair/radial spoke structures in the presence of salts and organic compounds. Cell Motil Cytoskeleton. 2000;46:190–9. doi: 10.1002/1097-0169(200007)46:3<190::AID-CM4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Wakabayashi K, Yagi T, Kamiya R. Ca2+-dependent waveform conversion in the flagellar axoneme of Chlamydomonas mutants lacking the central-pair/radial spoke system. Cell Motil Cytoskeleton. 1997;38:22–8. doi: 10.1002/(SICI)1097-0169(1997)38:1<22::AID-CM3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Bessen M, Fay RB, Witman GB. Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J Cell Biol. 1980;86:446–55. doi: 10.1083/jcb.86.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brokaw CJ, Josslin R, Bobrow L. Calcium ion regulation of flagellar beat symmetry in reactivated sea urchin spermatozoa. Biochem Biophys Res Commun. 1974;58:795–800. doi: 10.1016/S0006-291X(74)80487-0. [DOI] [PubMed] [Google Scholar]
- 36.King SM. Axonemal dyneins winch the cilium. Nat Struct Mol Biol. 2010;17:673–4. doi: 10.1038/nsmb0610-673. [DOI] [PubMed] [Google Scholar]
- 37.Brokaw CJ. Thinking about flagellar oscillation. Cell Motil Cytoskeleton. 2009;66:425–36. doi: 10.1002/cm.20313. [DOI] [PubMed] [Google Scholar]
- 38.Lindemann CB. The geometric clutch as a working hypothesis for future research on cilia and flagella. Ann N Y Acad Sci. 2007;1101:477–93. doi: 10.1196/annals.1389.024. [DOI] [PubMed] [Google Scholar]
- 39.Riedel-Kruse IH, Hilfinger A, Howard J, Jülicher F. How molecular motors shape the flagellar beat. HFSP J. 2007;1:192–208. doi: 10.2976/1.2773861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat Struct Mol Biol. 2010;17:761–7. doi: 10.1038/nsmb.1832. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell DR, Nakatsugawa M. Bend propagation drives central pair rotation in Chlamydomonas reinhardtii flagella. J Cell Biol. 2004;166:709–15. doi: 10.1083/jcb.200406148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue Y, Shingyoji C. The roles of noncatalytic ATP binding and ADP binding in the regulation of dynein motile activity in flagella. Cell Motil Cytoskeleton. 2007;64:690–704. doi: 10.1002/cm.20216. [DOI] [PubMed] [Google Scholar]
- 43.Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for Flagellar function. Cell. 1982;28:115–24. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 44.Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118:1163–76. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J, Heuser T, Carbajal-González BI, Song K, Nicastro D. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 2012;69:88–100. doi: 10.1002/cm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess SA, Dover SD, Woolley DM. Architecture of the outer arm dynein ATPase in an avian sperm flagellum, with further evidence for the B-link. J Cell Sci. 1991;98:17–26. doi: 10.1242/jcs.98.1.17. [DOI] [PubMed] [Google Scholar]
- 47.Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 2011;68:389–400. doi: 10.1002/cm.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohno T, Wakabayashi K-I, Diener DR, Rosenbaum JL, Kamiya R. Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken) 2011;68:237–46. doi: 10.1002/cm.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueno H, Iwataki Y, Numata O. Homologues of radial spoke head proteins interact with Ca2+/calmodulin in Tetrahymena cilia. J Biochem. 2006;140:525–33. doi: 10.1093/jb/mvj182. [DOI] [PubMed] [Google Scholar]